Abstract
The Rep protein of tomato yellow leaf curl Sardinia virus (TYLCSV), a single-stranded DNA virus of plants, is the replication initiator essential for virus replication. TYLCSV Rep has been classified among ATPases associated with various cellular activities (AAA+ ATPases), in superfamily 3 of small DNA and RNA virus replication initiators whose paradigmatic member is simian virus 40 large T antigen. Members of this family are DNA- or RNA-dependent ATPases with helicase activity necessary for viral replication. Another distinctive feature of AAA+ ATPases is their quaternary structure, often composed of hexameric rings. TYLCSV Rep has ATPase activity, but the helicase activity, which is instrumental in further characterization of the mechanism of rolling-circle replication used by geminiviruses, has been a longstanding question. We present results showing that TYLCSV Rep lacking the 121 N-terminal amino acids has helicase activity comparable to that of the other helicases: requirements for a 3′ overhang and 3′-to-5′ polarity of unwinding, with some distinct features and with a minimal AAA+ ATPase domain. We also show that the helicase activity is dependent on the oligomeric state of the protein.
Initiation of DNA replication is a highly specific and controlled event which depends on the coordinated assembly of large protein complexes at the origin of replication. This process, which is universal, is activated by recognition and binding of the initiator protein to its cognate replication origin on the chromosome. Other steps follow, such as melting at the origin of replication and further unwinding carried out by replicative helicases recruited at the site of replication (31). In the case of extrachromosomal elements, such as viruses and plasmids, the initiator protein is often multifunctional and performs several of these steps, as well as interacting with host factors necessary for replication (42).
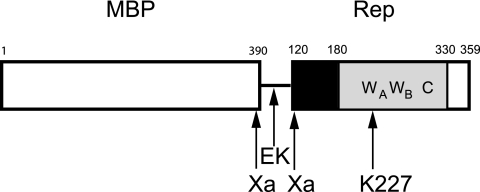
The Rep protein is the initiator protein of tomato yellow leaf curl Sardinia virus (TYLCSV), a begomovirus of the Geminiviridae family of plant single-stranded DNA (ssDNA) viruses. The Rep proteins of geminiviruses are closely related and show substantial sequence conservation. Rep, also named C1 and AL1, is a multifunctional protein and the only viral protein absolutely required for virus replication. Four functional domains have been delineated for begomovirus Rep: the N-terminal domain (amino acids [aa] 1 to 120), which is involved in initiation of the rolling-circle replication (RCR) utilized by geminiviruses (4, 34, 35); the oligomerization domain (aa 121 to 180), leading to interactions with itself (34) and with host factors (17); the ATPase domain (aa 181 to 330), which is characterized by the presence of a P loop (discussed in more detail below); and a carboxyl-terminal domain (aa 331 to 359) of unknown function but shown to be required for viral replication in the case of tomato golden mosaic virus (35) (Fig. 1).
FIG. 1.
Map of MBP-Rep120-359. The white box (aa 1 to 390) represents MBP, the line corresponds to the linker region with the enterokinase (EK) cleavage site, and the two factor Xa cleavage sites (121/122 in Rep) are indicated. The Rep120-359 region is constituted by the oligomerization domain (aa 120 to 180, black box), the ATPase domain (aa 181 to 330, gray box), and the C-terminal region (aa 331 to 359, white box) of unknown function. The conserved Walker A (WA, aa 217 to 230), Walker B (WB, aa 257 to 263), and C (aa 300 to 305) are indicated, as is the position of the mutated K at position 227 in the P loop.
The ATPase domain of geminivirus Rep proteins was identified as a common element among proteins encoded by small DNA and RNA viruses (16) and is characterized by three conserved motifs: Walker A in the P loop, Walker B, and motif C. The presence of simian virus 40 (SV40) large T antigen (LTag) in this family, being the only bona fide helicase of the group, led those authors to postulate that all the members were putative helicases, which were later classified as superfamily 3 (SF3) (15). More recently, classification of the SF3 helicases among the very large family of AAA+ ATPases was proposed based on the presently available tertiary structures of other well-established helicases in this family (13, 21). The SF3 helicases form a distinct clade among the AAA+ ATPase “pre-sensor 1-β hairpin” superclade, since it is constituted only by viral RNA or DNA helicases with distinct features. The AAA+ ATPase domain of SF3 helicases is about 200 amino acids long and is formed by two domains. The first domain is a α/β-fold formed by a five-stranded β-sheet (β5-β1-β4-β3-β2) flanked by four α-helices. Strands β1, β3, and β4 carry the three highly conserved motifs. Also present are a β-hairpin, which is the distinctive feature of the pre-sensor 1-β hairpin superclade, and an arginine finger coordinating ATP hydrolysis and conformational changes of the protein (33). Last, a B′ box involved in the coupling of DNA binding and ATP hydrolysis is present only in SF3 helicases (40, 43). The second domain, more variable in length, is constituted by α-helices; there are four α-helices in the case of SV40 LTag but only one for adeno-associated virus type 2 (AAV2) Rep40.
Most of the helicases are oligomeric, and a majority are found as hexamers (reviewed in references 19, 27, and 36). This is also the case for several SF3 helicases. In general the conserved AAA+ ATPase domain alone is not sufficient to allow oligomerization; if expressed as such, only inactive monomeric forms are observed. In the case of SF3 helicases, multimerization results from the presence of an oligomerization domain neighboring the AAA+ ATPase and unrelated among the different helicases. Furthermore, complete oligomerization may be achieved only in conjunction with nucleotide and/or DNA binding (26).
Up to now, helicase activity of geminivirus Rep proteins has not been demonstrated. Oligomerization (32, 35) and ATPase activity independent of DNA (8), both of which are essential for viral replication, were established. In this report, we present results showing that TYLCSV Rep is indeed a helicase. To reveal this activity, the major difficulty encountered was the insolubility and the highly aggregated state of the full-length Rep, which was overcome by deleting the N-terminal domain involved in replication initiation. TYLCSV Rep122-359 helicase activity is dependent on a functional Walker A motif, as demonstrated by the lack of activity of Rep122-359K227A with a mutated lysine in Walker A. The helicase activity is also dependent on the oligomerization status but otherwise is similar to that of the other helicases, even though the AAA+ ATPase domain is reduced compared to that in other members of the family. The role of Rep as the replicative helicase is also discussed.
MATERIALS AND METHODS
Reagents.
New England Biolabs was the supplier of M13mp18, pMal-c2 vector, amylose, factor Xa, enterokinase, restriction enzymes, kinase, ligase, polymerase, and molecular weight markers for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). [γ-33P]ATP (111 TBq/mmol) was purchased from ICN and Perkin-Elmer. Bovine serum albumin, native molecular weight markers, and Sephacryl S300-HR were purchased from Sigma; stabilized proteinase K solution was from Eurobio; MicroSpin G-50 columns and Superdex 200 were from Amersham Biosciences; and DEAE Sepharose Fast Flow was from Pharmacia.
Construction and expression of MBP fusion proteins MBP-Rep120-359 and MBP-Rep120-359K227A.
Rep120-359 was first expressed as a fusion protein with MBP to improve its solubility and to facilitate purification. In order to separate MBP from the Rep domain, an enterokinase cleavage site was introduced in addition to the Xa factor sites already present in the vector and Rep120-359. A fragment of about 750 bp corresponding to the rep sequence (positions 358 to 1080, flanked by EcoRI and XbaI sites), obtained by PCR, was inserted into the vector pMAL-c2. For the PCR, the forward oligonucleotide was 5′CGgaattcGATGACGATGACAAAGGACGATCTGCTAGGGGAGG3′ (the EcoRI site is in lowercase, the sequence yielding the site for subsequent enterokinase cleavage is underlined, and the last 20 nucleotides [nt] correspond to position 358 to 377 on the rep gene), and the reverse oligonucleotide was 5′GCtctagaTTACGCCTCACTTGCCTCTTCTTG3′ (the XbaI site is in lowercase, and the remaining 24 nt correspond to the 3′ end of the rep gene between positions 1057 and 1077 plus a stop codon). The template used for amplification was the full-length rep gene of TYLCSV (GenBank accession no. X61153) cloned in the pQE60 vector (QIAGEN) (J. Brevet and D. Clérot, unpublished data). The construct pMAL-c2-rep358-1077 was transformed in Escherichia coli strain SCS1 (Stratagene).
To produce the K227A mutant form, a fragment of 460 bp between AgeI position 639 in the rep gene and HindIII in the linker region of pMAL-c2 was exchanged with an equivalent fragment of the pQE32-RepK227A mutant (8), AgeI position 639 in the mutant rep gene and HindIII in the linker region. The sequences of the constructs were verified.
To express and purify the two MBP-Rep fusion proteins, the New England Biolabs protocols were applied, with the following modifications. After growth at 37°C to an absorbance of 0.8 at 600 nm, the cultures were shifted to 18°C and expression was induced with 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 16 h. The buffer used for the purification was 20 mM HEPES (pH 7.5), 250 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol (DTT). The absorbance at 280 nm and the ATPase activity of the peak fractions were determined, only MBP-Rep120-359 has ATPase activity. The purity of the proteins was assessed by SDS-PAGE (see Fig. 2A and C). The pooled fractions were stored at −20°C. The yields of MBP-Rep120-359 and MBP-Rep120-359K227A were ca. 20 mg and 10 mg, respectively, of protein per liter of culture.
FIG. 2.
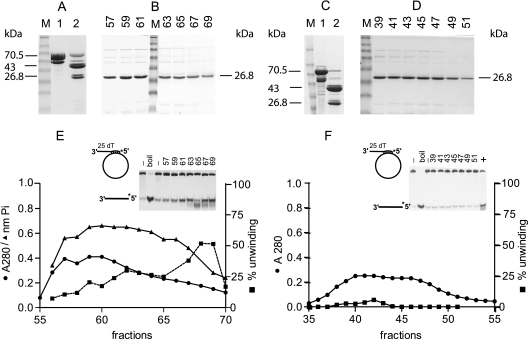
Purification of Rep122-359 and Rep122-359K227A. Panels A, B, and E display the purification steps for Rep122-359, and panels C, D, and F correspond to Rep122-359K227A purification. In panels A and C, lanes 1 are MBP-Rep120-359 and MBP-Rep120-359K227A, respectively; in lanes 2 are factor Xa digestion products of the corresponding proteins. The molecular masses of the different purified proteins are indicated. Panel B shows SDS-PAGE of DEAE fractions 57 to 69 of Rep122-359, and panel F shows the corresponding fractions 39 to 51 of Rep122-359K227A. Lanes M, molecular mass markers in kDa (175, 83, 62, 47.5, 32.5, 25, 16.5, and 6.5). Panel E shows the absorbance profile at 280 nm (•), the ATPase activity (nanomoles of Pi released) determined with 1 μl of the fraction for a 50-μl reaction mixture (▴), and the percentage unwinding with 0.5 μl of the fractions per 10 μl of assay mixture (▪). The M13-derived substrate used is shown, and the inset shows the gel used to quantify helicase activity; the two first lanes correspond to the substrate alone, with and without boiling, and numbers on top of the gel correspond to fraction numbers. Panel F shows Rep122-359K227A absorbance and activity profiles; details are as for panel E. Lane + in the inset corresponds to a positive control with Rep122-359.
ATPase assay.
After incubation of the protein (at the indicated concentration) with 1 mM ATP and 5 mM MgCl2, in buffer A (20 mM HEPES [pH 7.5], 100 mM NaCl, 1 mM DTT) at 25°C, the released Pi was detected as a colorimetric complex absorbing at 650 nm by a procedure described previously (23) and modified (2). GraphPadPrism (GraphPad) was used to analyze and plot the results.
Helicase substrates.
Purified oligonucleotides were obtained from MWG-Biotech. The oligonucleotides are derived from the M13mp18 sequence, the −20 M13 sequencing primer (17 nt; New England Biolabs), and 5′ GGACCGCTTGCTGCAACT 3′ (18 nt; positions 5913 to 5930), in either the sense or antisense orientation in order to hybridize to M13mp18 or between themselves. In addition, the same oligonucleotides have 3′ or 5′ extensions of dT of various lengths as indicated.
Oligonucleotides were labeled at the 5′ end with [γ-33P]ATP and T4 polynucleotide kinase, and nonincorporated [γ-33P]ATP was removed using MicroSpin G-50 columns. A specific activity of at least 1,000 cpm per fmol was obtained. Complementary oligonucleotides in equimolar amounts were hybridized either among themselves or with M13mp18 ssDNA in annealing buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, and 1 mM EDTA). The mixture was incubated at 75°C for 10 min and cooled slowly to room temperature. When necessary, the M13-derived substrates were purified on a Sephacryl S300-HR column to remove nonhybridized oligonucleotides.
Helicase assay.
The helicase assay was performed at 37°C in buffer A supplemented with 1 mM ATP, 5 mM MgCl2, and 0.1 mg/ml bovine serum albumin in a volume of 10 μl. The amount of protein was in the picomole range (as monomers) as indicated, and 2 fmol of substrates was used per assay. After 1 h of incubation (or as indicated otherwise), the reaction was stopped by addition of (final concentrations) 0.025% SDS, 1 μg of proteinase K, and 1 mM CaCl2, and the mixture was further incubated for 10 min. After addition of 2 μl of sample loading buffer (final concentrations of 20 mM EDTA [pH 8], 0.25% SDS, 5% glycerol, and 0.025% each of xylene cyanol, bromophenol blue, and orange G), the samples were analyzed on native polyacrylamide gels (8% for the M13-derived substrates and 12% for the substrates constituted by oligonucleotides only) and run at 16 V/cm for 50 min. The gels were dried on DE81 paper and exposed to a STORM phosphorimager screen. The scans were quantified with ImageQuant (Molecular Dynamics).
Polyacrylamide gel electrophoreses.
Polyacrylamide gels at various concentrations were prepared with acrylamide-bisacrylamide (29:1). The native gels for the helicase assay were run in 40 mM Tris-acetate-1 mM EDTA, and the running buffer for SDS-PAGE was 25 mM Tris 192 mM glycine, and 0.1% SDS.
Blue-native (BN) gradient gels were 4% to 12% bis-Tricine (Invitrogen), and electrophoresis was carried out in 50 mM Tris-HCl (pH 7.7)-50 mM MOPS (morpholinepropanesulfonic acid)-0.002% Coomassie blue as described by Schägger et al. and modified by Schülke et al. (37, 38). Native molecular weight markers were run in parallel.
Glycerol gradient centrifugation.
Glycerol concentrations were 10% to 40% in 20 mM Tris-HCl (pH 7.5), 250 mM NaCl, and 1 mM DTT, and 50 μg to 200 μg of protein was loaded on the gradients. Centrifugation was performed in a Beckman SW 55 Ti rotor at 45,000 rpm/min for 16 h at 4°C. Fractions were collected, and further analyzed by SDS-PAGE, and assayed for ATPase activity. The sedimentation coefficient S was determined using bovine serum albumin, β-amylase, and β-galactosidase as markers on a gradient run separately, or β-galactosidase was also used as an internal marker and detected by its enzymatic activity.
RESULTS
Purification of Rep122-359 and Rep122-359K227A.
The amylose-purified MBP-Rep120-359 and MBP-Rep120-359K227A are shown in lanes 1 of Fig. 2A and C, respectively. For each protein there is one major band with a molecular mass of 70.5 kDa corresponding to the expected protein and a contaminating band of 58 kDa resulting from a carboxyl-terminal truncation produced during bacterial growth; both proteins migrate with lower apparent molecular mass. In order to remove MBP, MBP-Rep120-359 and MBP-Rep120-359K227A enterokinase digestion was tested and showed a very low efficiency of cleavage; therefore, we resorted to factor Xa, which has two sites in the fusion proteins: one built in in the linker region between MBP and the fused protein and the second one in the Rep protein (Xa cleavage at position 121/122 [Fig. 1]). For factor Xa cleavage, the proteins were dialyzed against buffer A and after dialysis were supplemented with 2 mM CaCl2 and 1 mM ATP; the weight ratio of factor Xa to protein was between 1/150 and 1/200, and incubation was for 16 h at 0°C. The results are shown in lanes 2 of Fig. 2A and C; MBP (43 kDa) and Rep122-359 (26.8 kDa) are the two major bands, present as doublets resulting from the presence of the two Xa cleavage sites (Fig. 1).
The protein mixtures (14 mg for MBP-Rep122-359 or 6.5 mg for MBP-Rep122-359K227A) were centrifuged to remove insoluble material and were purified on a DEAE-Sepharose Fast Flow column (column volumes were 6 ml and 4 ml, respectively) in buffer A plus 5 mM MgCl2. All purification steps were carried out at 4°C. MBP was eluted in the flowthrough, and Rep122-359 or Rep122-359K227A was eluted with an NaCl gradient between 100 and 500 mM in the same buffer (the gradient volumes were 120 ml with a 1.5-ml fraction volume and 80 ml with 1-ml fraction volume, respectively). The two Rep proteins eluted at a concentration of about 200 mM NaCl, and a single major band of 26.8 kDa was present (Fig. 2B and D), with similar elution profiles for the two proteins. The N termini of the cleaved proteins were at position 122 (determined by mass spectroscopy [not shown]), which corresponds to the Xa cleavage site of Rep. The DEAE chromatography also removed contaminating nucleic acids that eluted at higher NaCl concentrations (fractions 107 to 115 and 75 to 87, respectively, at about 300 mM NaCl [not shown]) and factor Xa, which is eluted at 400 mM NaCl (according to the New England Biolabs protocols). The fractions were stored at −80°C.
The fractions were tested for ATPase and helicase activities by using an M13-derived substrate as described in Materials and Methods. No ATPase or helicase activities were detected in the case of Rep122-359K227A (Fig. 2F), demonstrating that the activities associated with Rep122-359 do not result from contamination of bacterial origin. The DEAE elution profiles of Rep122-359 at 280 nm and ATPase and helicase activities are presented in Fig. 2E. Whereas absorbance and ATPase activity were broadly distributed over 15 fractions, substantial helicase activity of Rep122-359 was highest in fractions 68 to 69. Degradation of the substrate was also observed in fraction 65 (inset in Fig. 2E). Further characterization of the helicase activity was therefore performed using the two most active fractions of Rep122-359.
Helicase activity of Rep122-359.
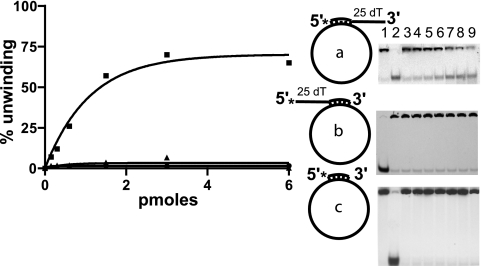
To achieve an initial characterization of Rep122-359 helicase activity, different substrates were tested as shown in Fig. 3 and 4. The main requirement for Rep122-359 helicase activity is the presence of a 3′ ss tail. All the substrates with a 3′ ss overhang (substrate a [Fig. 3], all the forked substrates [Fig. 4A], and substrates h and i, which lack a 5′ tail [Fig. 4B]), are unwound, indicating that a complete fork is not necessary.
FIG. 3.
Rep122-359 helicase activity on M13-derived substrates. The percentage of unwinding as a function of Rep122-359 concentration in picomoles per assay is shown with three different substrates (a [▪], b [▴], and c [•]), as are the corresponding gels. For a and c, lanes 1 and 2 are substrate alone nonboiled and boiled, respectively; for b, lanes 1 and 2 are boiled and native substrate, respectively. Lanes 3 to 9 show increasing concentration of Rep122-359 as indicated on the graph.
FIG. 4.
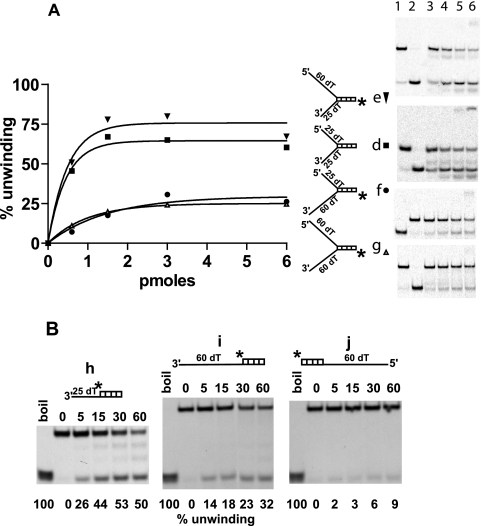
Rep122-359 helicase activity on forked and flap substrates. A, percentage of unwinding of four different forked substrates (d[▪], e [▾], f [•], and g [▵]), with different lengths of ss tails as indicated, presented as a function of Rep122-359 concentration. The corresponding gels are displayed; for d, e, and g, lanes 1 and 2 show the substrate alone native and boiled, respectively, and for f, lanes 1 and 2 show boiled and native substrate, respectively. B, unwinding of three flap substrates h, i, and j with different length of overhang as indicated and as a function of time. Each reaction mixture contained 2 fmol of substrate and 3 pmol of Rep122-359. The percentage of unwinding is indicated at the bottom of the gel, and the time intervals are indicated above the gel.
Increasing the length of the 3′ overhang from 25 to 60 dTs leads to a reduction in unwinding activity of more than 50%. Comparing substrates with the same 5′ tail, such as d and f (5′ tail of 25 dTs), one observes a decrease in the percentage of unwinding from 65% to 31%; for e and g (5′ tail of 60 dTs) there is a decrease from 80% to 24% (Fig. 4A). In contrast, the presence of a 5′ ss tail enhances the helicase activity; for substrates having the same 3′ overhang (25 dTs), the percentage of unwinding increases from 50% with substrate h lacking a 5′ tail to 65% with 25 dTs (substrate d) (Fig. 4A), 80% with 60 dTs (substrate e) (Fig. 4A), and 70% with a circular ssM13 (substrate a) (Fig. 3).
No unwinding was observed with the fully annealed oligonucleotide of substrate c (Fig. 3) or with substrates having only a 5′ ss extension, such as substrate b (Fig. 3) and substrate j (Fig. 4B). The latter results indicate that the polarity of unwinding is 3′ to 5′. Furthermore if the substrate commonly used to determine polarity (27), i.e., a DNA strand with two labeled oligonucleotides of different sizes annealed at each extremity with no overhang, was used, no unwinding was observed (not shown). Considering also the lack of unwinding for substrates b and c (Fig. 3), it appears that Rep122-359 may require a free 3′ end for helicase activity. Indeed most of the 3′→5′ helicases requiring a 3′ ssDNA are able to unwind substrates a, b, and c, as the circular ssM13 DNA provides a single strand of the required polarity. Alternatively, the length of ssM13 may be inhibitory, as we have shown that increasing the length of the 3′overhang hinders the helicase activity. Taken together, the properties of Rep122-359 are similar to those of other helicases: necessity for a 3′ ss tail and 3′-to-5′ unwinding, with dependence on the length of the tails and the possible requirement for a free 3′ end.
In addition to the unwound free oligonucleotide resulting from helicase activity, additional discrete bands of different sizes are observed with substrates d, e, and h (Fig. 4A and B) and for substrate a (Fig. 2, fraction 65). These bands appear at prolonged incubation times or when the amount of Rep122-359 is increased; they are not produced in the absence of helicase activity, particularly with Rep122-359K227A. These additional products seem to be produced by a cleavage in the 3′ ss extension. Whether the production of these fragments represents another activity associated with Rep122-359 will have to be assessed.
Oligomerization of the different Rep proteins.
The oligomeric states of the different Rep proteins were determined using two techniques, density gradient centrifugation and BN-PAGE, as described in Materials and Methods. The sedimentation profile of MBP-Rep120-359 is shown in Fig. 5A, and its sedimentation coefficient is 12.9 S. The Stokes radius of MBP-Rep120-359 determined by gel filtration on Superdex 200 (not shown) is 7.5 nm, leading to the calculated molecular mass of 406 kDa (9, 41). With a molecular mass of 70.5 kDa for the monomer, and not taking into account the presence in the oligomers of cleaved protein (visible on the gel in Fig. 5A), we find that oligomers of MBP-Rep120-359 are constituted by 5.8 monomers; MBP-Rep120-359 is therefore likely to form a hexamer, as do many helicases. MBP-Rep120-359K227A had a similar sedimentation profile (not shown) and has the same oligomeric structure as the wild-type protein.
FIG. 5.
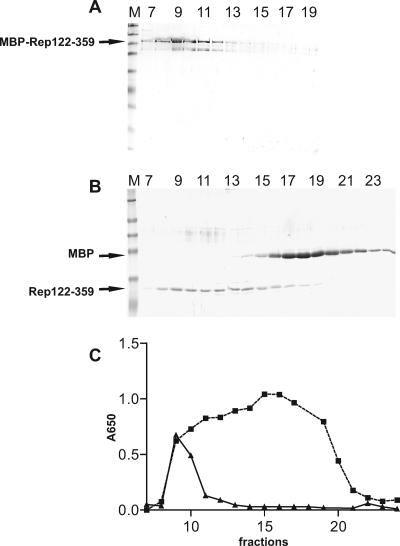
Glycerol gradient sedimentation of MBP-Rep120-359 and Rep122-359. A, SDS-PAGE of fractions 7 to 19 (from bottom to top of the gradient) corresponding to MBP-Rep122-359 sedimentation. B, SDS-PAGE of MBP and Rep122-359 after factor Xa cleavage (fractions 7 to 23). Lanes M, molecular mass markers (same as in Fig. 2). C, ATPase activity expressed as absorbance at 650 nm for MBP-Rep122-359 (▴) and Rep122-359 (▪) for the same fractions as in A and B. β-Galactosidase (15.9 S) migrates in fraction 6 in both gradients (not shown).
Sedimentation of MBP and Rep122-359 obtained after factor Xa cleavage reveals a broad distribution of Rep122-359 (Fig. 5B). The highest ATPase activity occurs in fractions 15 and 16 (Fig. 5C), corresponding to oligomers with an estimated sedimentation coefficient of 7.1 S. The ATPase shoulder in fraction 11 corresponds to complexes with a sedimentation coefficient of 12.1 S. The unavailability of the Stokes radius for these components led us to calculate the molecular mass as described previously (29), giving estimates of 307 kDa and 151 kDa for oligomers in fractions 15 to 16 and 11, respectively. With a molecular mass of 26.8 kDa for Rep122-359, the distribution of the oligomer size is therefore between dodecamers and hexamers.
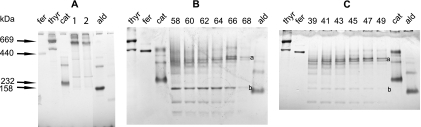
As a second method to determine the oligomeric state of the Rep proteins, we used BN-PAGE. This technique is convenient for the analysis of many samples at a time. Figure 6A shows a comparison of the migrations of MBP-Rep120-359 and MBP-Rep120-359K227A, which are identical, with a molecular mass of approximately 660 kDa for the main species, corresponding to an oligomerization degree of ca. 9. The migrations of Rep122-359 and Rep122-359K227A present in the fractions of the respective DEAE chromatographies (same as in Fig. 2) are presented in Fig. 6B and C, respectively. First, one observes that the active fraction 68 of Rep122-359 (Fig. 6B) and the equivalent fraction 49 of Rep122-359K227A (Fig. 6C) show two major components: a double band (band a) with an estimated molecular mass of 400 kDa, corresponding to an oligomer of 14 or 15 monomers, and band b, corresponding to a hexamer (Fig. 5B and C). Second, the results are in agreement with those from the glycerol gradients; after removal of MBP, Rep122-359 and Rep122-359K227A are composed of a population of oligomers of various sizes, ranging from hexamers up to dodecamers or higher. As Rep122-359 and its inactive counterpart Rep122-359K227A have the same pattern of oligomerization, ATP hydrolysis does not influence oligomerization.
FIG. 6.
BN-PAGE of MBP-Rep120-359, MBP-Rep120-359K227A, Rep122-359, and Rep122-359K227A. A, migration of the two fusion proteins MBP-Rep120-359 (lane 1) and MBP-Rep120-359K227A (lane 2); the major band in both cases has an estimated molecular mass of 660 kDa. B, migration of the DEAE fractions of Rep122-359. C, migration of the DEAE fractions of Rep122-359K227A. In B and C, the corresponding fraction numbers are indicated above the gel and 25 μl of the fractions was loaded. Products a and b are 400 kDa and 180 kDa, respectively. The molecular mass markers used are thyroglobulin (thyr) (669 kDa), ferritin (fer) (440 kDa), catalase (cat) (232 kDa), and aldolase (ald) (158 kDa).
The degrees of oligomerization determined by BN-PAGE and by sedimentation in glycerol gradients for the different proteins are at variance. The migration of proteins in BN gels depends on the charge shift induced by binding of Coomassie blue to the protein, and all proteins do not behave the same way; up to a 20% error can occur, compared to 10% for determination by combined density gradient and gel filtration. The general conclusions from these experiments are that (i) the MBP fusion proteins are composed of a major hexameric species (about a 9-mer in BN gels) and (ii) removal of MBP leads to the formation of oligomers of different sizes with ATPase activity, but only the proteins forming higher oligomers, dodecamers or larger, are helicases.
DISCUSSION
In this report we present results showing that the amino-terminally truncated Rep protein (aa 122 to 359) of the geminivirus TYLCSV has helicase activity in vitro and that this activity is dependent on the oligomerization state of the protein. We have therefore experimentally demonstrated that another putative member of the SF3 family (15) is a bona fide helicase. To establish the helicase activity of TYLCSV Rep, conditions for producing a soluble protein had to be found. Even though experimental conditions (MBP carrier and growth conditions) improved solubility, removal of the N-terminal domain (aa 1 to 121) was essential to obtain a workable protein.
The N-terminal domain of TYLCSV Rep carries the origin binding and endonuclease domains essential for initiation of RCR (24). Origin recognition is a key element of SF3 replication-associated helicases and is dispensable for helicase activity in vitro. This is a common feature exemplified by SV40 LTag (11) and the AAV2 Rep 40 helicase, which consists of only the helicase domain (6). Despite the fact that the DNA binding domain is nonessential for the helicase activity, it is involved in the proper assembly of the SF3 helicases at the origin of replication (reviewed in reference 18). At present, the nicking/closing activity of Rep, which is the first step in RCR, acts efficiently only an ssDNA in vitro (24) and is not naturally present on the double-stranded DNA replicative form; origin melting, producing the appropriate structure, could be another aspect of Rep helicase activity.
Data on the oligomerization state of geminivirus Rep protein are scarce. The presence and localization of the oligomerization domains for different Rep proteins were determined using the yeast double-hybrid technique or pull-down experiments (20, 35); these techniques, however, do not provide information on the size of the oligomers. Tomato golden mosaic virus AL1 has been estimated to be an octamer by gel filtration (35). In the case of wheat dwarf virus, Missich et al. (32) have characterized self-interactions of Rep by using chemical cross-linking and have shown that free monomers assemble into octamers depending on the pH; in addition, the octamers were shown to have low affinity for double-stranded DNA. TYLCSV Rep122-359 is always found as an oligomer; monomeric forms of Rep122-359 and Rep122-359K227A, free or as MBP fusions, in the absence of ATP and/or ssDNA (conditions which favor the monomers [36]), were never observed. In addition Rep122-359 and Rep122-359K227A behave exactly the same way as far as oligomerization is concerned, indicating that ATP hydrolysis is not involved in oligomerization and that the loss of helicase activity of Rep122-359K227A results only from loss of ATP hydrolysis. Partially deleting the oligomerization domain in MBP-Rep120-359 leads to a smaller oligomeric form (a tetramer on BN gels) that is totally inactive with regard to ATPase activity and nonspecific ssDNA binding, and supposedly to an inactive helicase, further underlining the importance of oligomerization for ATPase and helicase activities of Rep122-359 (F. Bernardi, unpublished data).
Our attempt to determine the precise oligomeric state of Rep122-359 with two different techniques has led to discrepant results, probably as a result of experimental conditions. We observe that the helicase activity is associated with dodecamers, which in the form of double hexamers have been shown to be the active form of several helicases, and this seems to be an emerging feature for replicative helicases that are well adapted to bidirectional replication (39). In the case of Rep122-359 this degree of oligomerization is achieved only after removal of MBP, which has two effects on Rep122-359 oligomerization. First, its presence reduces the degree of oligomerization, giving a major species of 6 to 9 oligomers for MBP-Rep120-359, and second, its removal increases the complexity of the oligomers (compare MBP-Rep120-359 in Fig. 5A and 6A and Rep122-359 in Fig. 5B and 6B and C), with formation of as high as 12 to 14 oligomers for Rep122-359. This complexity is not usually found for other helicases. Misfolding during expression in E. coli may account for the formation of some nonproductive complexes; however, complexity arises mainly after removal of MBP and therefore during assembly of the hexamers. Building of double hexamers primarily from monomeric subunits requires several steps, such as ATP binding and hydrolysis as well as DNA binding, these processes have been reviewed for some SF3 helicases (18). We never observed monomeric forms of Rep (F. Bernardi, unpublished data) or Rep122-359, which better resembles minichromosome maintenance (MCM) proteins, which are AAA+ helicases and the functional cellular homologs of the viral SF3 helicases. Particularly interesting is the archaeal MCM of Methanobacterium thermoautotrophicum, which shows structural polymorphism, hexamers, heptamers, dodecamers, open rings, and filaments, as well as a prevalence of double hexamers under physiological conditions (14). Oligomerization of M. thermoautotrophicum MCM involves the oligomerization and DNA binding domains, and the latter is absent in Rep122-359; consequently, improper interactions may occur, leading to the formation of anomalous oligomers. On the other hand, it cannot be ruled out that some of these complexes may have other activities in the virus life cycle.
The initial characterization of the helicase activity of Rep122-359 presented here shows that it shares common features with other helicases, such as the need for a free 3′ ssDNA tail and an unwinding polarity in the 3′-to-5′ direction; differences exist in the precise substrate requirements. A forked substrate is not needed; since the presence of a 3′ ssDNA is sufficient for unwinding (substrates h and i in Fig. 4B), increasing the length of the 3′overhang reduces the unwinding activity, contrary to what is observed for SV40 LTag, which requires an optimal ssDNA length for cooperative binding of hexamers (1). On the other hand, the presence of the dispensable 5′ ss tail enhances Rep122-359 activity, resembling the requirements of other helicases, among them MCM, where increasing the length of the 5′ tail as well as that of the 3′ tail enhances processivity (25). A similar behavior has been described for DnaB, the hexameric replicative helicase of prokaryotes (12). This observation indicates that Rep122-359 may interact with both strands. We have also observed a possible requirement for a free 3′ end which is not observed for the other SF3 helicases; other interpretations are possible, however, and a more detailed characterization will be necessary to clarify this point. These distinctive features of TYLCSV Rep122-359 may be related to the high oligomeric state formed in the absence of substrate and not found with other SF3 helicases.
The 3′-to-5′ polarity of unwinding by Rep is well suited for a replicative helicase participating in elongation during RCR. Motion of Rep on the minus strand would allow release of the viral plus strand as well as leading-strand synthesis, producing the new viral strand. In view of our results, the main difficulty that remains to be solved is loading of Rep122-359 on the minus strand, as we have shown that the lack of unwinding of the M13-derived substrates could result either from the lack of a free 3′ end or from the presence of a long stretch of ssDNA. The latter would probably not exist in the case of the nicked replicative form, allowing the helicase to be loaded and active. Other possibilities may exist; in particular, the N-terminal domain absent in Rep122-359 could control Rep loading, or other cellular factors binding to the Rep oligomerization domain may be needed (for instance, a loading factor) (7).
Dimerization of hexamers is associated with bidirectional replication (39), whereas RCR is thought to be mainly unidirectional, a point that remains to be clarified. An interesting hypothesis has been proposed by Christensen and Tattersall (5) for the rolling-hairpin replication of a parvovirus, the minute virus of mice (MVM). Rolling-hairpin replication is closely related to RCR and acts on a linear ssDNA genome instead of a closed ssDNA for RCR. MVM initiator protein NS1 (a dimer functional homolog of Rep for MVM), which has a 3′-to-5′ unwinding activity, is inactive as helicase when covalently bound to the genome at the initiation step, and these authors postulate that unwinding occurs through the association with another NS1 oligomer, yielding a double oligomer with only one active helicase component. It is conceivable that a similar situation may occur for Rep, associating a bound and inactive oligomer with a second, active oligomer, spatially coordinating the different steps of RCR at the origin of replication.
It is also significant that Rep interacts with several components of the replisome (proliferating cell nuclear antigen [3], and in the case of a mastrevirus Rep, also replication factor C [28]), indicating that the replisome builds around Rep; the polymerase remains to be identified yet, as well as the ssDNA binding protein essential for other well-characterized viral replication (5, 10).
The overall ATPase domain of TYLCSV Rep122-359 is 140 aa, that of AAV2 Rep40 is 180 aa, and that of SV40 LTag is 227 aa. A comparison of the secondary structures of SV40 LTag and AAV2 Rep40 with a predicted secondary structure of TYLCSV Rep122-359 (30) is presented in Fig. 7. The secondary structure prediction for Rep122-359 fits well with the known secondary elements of the two other helicases; the core ATPase domain with the conserved motifs is well preserved, but several structural elements are missing. The ATPase core domain formed by the five-stranded β-sheet is reduced to three β-strands (β1-β4-β3, carrying Walker A, C/sensor-1, and Walker B, respectively). Strand β2 and the cognate α2 helix are substituted by a loop or alternatively have not been correctly predicted, α2 is also missing in AAV2 Rep40. Last, helix α4 and strand β5 with the corresponding arginine finger are also missing. In TYLCSV Rep the second α-helical domain is limited to a single α-helix, as in the case of AAV2 Rep40. The arginine finger is an important structural element present in almost all AAA+ ATPases (21); it extends from one monomer to the neighboring one to form the nucleotide binding pocket and therefore often participates in ATP hydrolysis and association of the monomers; its role in the activity of several AAA+ ATPases has been reviewed (33).
FIG. 7.
Sequence alignment of the ATPase domains of SV40 LTag, AAV2 Rep40, and TYLCSV Rep122-359. Amino acids in blue are for α-helices, amino acids in orange are β-strands, conserved amino acids of AAA+ ATPases are in red, and asterisks indicate conserved amino acids in the B′ box of the SF3 helicases. The conserved motifs are underlined and labeled. The SV40 LTag sequence and secondary structure are from reference 26, and the AAV2 Rep40 sequence and secondary structure are from reference 22. The numbering of α-helices and β-strands is according to reference 21.
It will be of interest to further characterize TYLCSV Rep helicase, particularly in view of the much reduced size of its AAA+ ATPase domain, and to determine the precise function of the structural elements present in this domain in the helicase activity and in the mechanism of RCR in geminivirus.
Acknowledgments
We thank Bruno Gronenborn for his support during this work and for critical reading of the manuscript, and we are grateful to Tatiana Timchenko for comments on the manuscript. We thank Sylvie Lazereg for performing the mass spectroscopy experiments.
Footnotes
Published ahead of print on 30 August 2006.
REFERENCES
- 1.Alexandrov, A. I., M. R. Botchan, and N. R. Cozzarelli. 2002. Characterization of simian virus 40 T-antigen double hexamers bound to a replication fork. The active form of the helicase. J. Biol. Chem. 277:44886-44897. [DOI] [PubMed] [Google Scholar]
- 2.Bronstein, J. C., and P. C. Weber. 2001. Purification of a bacterially expressed herpes simplex virus type 1 origin binding protein for use in posttranslational processing studies. Protein Expr. Purif. 22:276-285. [DOI] [PubMed] [Google Scholar]
- 3.Castillo, A. G., D. Collinet, S. Deret, A. Kashoggi, and E. R. Bejarano. 2003. Dual interaction of plant PCNA with geminivirus replication accessory protein (Ren) and viral replication protein (Rep). Virology 312:381-394. [DOI] [PubMed] [Google Scholar]
- 4.Choi, I. R., and D. C. Stenger. 1995. Strain-specific determinants of beet curly top geminivirus DNA replication. Virology 206:904-912. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, J., and P. Tattersall. 2002. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 76:6518-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collaco, R. F., V. Kalman-Maltese, A. D. Smith, J. D. Dignam, and J. P. Trempe. 2003. A biochemical characterization of the adeno-associated virus Rep40 helicase. J. Biol. Chem. 278:34011-34017. [DOI] [PubMed] [Google Scholar]
- 7.Davey, M. J., and M. O'Donnell. 2003. Replicative helicase loaders: ring breakers and ring makers. Curr. Biol. 13:R594-R596. [DOI] [PubMed] [Google Scholar]
- 8.Desbiez, C., C. David, A. Mettouchi, J. Laufs, and B. Gronenborn. 1995. Rep protein of tomato yellow leaf curl geminivirus has an ATPase activity required for viral DNA replication. Proc. Natl. Acad. Sci. USA 92:5640-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erickson, H. P. 2004. Protein-protein interactions. Chapter 1. Protein structure-size and shape at the nm level. http://www.cellbio.duke.edu/Faculty/Erickson/pdf%27s/Prot-p%20Ch1.pdf.
- 10.Fairman, M. P., and B. Stillman. 1988. Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J. 7:1211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gai, D., D. Li, C. V. Finkielstein, R. D. Ott, P. Taneja, E. Fanning, and X. S. Chen. 2004. Insights into the oligomeric states, conformational changes, and helicase activities of SV40 large tumor antigen. J. Biol. Chem. 279:38952-38959. [DOI] [PubMed] [Google Scholar]
- 12.Galletto, R., M. J. Jezewska, and W. Bujalowski. 2004. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: the effect of the 3′ arm and the stability of the dsDNA on the unwinding activity of the Escherichia coli DnaB helicase. J. Mol. Biol. 343:101-114. [DOI] [PubMed] [Google Scholar]
- 13.Giraldo, R. 2003. Common domains in the initiators of DNA replication in Bacteria, Archaea and Eukarya: combined structural, functional and phylogenetic perspectives. FEMS Microbiol. Rev. 26:533-554. [DOI] [PubMed] [Google Scholar]
- 14.Gomez-Llorente, Y., R. J. Fletcher, X. S. Chen, J. M. Carazo, and C. San Martin. 2005. Polymorphism and double hexamer structure in the archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. J. Biol. Chem. 280:40909-40915. [DOI] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419-429. [Google Scholar]
- 16.Gorbalenya, A. E., E. V. Koonin, and Y. I. Wolf. 1990. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 262:145-148. [DOI] [PubMed] [Google Scholar]
- 17.Hanley-Bowdoin, L., S. B. Settlage, and D. Robertson. 2004. Reprogramming plant gene expression: a prerequisite to geminivirus DNA replication. Mol. Plant Pathol. 5:149-156. [DOI] [PubMed] [Google Scholar]
- 18.Hickman, A. B., and F. Dyda. 2005. Binding and unwinding: SF3 viral helicases. Curr. Opin. Struct. Biol. 15:77-85. [DOI] [PubMed] [Google Scholar]
- 19.Hingorani, M. M., and M. O'Donnell. 2000. A tale of toroids in DNA metabolism. Nat. Rev. Mol. Cell. Biol. 1:22-30. [DOI] [PubMed] [Google Scholar]
- 20.Horvath, G. V., A. Pettko-Szandtner, K. Nikovics, M. Bilgin, M. Boulton, J. W. Davies, C. Gutierrez, and D. Dudits. 1998. Prediction of functional regions of the maize streak virus replication-associated proteins by protein-protein interaction analysis. Plant Mol. Biol. 38:699-712. [DOI] [PubMed] [Google Scholar]
- 21.Iyer, L. M., D. D. Leipe, E. V. Koonin, and L. Aravind. 2004. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146:11-31. [DOI] [PubMed] [Google Scholar]
- 22.James, J. A., C. R. Escalante, M. Yoon-Robarts, T. A. Edwards, R. M. Linden, and A. K. Aggarwal. 2003. Crystal structure of the SF3 helicase from adeno-associated virus type 2. Structure 11:1025-1035. [DOI] [PubMed] [Google Scholar]
- 23.Lanzetta, P. A., L. J. Alvarez, P. S. Reinach, and O. A. Candia. 1979. An improved assay for nanomole amounts o f inorganic phosphate. Anal. Biochem. 100:95-97. [DOI] [PubMed] [Google Scholar]
- 24.Laufs, J., W. Traut, F. Heyraud, V. Matzeit, S. G. Rogers, J. Schell, and B. Gronenborn. 1995. In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc. Natl. Acad. Sci. USA 92:3879-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, J. K., and J. Hurwitz. 2001. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc. Natl. Acad. Sci. USA 98:54-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, D., R. Zhao, W. Lilyestrom, D. Gai, R. Zhang, J. A. DeCaprio, E. Fanning, A. Jochimiak, G. Szakonyi, and X. S. Chen. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423:512-518. [DOI] [PubMed] [Google Scholar]
- 27.Lohman, T. M., and K. P. Bjornson. 1996. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 65:169-214. [DOI] [PubMed] [Google Scholar]
- 28.Luque, A., A. P. Sanz-Burgos, E. Ramirez-Parra, M. M. Castellano, and C. Gutierrez. 2002. Interaction of geminivirus Rep protein with replication factor C and its potential role during geminivirus DNA replication. Virology 302:83-94. [DOI] [PubMed] [Google Scholar]
- 29.Martin, R. G., and B. N. Ames. 1961. A method for determining the sedimentation behavior of enzymes: application to protein mixture. J. Biol. Chem. 236:1372-1379. [PubMed] [Google Scholar]
- 30.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 31.Mendez, J., and B. Stillman. 2003. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays 25:1158-1167. [DOI] [PubMed] [Google Scholar]
- 32.Missich, R., E. Ramirez-Parra, and C. Gutierrez. 2000. Relationship of oligomerization to DNA binding of wheat dwarf virus RepA and Rep proteins. Virology 273:178-188. [DOI] [PubMed] [Google Scholar]
- 33.Ogura, T., S. W. Whiteheart, and A. J. Wilkinson. 2004. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J. Struct. Biol. 146:106-112. [DOI] [PubMed] [Google Scholar]
- 34.Orozco, B. M., L. J. Kong, L. A. Batts, S. Elledge, and L. Hanley-Bowdoin. 2000. The multifunctional character of a geminivirus replication protein is reflected by its complex oligomerization properties. J. Biol. Chem. 275:6114-6122. [DOI] [PubMed] [Google Scholar]
- 35.Orozco, B. M., A. B. Miller, S. B. Settlage, and L. Hanley-Bowdoin. 1997. Functional domains of a geminivirus replication protein. J. Biol. Chem. 272:9840-9846. [DOI] [PubMed] [Google Scholar]
- 36.Patel, S. S., and K. M. Picha. 2000. Structure and function of hexameric helicases. Annu. Rev. Biochem. 69:651-697. [DOI] [PubMed] [Google Scholar]
- 37.Schagger, H., W. A. Cramer, and G. von Jagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220-230. [DOI] [PubMed] [Google Scholar]
- 38.Schulke, N., M. S. Vesanen, R. W. Sanders, P. Zhu, M. Lu, D. J. Anselma, A. R. Villa, P. W. H. I. Parren, J. M. Binley, K. H. Roux, P. J. Maddon, J. P. Moore, and W. C. Olson. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J. Virol. 76:7760-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sclafani, R. A., R. J. Fletcher, and X. S. Chen. 2004. Two heads are better than one: regulation of DNA replication by hexameric helicases. Genes Dev. 18:2039-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen, J., D. Gai, A. Patrick, W. B. Greenleaf, and X. S. Chen. 2005. The roles of the residues on the channel beta-hairpin and loop structures of simian virus 40 hexameric helicase. Proc. Natl. Acad. Sci. USA 102:11248-11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siegel, L. M., and K. J. Monty. 1966. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta 112:346-362. [DOI] [PubMed] [Google Scholar]
- 42.Stenlund, A. 2003. Initiation of DNA replication: lessons from viral initiator proteins. Nat. Rev. Mol. Cell. Biol. 4:777-785. [DOI] [PubMed] [Google Scholar]
- 43.Yoon-Robarts, M., A. G. Blouin, S. Bleker, J. A. Kleinschmidt, A. K. Aggarwal, C. R. Escalante, and R. M. Linden. 2004. Residues within the B′ motif are critical for DNA binding by the superfamily 3 helicase Rep40 of adeno-associated virus type 2. J. Biol. Chem. 279:50472-50481. [DOI] [PubMed] [Google Scholar]