Abstract
Despite the high degree of sequence homology between adeno-associated virus (AAV) serotype 1 and 6 capsids (99.2%), these viruses have different liver transduction profiles when tested as vectors. Examination of the six amino acid residues that differ between AAV1 and AAV6 revealed that a lysine-to-glutamate change (K531E) suppresses the heparin binding ability of AAV6. In addition, the same mutation in AAV6 reduces transgene expression to levels similar to those achieved with AAV1 in HepG2 cells in vitro and in mouse liver following portal vein administration. In corollary, the converse E531K mutation in AAV1 imparts heparin binding ability and increases transduction efficiency. Extraction of vector genomes from liver tissue suggests that the lysine 531 residue assists in preferential transduction of parenchymal cells by AAV6 vectors in comparison with AAV1. Lysine 531 is unique to AAV6 among other known AAV serotypes and is located in a basic cluster near the spikes that surround the icosahedral threefold axes of the AAV capsid. Similar to studies with autonomous parvoviruses, this study describes the first example of single amino acid changes that can explain differential phenotypes such as viral titer, receptor binding, and tissue tropism exhibited by closely related AAV serotypes. In particular, a single lysine residue appears to provide the critical minimum charged surface required for interacting with heparin through electrostatic interaction and simultaneously plays an unrelated yet critical role in the liver tropism of AAV6 vectors.
The diverse tissue tropisms exhibited by different serotypes of adeno-associated virus (AAV) vectors have enabled their widespread application in therapeutic gene transfer in vitro and in vivo (for reviews, see references 1 and 24). Such preferential transduction of select tissue types by AAV serotypes can possibly be explained, at least in part, by initial cell surface binding of the viral capsid to complex cell surface glycosaminoglycans (2). For example, AAV serotype 2 has been shown to utilize heparan sulfate as a primary receptor for cell attachment (18). Intriguingly, the ability of AAV2 to transduce different types of myofibers, such as slow and fast twitch muscle, has been shown to correlate with the expression of heparan sulfate (14). Similarly, the tropism of AAV serotype 5 for airway epithelial cells appears to correlate with its ability to utilize sialic acid for cell surface binding (22).
Despite the high degree of sequence homology between AAV1 and AAV6 capsids (∼99.2%), the two serotypes display several unique characteristics. For example, previous studies have shown that AAV6 vectors display increased liver transduction compared to AAV1 (3). Another distinguishing feature of AAV1 and AAV6 vectors is the ability of the latter serotype to bind heparin with moderate affinity (4). However, the AAV6 capsid does not possess R585 and R588 residues involved in heparin binding by AAV2 (8, 12). These observations suggest that the molecular determinants of heparin binding and possibly liver tropism might lie within the six amino acid residues that differ between AAV6 and AAV1.
To identify a critical residue(s) involved in the aforementioned features unique to AAV6, we generated a series of AAV1 and AAV6 mutants by site-directed mutagenesis (QuikChange; Stratagene) on plasmids pXR1 (15) and pXR6 (a kind gift from Joseph Rabinowitz at Thomas Jefferson University). Positions of the six amino acids (five located in the VP3 subunit and one in VP1) that were swapped between AAV1 and AAV6 capsids, genome titers of AAV1/AAV6 mutant vectors packaging the firefly luciferase transgene cassette determined as described previously (19) (Table 1), and their respective heparin binding profiles (Fig. 1a and b) are shown. Of the six amino acids studied, swapping amino acids at position 129, 584, or 598 between AAV6 and AAV1 independently and reproducibly affected viral genome titers (Table 1). The increase in titer from AAV1 recombinants 129, 584, and 598 was of keen interest, since these vector yields exceeded those of both parent serotypes and suggested that further improvements in other serotype-specific titers may also be improved by single amino acid changes (Table 1). Similar outcomes have been observed in mutants isolated from combinatorial libraries of AAV capsids generated by a method analogous to DNA shuffling, an approach independent of rational design (11). The increased titers in the case of the aforementioned AAV1 mutants can possibly be explained by improved folding/stability of capsid protein subunits and/or packaging efficiency (9, 23).
TABLE 1.
Virus titers for mutants based on AAV1 and AAV6
| Virus | Titer (v.g./ml)a |
|---|---|
| AAV1 | 6.44E × 1011 |
| AAV1-L129F | 1.91E × 1012 |
| AAV1-E418D | 7.04E × 1011 |
| AAV1-E531K | 4.51E × 1011 |
| AAV1-F584L | 1.18E × 1012 |
| AAV1-A598V | 1.03E × 1012 |
| AAV1-N642H | 6.02E × 1011 |
| AAV6 | 4.05E × 1011 |
| AAV6-F129L | 1.95E × 1011 |
| AAV6-D418E | 4.78E × 1011 |
| AAV6-K531E | 5.13E × 1011 |
| AAV6-L584F | 2.20E × 1011 |
| AAV6-V598A | 2.78E × 1011 |
| AAV6-H642N | 4.44E × 1011 |
v.g., virus genome.
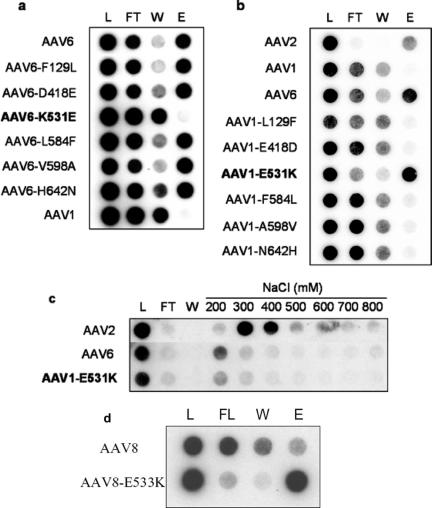
FIG. 1.
(a) Heparin binding profile of AAV6 mutants. (b) Heparin binding profile of AAV1 mutants. (c) Elution profiles of AAV2, AAV6, and AAV1-E531K at different salt concentrations. Mutants at the 531 position are shown in boldface letters. (d) Heparin binding profile of AAV8 and AAV8-E533K. L, load; FT, flowthrough; W, wash; E, eluate.
With respect to loss or gain of affinity to heparin, our mutant collection further helped identify a single amino acid change responsible for such binding activity. Binding of parental AAV1, AAV2, AAV6, and AAV1/AAV6 mutants packaging a firefly luciferase transgene to heparin-conjugated agarose type I (H-6508; Sigma) was analyzed by loading affinity columns (Bio-Rad microspin column) with 5 × 1010 particles of each viral stock in 500 μl Ringer's saline solution (RSS) followed by the sequential collection of fractions from flowthrough, wash with RSS, and elution with RSS containing 800 mM NaCl. The number of mutant or parental AAV particles present in each fraction was determined by dot blot hybridization.
As shown in Fig. 1a, the heparin column elution profile of the AAV6-K531E mutant is identical to that of AAV1, suggesting an attenuation of heparin binding ability. The remaining five amino acid changes, however, fail to alter the ability of AAV6 to bind heparin. These results are further corroborated in Fig. 1b, where the reciprocal change in AAV1-E531K imparts heparin-binding characteristics onto the AAV1 capsid, similar to wild-type AAV2 and AAV6. Swapping other amino acid residues from AAV6 onto AAV1 does not impart heparin binding. Further, as shown in Fig. 1c, the elution profile of AAV capsids from heparin agarose columns at different salt concentrations suggests that AAV6 and AAV1-E531K bind heparin with slightly lower affinity (200 to 300 mM) compared to AAV2 (300 to 400 mM). Taken together, these data suggest that lysine 531 is the core residue at a low-affinity heparin binding site on the AAV6 capsid. The K531 residue is unique to AAV6 among currently known serotypes, which contain a conserved glutamate or aspartate residue at the corresponding 531 position (13, 16). Identical alteration in serotype 8 also conferred heparin binding (Fig. 1d), supporting the importance of the K531 position in the type 6 VP3 capsid subunit. The ability to alter type 8 capsid heparin binding activity suggests that the heparin binding phenotype conferred by this single amino acid change is not restricted to closely related AAV1 and AAV6 serotypes.
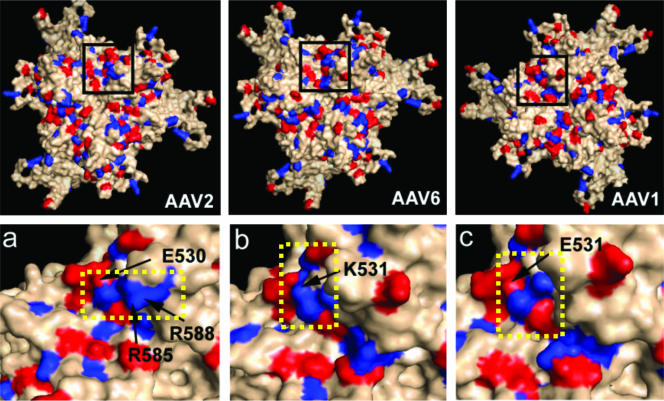
The potential contribution of lysine 531 to surface properties of the AAV6 capsid (and of E531 to that of AAV1) was analyzed by generating three-dimensional models of the surface charge distribution on the VP3 subunit (Fig. 2) of either serotype utilizing Swiss Model (17) with available coordinates of AAV2 (20) (Protein Databank accession no. 1LP3) supplied as a template (13). As shown in Fig. 2, insets a, b, and c, we have compared the capsid surface disposition of equivalent residues to those previously mapped for AAV2 heparin binding as well as neighboring basic residues located on icosahedral threefold symmetry-related VP3 subunits. The key positive residues and the 531 residue are proximal to the spikes that surround this symmetry axis. When the capsid is viewed perpendicular to the threefold axis, the side chain for K531 is surface exposed and forms a continuation for the basic patch between K533 and R488 as well as K528 and R567 at the base of each threefold spike of the AAV6 capsid surface model (Fig. 2, inset b, continuous blue patch, boxed with a dotted yellow line). Such presentation of a continuous cationic patch is reminiscent of that seen in the AAV2 crystal structure in this capsid region, comprised of basic residues R487, R527, R566, R585, and R588 (Fig. 2, inset a, continuous blue patch, boxed with a dotted yellow line), even though it contains E530 at the position equivalent to K531 in AAV6. In contrast, the negatively charged E531 in AAV1 forms a discontinuous positively charged surface (Fig. 2, inset c, broken blue patch, boxed with a dotted yellow line) which is incapable of heparin binding. Thus, a minimum continuous positive surface appears to be necessary for the electrostatic interaction of the AAV capsid with heparin rather than the exact disposition of the residues.
FIG. 2.
Three-dimensional surface maps and charge distribution of a trimer of VP3 subunits viewed perpendicular to the icosahedral threefold symmetry axis for the AAV2 crystal structure as well as AAV6 and AAV1 models. All basic residues (lys, arg, his) are shown in blue, and the acidic residues (glu, asp) are shown in red. The insets (zoomed in) below represent magnification of black boxed regions showing (a) the position of E530 residue at the AAV2 threefold axis with heparin binding residues R585 and R588 forming a continuous basic blue patch (boxed inside a yellow dotted line); (b) the position of the K531 residue at the AAV6 threefold connecting residues R487, K528, K533, and K567 to form a continuous basic blue patch similar to AAV2 (boxed inside a yellow dotted line), albeit proximal to the threefold spikes; and (c) position of the E531 residue at the AAV1 threefold axis forming a discontinuous basic patch (boxed inside a yellow dotted line). Models were generated as described in the text, and images were rendered using Pymol.
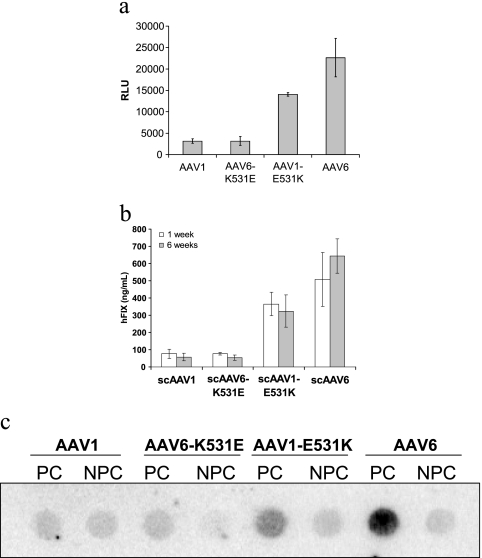
We next investigated whether lysine 531, which appears to be a critical distinguishing feature between AAV6 and AAV1, might possibly explain the enhanced transduction potential of AAV6 compared to AAV1 in hepatocytes in vitro and in vivo. The human coagulation factor IX (hFIX) expression cassette driven by a liver-specific transthyretin promoter was packaged into AAV1, AAV1-E531K, AAV6, and AAV6-K531E capsids to generate self-complementary AAV vectors. In addition, conventional AAV vectors packaging the firefly luciferase transgene were also generated, and vector genome titers were determined by dot blot hybridization. As shown in Fig. 3a, the luciferase transgene expression level achieved using AAV6 vectors is nearly 1 log unit higher than that obtained with AAV1 vectors in HepG2 cells in vitro. The K531E mutation in AAV6 reduces the luciferase expression level to that of AAV1, while the converse E531K change in AAV1 appears to substantially enhance the transduction efficiency of AAV1 in vitro.
FIG. 3.
(a) Luciferase transgene expression levels achieved using AAV1 and AAV6 compared with mutants AAV1-E531K and AAV6-K531E in HepG2 cells (n = 3). (b) Serum hFIX levels at 1 week and 6 weeks following portal vein infusion of wild-type and mutant AAV1 and AAV6 self-complementary (sc) vectors (5 × 1010/mouse) packaging the hFIX transgene driven by the transthyretin promoter (n = 4). (c) Vector genome levels in parenchymal (PC) and nonparenchymal cells (NPC) at 6 weeks following portal vein infusion of wild-type and mutant AAV1 and AAV6 vectors. Total DNA from PC and NPC isolated from collagenase-treated mouse livers was extracted using the DNeasy kit (QIAGEN). RLU, relative light units.
For assessment of in vivo transduction, vectors packaging the hFIX expression cassette were infused into the portal vein of 6-week-old male C57BL/6 mice (Jackson Laboratories, Bar Harbor, ME) at a dose of 5 × 1010 particles per mouse. Blood was collected at 1 week and 6 weeks postinfusion, and hFIX levels in plasma were determined by hFIX enzyme-linked immunosorbent assay (21). As shown in Fig. 3b, serum hFIX levels at 1 and 6 weeks following administration of AAV6 vectors is approximately an order of magnitude higher than those achieved with AAV1 vectors. In the case of AAV6-K531E vectors, hFIX expression is reduced to levels obtained with AAV1, identical to in vitro results and suggesting that K531 is critical for enhanced liver transduction by AAV6. These results are supported further by the increased hFIX levels obtained with AAV1-E531K vectors, albeit at lower levels than those achieved with AAV6. The aforementioned observation can possibly be attributed to the potential role(s) played by the remaining five residues that differ between AAV1 and AAV6 in facilitating favorable interactions between the AAV6 capsid and liver tissue. It is also noteworthy to mention that preincubation of AAV6 vectors with heparin does not block AAV6 transduction in HepG2 cells in vitro (data not shown). These results suggest that the heparin binding ability and liver tropism of AAV6 might constitute two unrelated phenotypic characteristics caused by the same lysine 531 residue.
To further characterize AAV capsid interactions with hepatocytes, mouse liver parenchymal (PC) and nonparenchymal cells (NPC) were isolated at 6 weeks postadministration of different vectors using the method described by Kawakami et al. (7), with some modifications. Briefly, mice were sacrificed, and their livers were perfused with phosphate-buffered saline (PBS) containing 0.05% (wt/vol) collagenase for about 20 min. Following excision of the liver, cells were dispersed by gentle stirring in ice-cold PBS, filtered through cotton mesh sieves, and centrifuged for 1 min at 50 × g to separate liver PC and NPC in the pellet and supernatant, respectively. This process was repeated several times with the PC pellet and NPC supernatant fractions to remove contaminating cell types. Total DNA from PC and NPC was extracted using the DNeasy kit (QIAGEN). Dot blot DNA hybridization was performed using a radiolabeled hFIX cDNA-derived probe. As shown in Fig. 3c, the amounts of accumulated DNA in PC from different vectors correlate well with their hFIX expressions. Significantly larger amounts of vector DNA from AAV6 were detected in PC compared to those of AAV1. The K-to-E change in the AAV6 capsids reduced the amount of vector DNA in PC to a level similar to that of AAV1, while the converse E-to-K change in AAV1 capsids markedly increased the amount of vector DNA accumulated in PC. These data suggest that, in addition to heparin binding, the lysine 531 residue plays a key role in uptake of AAV6 in PC.
In summary, of the six amino acids that differ between AAV types 1 and 6, residues at positions 129, 584, and 598 appear critical in determining viral genome titer. In addition, we have identified a single amino acid (K531) that is essential for conferring the heparin binding characteristics of AAV6. Introduction of an E531K change in AAV1 imparts a heparin binding ability similar to that of AAV6. These observations suggest that a minimum basic footprint is required to facilitate heparin binding through electrostatic interactions. Introduction of K531 in AAV type 8 capsid resulted in recombinant vector particles with heparin binding activity similar to that of AAV6 (Fig. 1d). These data not only speak to the importance of the K531 position but also illustrate the ability of a single amino acid change to confer a serotype-specific trait in another distantly related serotype. We also found that lysine 531 is important for higher transduction of hepatocytes by AAV6 than by AAV1. Although competitive inhibition study with heparin suggests that heparan sulfate is not a receptor for AAV6 in the cell types tested in this study, it is likely that the lysine 531 residue might mediate uptake through an unknown alternative mechanism(s). Nevertheless, the results described herein allow, for the first time, a partial mechanistic interpretation of the molecular determinants of liver tropism in AAV6 vectors in comparison with AAV1.
It should be noted that similar studies with autonomous parvoviruses suggest that the molecular determinants controlling host range and tropism in autonomous parvoviruses can often be narrowed down to a few critical amino acid residues in the capsid. For example, the feline panleukopenia virus (FPV) and its host range variant, canine parvovirus (CPV), can both bind the feline transferrin receptor, while only CPV binds to the canine transferrin receptor (cTfR). Introducing two CPV-specific changes into FPV allowed FPV to bind the canine transferrin receptor-infected canine cells, although neither change alone altered such phenotypes (5). More recently, studies with the minute virus of mice suggested that single amino acid changes at the sialic acid binding pocket of the capsid can modulate tropism and determine virulence (10). In the case of AAV, only two types of glycosaminoglycan-based primary receptors, heparan sulfate and sialic acid, have been identified thus far. While AAV2 binds heparan sulfate, AAV4 and AAV5 as well as AAV1 and AAV6 have been shown to exploit sialic acid residues with different linkage specificities for cellular entry (6, 22, 25). We anticipate that further studies mapping the potential role of single amino acid changes in closely related AAV serotypes and variants, as described above for AAV1 and AAV6, are likely to shed light on molecular determinants of capsid-receptor interactions, tissue tropism, immune profile, and other aspects of the AAV life cycle in general. Such structure-function studies will lay the foundation for rational engineering of cell type-specific AAV vectors in targeted gene delivery applications.
Acknowledgments
We thank Alda Fernandez for technical assistance.
This work was supported by NIH research grants HL051818, HL069973, and GM059299 and a Brain Tumor Research Award from the Goldhirsh Foundation (R.J.S.) as well as NIH research grants HL59412 and HL51811 (M.A.-M). Zhijian Wu is a recipient of the Judith Graham Poole postdoctoral fellowship from the National Hemophilia Foundation.
Footnotes
Published ahead of print on 30 August 2006.
REFERENCES
- 1.Choi, V. W., D. M. McCarty, and R. J. Samulski. 2005. AAV hybrid serotypes: improved vectors for gene delivery. Curr. Gene Ther. 5:299-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding, W., L. Zhang, Z. Yan, and J. F. Engelhardt. 2005. Intracellular trafficking of adeno-associated viral vectors. Gene Ther. 12:873-880. [DOI] [PubMed] [Google Scholar]
- 3.Grimm, D., S. Zhou, H. Nakai, C. E. Thomas, T. A. Storm, S. Fuess, T. Matsushita, J. Allen, R. Surosky, M. Lochrie, L. Meuse, A. McClelland, P. Colosi, and M. A. Kay. 2003. Preclinical in vivo evaluation of pseudotyped adeno-associated virus vectors for liver gene therapy. Blood 102:2412-2419. [DOI] [PubMed] [Google Scholar]
- 4.Halbert, C. L., J. M. Allen, and A. D. Miller. 2001. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J. Virol. 75:6615-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hueffer, K., L. Govindasamy, M. Agbandje-McKenna, and C. R. Parrish. 2003. Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J. Virol. 77:10099-10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaludov, N., K. E. Brown, R. W. Walters, J. Zabner, and J. A. Chiorini. 2001. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J. Virol. 75:6884-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawakami, S., F. Yamashita, M. Nishikawa, Y. Takakura, and M. Hashida. 1998. Targeted delivery of plasmid DNA to hepatocytes in vivo: optimization of the pharmacokinetics of plasmid DNA/galactosylated poly(L-lysine) complexes by controlling their physicochemical properties. J. Pharmacol. Exp. Ther. 287:408-415. [PubMed] [Google Scholar]
- 8.Kern, A., K. Schmidt, C. Leder, O. J. Muller, C. E. Wobus, K. Bettinger, C. W. Von der Lieth, J. A. King, and J. A. Kleinschmidt. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lochrie, M. A., G. P. Tatsuno, B. Christie, J. W. McDonnell, S. Zhou, R. Surosky, G. F. Pierce, and P. Colosi. 2006. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J. Virol. 80:821-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez-Bueno, A., M. P. Rubio, N. Bryant, R. McKenna, M. Agbandje-McKenna, and J. M. Almendral. 2006. Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J. Virol. 80:1563-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maheshri, N., J. T. Koerber, B. K. Kaspar, and D. V. Schaffer. 2006. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat. Biotechnol. 24:198-204. [DOI] [PubMed] [Google Scholar]
- 12.Opie, S. R., K. H. Warrington, Jr., M. Agbandje-McKenna, S. Zolotukhin, and N. Muzyczka. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 77:6995-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Padron, E., V. Bowman, N. Kaludov, L. Govindasamy, H. Levy, P. Nick, R. McKenna, N. Muzyczka, J. A. Chiorini, T. S. Baker, and M. Agbandje-McKenna. 2005. Structure of adeno-associated virus type 4. J. Virol. 79:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruchnic, R., B. Cao, Z. Q. Peterson, X. Xiao, J. Li, R. J. Samulski, M. Epperly, and J. Huard. 2000. The use of adeno-associated virus to circumvent the maturation-dependent viral transduction of muscle fibers. Hum. Gene Ther. 11:521-536. [DOI] [PubMed] [Google Scholar]
- 15.Rabinowitz, J. E., F. Rolling, C. Li, H. Conrath, W. Xiao, X. Xiao, and R. J. Samulski. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rutledge, E. A., C. L. Halbert, and D. W. Russell. 1998. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 72:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao, X., J. Li, and R. J. Samulski. 1998. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 72:2224-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter, J., Q. You, J. N. Hagstrom, M. Sands, and K. A. High. 1996. Successful expression of human factor IX following repeat administration of adenoviral vector in mice. Proc. Natl. Acad. Sci. USA 93:3056-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walters, R. W., S. M. Yi, S. Keshavjee, K. E. Brown, M. J. Welsh, J. A. Chiorini, and J. Zabner. 2001. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J. Biol. Chem. 276:20610-20616. [DOI] [PubMed] [Google Scholar]
- 23.Wu, P., W., Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu, Z., A. Asokan, and R. J. Samulski. 2006. Adeno-associated virus (AAV) serotypes: vector toolkit for human gene therapy. Mol. Ther. 14:316-327. [DOI] [PubMed] [Google Scholar]
- 25.Wu, Z., E. Miller, M. Agbandje-McKenna, and R. J. Samulski. 2006. α-2,3 and α-2,6 N-linked sialic acid facilitate efficient binding and transduction by adeno-associated virus type 1 and 6. J. Virol. 80:9093-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]