Abstract
Swine influenza viruses (SIV) naturally infect pigs and can be transmitted to humans. In the pig, genetic reassortment to create novel influenza subtypes by mixing avian, human, and swine influenza viruses is possible. An SIV vaccine inducing cross-protective immunity between different subtypes and strains circulating in pigs is highly desirable. Previously, we have shown that an H3N2 SIV (A/swine/Texas/4199-2/98 [TX98]) containing a deleted NS1 gene expressing a truncated NS1 protein of 126 amino acids, NS1▴126, was attenuated in swine. In this study, 4-week-old pigs were vaccinated with the TX98 NS1▴126 modified live virus (MLV). Ten days after boosting, pigs were challenged with wild-type homologous H3N2 or heterosubtypic H1N1 SIV and sacrificed 5 days later. The MLV was highly attenuated and completely protected against challenge with the homologous virus. Vaccinated pigs challenged with the heterosubtypic H1N1 virus demonstrated macroscopic lung lesions similar to those of the unvaccinated H1N1 control pigs. Remarkably, vaccinated pigs challenged with the H1N1 SIV had significantly lower microscopic lung lesions and less virus shedding from the respiratory tract than did unvaccinated, H1N1-challenged pigs. All vaccinated pigs developed significant levels of hemagglutination inhibition and enzyme-linked immunosorbent assay titers in serum and mucosal immunoglobulin A antibodies against H3N2 SIV antigens. Vaccinated pigs were seronegative for NS1, indicating the potential use of the TX98 NS1▴126 MLV as a vaccine to differentiate infected from vaccinated animals.
Swine influenza virus (SIV) is an important swine pathogen involved in the porcine respiratory disease complex in most swine-producing countries. Serological and virological studies demonstrated 23 to 28% prevalence of SIV in swine populations in the midwestern and north central United States (4, 35, 56). Mortality of SIV in pigs is low, while morbidity may approach 100%. SIV-affected pigs show clinical signs of anorexia and weight loss, fever, respiratory distress, coughing, and nasal discharge (18). SIVs currently circulating in North American swine are subtypes H1N1, H3N2, and H1N2 (36). From 1930 to 1998, classical H1N1 viruses were the subtype predominantly isolated from U.S. swine. In 1998, a new SIV subtype, H3N2, emerged from reassortment of swine, human, and avian influenza viruses (19, 56, 58). The H3N2 SIV acquired PB1, hemagglutinin (HA), and NA genes from recent human viruses, PB2 and PA from avian viruses, and NP, M, and NS genes from the classical H1N1 swine virus (19, 56, 58). A year later, reassortment between the H3N2 and classical H1N1 SIVs resulted in a new subtype, H1N2, where the HA of the H3N2 subtype was replaced by the HA from the classical H1N1 virus (3, 20).
SIV is classified in the genus Influenzavirus A, containing eight segments of a single-stranded RNA genome of negative polarity, PB1, PB2, PA, HA, NP, NA, M, and NS. The NS gene of influenza A virus encodes two different proteins, NS1 and nuclear export protein (NEP) (22). NS1 of influenza A viruses contains 202 to 237 amino acid residues. NEP is a product of the spliced NS mRNA and shares the first 10 amino acids with NS1. NS1 is the most abundant nonstructural protein of influenza A viruses expressed in infected cells. NS1 is responsible for virus virulence due to its immunomodulatory functions (13, 21).
One of the front lines of the innate host defense mechanism against viral infection is the production of type I interferon (IFN). Viral infection induces the activation of several transcription factors, including NF-κB, AP-1, and members of the IFN regulatory factor family that bind to the IFN-β promoter, resulting in transcriptional activation of the IFN-β gene and production of IFN-β (13). Binding of secreted IFN-β to the IFN receptor switches on a cascade of events which turns on the Jak-STAT pathway, resulting in the production of IFN-inducible proteins with antiviral activity, including Mx, protein kinase R (PKR), and 2′,5′-oligoadenylate synthetase (OAS), which inhibit viral replication by a variety of mechanisms. The main function of the influenza virus NS1 protein appears to be inhibition of the IFN system, since a recombinant influenza virus lacking the NS1 gene is able to replicate in IFN-deficient systems but not in IFN-competent systems (14).
Currently available swine influenza vaccines are based on killed viruses containing both H1N1 and H3N2 subtypes. Phylogenetic analysis of the HA1 region of HA genes from various H3N2 SIVs revealed three different virus clusters: I, II, and III (56). Viruses in cluster II had limited cross-reactivity with viruses of other clusters in hemagglutination inhibition (HI) and serum neutralization tests (42). Similarly, there were only low to moderate degrees of cross-reactivity among recent H1N1 and H1N2 SIV isolates from the United States (53). These studies indicate that genetic and antigenic heterogeneities that might reduce vaccine effectiveness occur within each subtype of SIV (5, 42). In addition, killed-virus vaccine formulations induce mainly humoral immune responses while complete protection from SIV infections might require both humoral and cell-mediated immunity (16). Modified live-virus (MLV) vaccines may induce both types of defense mechanisms and provide a substantial level of heterosubtypic immunity (Het-I), resulting in cross-protection among different subtypes of influenza viruses (15, 50).
In this study, a modified live-influenza vaccine based on an H3N2 SIV containing a deleted NS1 gene expressing a truncated NS1 protein of 126 amino acids (NS1▴126) was produced using reverse genetics (46). Previously, we demonstrated that this virus is highly attenuated in swine due to an impaired ability to inhibit the induction of type I IFN (46). We now report that vaccination of pigs with NS1▴126 virus resulted in complete protection against challenge with a homosubtypic H3N2 virus and partial protection against challenge with a heterosubtypic H1N1 virus.
MATERIALS AND METHODS
Cells and viruses.
MDCK cells were cultured in a growth medium containing minimal essential medium (MEM) and 5% fetal bovine serum. 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. The H3N2 SIVs used in our studies were generated from plasmid DNA by reverse-genetics techniques, as previously described (46). Viruses generated include a recombinant wild-type A/swine/Texas/4199-2/98 (rTX98) virus and the MLV TX98 NS1▴126 virus, containing a mutated NS gene plus the remaining seven genes of wild-type TX98. These two viruses were propagated in the allantoic cavity of 7-day-old embryonated chicken eggs. The rTX98 and TX98 NS1▴126 virus stocks had titers of 3 × 107 and 5.3 × 107 PFU/ml, respectively. The other isolate used for the challenge experiments was A/swine/Minnesota/37866/99 (MN99), a classical H1N1 SIV isolate. The MN99 isolate used in this study was derived from the bronchoalveolar lavage fluid (BALF) from a pig experimentally inoculated with egg-derived virus and had a titer of 5 × 105 PFU/ml.
Experimental design for vaccination and challenge.
At 10 days of age, 48 weaned piglets from an SIV-negative herd were randomly numbered and treated with 0.5 ml ceftiofur hydrochloride (broad-spectrum cephalosporin antibiotic, Excenel RTU; Pfizer) once daily for 3 days at the time of weaning. The piglets were assigned to six groups of eight or nine pigs each (Table 1). One week before inoculation, pigs in each group were housed separately in an isolation room. All piglets were seronegative for H1N1 and H3N2 SIVs in hemagglutination inhibition assays. At 4 weeks of age (−31 days postinfection [p.i.]), pigs in groups 1 to 3 were mock infected intratracheally with 1 ml of MEM, while those in groups 4 to 6 were inoculated intratracheally, as described elsewhere (42), with the TX98▴126 MLV diluted in MEM at a dose of 2 × 105 PFU/ml/pig. At 3 weeks after the first vaccination (−10 days p.i.), pigs in groups 1 to 3 received mock inoculum, whereas the vaccinated groups (groups 4 to 6) were boosted intranasally with 2 × 105 PFU/ml/pig of TX98▴126 MLV vaccine. Pigs were challenged intratracheally with different viruses 10 days (0 days p.i.) after the second dose of the respective vaccine inoculum. Pigs in groups 1 and 4 were mock infected, groups 2 and 5 received 2 × 105 PFU/ml of rTX98 (H3N2) per pig, and groups 3 and 6 were challenged with 2 × 105 PFU/ml of MN99 (H1N1) per pig (Table 1). We used the intratracheal route of inoculation in order to assure a consistent infection of the pigs with the MLV and challenge virus. During the 5-day observation period after SIV challenge, pigs were observed twice daily for clinical signs such as respiratory distress and weight loss, as well as nasal and ocular discharge. Body temperature of the animals was not determined.
TABLE 1.
Assignment of pigs in each group
| Group no. (n = 8 or 9) | Inoculum
|
||
|---|---|---|---|
| First vaccination (−31 days p.i.) | Second vaccination (−10 days p.i.) | Challenge (0 days p.i.) | |
| 1 | Mock inoculum | Mock inoculum | Mock inoculum |
| 2 | Mock inoculum | Mock inoculum | rTX98 (H3N2) |
| 3 | Mock inoculum | Mock inoculum | MN99 (H1N1) |
| 4 | TX98▴126 MLV | TX98▴126 MLV | Mock inoculum |
| 5 | TX98▴126 MLV | TX98▴126 MLV | rTX98 (H3N2) |
| 6 | TX98▴126 MLV | TX98▴126 MLV | MN99 (H1N1) |
Nasal swab samples were collected from all pigs at 2 and 5 days p.i. Pigs were euthanized at 5 days after challenge, and lungs were examined for the presence of macroscopic lesions in all seven lung lobes. Lungs and tracheal samples were fixed in 10% buffered formalin for further histopathological and immunohistochemical evaluations. Mean lung scores were calculated as the averages of percent consolidation present in seven lung lobes (42). BALF was collected at necropsy by lavaging the lungs with 50 ml of MEM for examination of antibody levels and virus titers. Serum samples were obtained before the first vaccination (−31 days p.i.), before the second vaccination (−10 days p.i.), at day of challenge (0 days p.i.), and at necropsy (5 days p.i.).
Histopathology.
Sections of trachea (upper and lower parts) and lung were routinely stained with hematoxylin and eosin and examined microscopically for histopathological changes. The trachea was evaluated for epithelial lesions and scored 0 for no lesions, 1 for occasional epithelial attenuation or degeneration, 2 for consistent epithelial attenuation or degeneration with cilia still present, and 3 for consistent epithelial attenuation or degeneration with no cilia present.
Sections of lungs were examined for bronchiolar epithelial changes and peribronchiolar inflammation in large, medium, and small or terminal bronchioles. Lesion severity was scored by the distribution or extent of lesions within the sections examined as follows: 0, no airways affected; 1, only a few isolated airways affected; 2, localized cluster of affected airways, often within one or two lobules; 3, several airways affected throughout entire section; and 4, many airways affected, often severely. One trained examiner was utilized for evaluation of tissue sections. The examiner did not know from which group of animals the tissues were derived.
IHC.
SIV-specific antigen was detected in lung tissues with a modified automated procedure using a previously described immunohistochemistry (IHC) method with minor alterations (54). Briefly, tissue sections were deparaffinized and hydrated in distilled water. Slides were quenched in 3% hydrogen peroxide for 10 min, rinsed three times in deionized water, and treated in 0.1% proteinase K for 2 min. Slides were then rinsed twice in deionized water and once in Tris-buffered saline. SIV-specific monoclonal antibody HB65 (ATCC), specific for the nucleoprotein of influenza A viruses, was applied at a 1:1,000 dilution, and slides were incubated at room temperature for 1 h. To detect positive signals, bound monoclonal antibodies were stained with peroxidase-labeled anti-mouse immunoglobulin G (IgG) followed by chromogen using a DAKO Envision IHC system according to the manufacturer's instructions. The slides then were rinsed in deionized water and counterstained with Gill's hematoxylin.
Positive IHC signals were scored according to the following criteria: 0, no signal present; 1, only a few cells were positive in an occasional airway; 2, only a few cells were positive in scattered airways; 3, moderate numbers of cells were positive in an occasional airway; and 4, moderate numbers of cells were positive in scattered airways and alveoli. The cardiac lobe was collected for IHC analysis because our previous experience with SIV infections in swine revealed that macroscopic and microscopic lesions are regularly present in this lung lobe.
Virus titration.
Quadruplicates of each BALF and nasal swab sample were 10-fold serially diluted in an inoculation medium containing MEM, 4% bovine serum albumin fraction V (Invitrogen), and 1 μg/ml TPCK [l-(tosylamido-2-phenyl) ethyl chloromethyl ketone]-treated trypsin (Worthington Biochemical) and inoculated onto a monolayer of MDCK cells grown in 96-well plates. Each sample was 10-fold serially diluted, starting at 10−1 and continuing to 10−10. Infected cells were incubated for 48 h prior to being fixed with methanol. An indirect immunofluorescence assay was performed to detect the presence of viral protein in the infected cells. Briefly, cells were incubated with anti-SIV hyperimmune serum (obtained from H3N2- and H1N1-infected pigs 14 days p.i.) for 30 min before being stained with fluorescein isothiocyanate-labeled anti-swine IgG (Sigma). Positive signals were visualized using fluorescence microscopy. Virus titers on MDCK cells were determined as 50% tissue culture infective dose (TCID50)/ml by the Reed and Muench method (40).
Serological assays. (i) HI test.
HI tests were performed in microtiter plates. Sera were incubated at 56°C for 30 min to inactivate complement prior to incubation with 20% kaolin suspension for 20 min to remove nonspecific inhibitors, followed by absorption with 0.5% chicken red blood cells for 20 min. Allantoic fluids containing wild-type TX98 (H3N2) and MN99 (H1N1) SIVs were titrated using the hemagglutination assay. Viruses were diluted to 8 HA units and used in a standard HI test (37). The first serum dilution tested was 1 to 10, and then each sample was twofold serially diluted.
(ii) Serum ELISA.
Serum antibodies against SIV were measured by commercially available enzyme-linked immunosorbent assays (ELISA) (HerdChek SIV H1N1 and H3N2; IDEXX Laboratories, Inc., Westbrook, Maine) according to the manufacturer's protocol. The presence or absence of antibodies was determined by calculating the total sample-to-positive sample (S/P) ratio, and the mean S/P ratios for the different treatment groups were compared. According to the manufacturer's protocol, a sample was considered to be positive for SIV antibodies if the S/P ratio was ≥0.4.
(iii) SIV-specific antibodies in bronchoalveolar lavage fluid.
The ELISA used to determine SIV-specific IgG and IgA mucosal antibodies present in the respiratory tract was performed as previously described (23), with slight modifications. The BALF samples at 5 days p.i. were incubated at 37°C for 1 h with an equal volume of 10 mM dithiothreitol to disrupt mucus present in the fluids. Independent assays were run using TX98 H3N2 and MN99 H1N1 as ELISA antigens. Concentrated wild-type virus was resuspended in Tris-EDTA buffer, pH 7.8, and diluted to a hemagglutination concentration of 100 HA units/50 μl. Immulon-2HB 96-well plates (Dynex) were coated with 100 μl of SIV antigen and incubated at room temperature overnight. Plates were blocked for 1 h with 100 μl of 10% BSA in PBS and washed three times with 0.05% Tween 20 in PBS. The assays were performed on each BALF sample in triplicate. Plates were incubated at room temperature for 1 h, washed three times with 0.05% Tween 20 in PBS, and then incubated with peroxidase-labeled goat anti-swine IgA (Bethyl) or IgG (Kirkegaard & Perry Laboratories) at 37°C for 1 h. ABTS [2,2′-azino-di-(3-ethylbenzthiazoline-6-sulfonate)]-peroxidase was added as the substrate (Kirkegaard & Perry Laboratories), and optical density (OD) was measured at a 405-nm wavelength. Antibody levels were reported as mean ODs, and the mean ODs of the different treatment groups were compared.
NS1 ELISA.
Serum samples at 5 days p.i. were evaluated for the presence of antibodies specific for the NS1 protein of SIV. Positive-control sera were obtained from piglets (n = 19) infected with the wild-type TX98 SIV at 4 weeks of age and euthanized at 7, 14, and 21 or 28 days p.i. An indirect ELISA was carried out in a 96-well format to detect the presence of antibodies specific for the NS1 protein of SIV in pigs. Recombinant NS1 protein was expressed as a glutathione S-transferase (GST) fusion protein in Escherichia coli. The purified NS1-GST fusion protein was used as the positive antigen, and GST protein was used as the negative-control antigen for coating ELISA plates. Plates were coated with 100 ng of each antigen per well. Serum samples were tested after being diluted 1:100. Each sample was run in duplicate, and the test was carried out at ambient temperature. Specific antigen-antibody reactions were visualized by adding an optimal dilution (1:2,500) of goat anti-swine IgG conjugated with horseradish peroxidase (Kirkegaard & Perry Laboratories) prior to incubation with 3,3′,5,5′-tetramethylbenzidine (TMB) substrate according to the manufacturer's instructions (Kirkegaard & Perry Laboratories). The OD of each sample was measured at a 450-nm wavelength. The reactions of serum samples with both antigens were determined, and the OD of the serum with GST alone was subtracted from the OD with the NS1-GST fusion protein. The cutoff value was evaluated by receiver operating characteristic analysis using MedCalc, version 8.1.1.0. A mean net OD of ≥0.15 was considered positive for antibodies against the NS1 protein.
Statistical analysis.
Raw data from macroscopic lung lesion scores from all lung lobes and microscopic lesion and IHC scores from right cardiac lobes as well as ELISA ODs were used directly for statistical calculations. Virus titers of 100 were used in calculations for samples with titers less than 101 and were transformed to a log10 scale. Statistical analyses of macroscopic and microscopic lesion scores, virus titers, and ELISA ODs were performed using Statistix8 (Analytical Software, Tallahassee, FL). Differences between means of each group in each assay were determined by using one-way analysis of variance methods. When the mean of at least one group significantly differed from others, with a P of <0.05, the difference between each pair of means was computed using a Tukey post-hoc comparison test.
RESULTS
MLV protects pigs from macroscopic lung lesions after homosubtypic but not heterosubtypic challenge.
At 5 days postchallenge, pigs were euthanized and necropsies were performed. No obvious respiratory signs were observed during 5 days of observation before the animals were euthanized. No significant differences between vaccinated and unvaccinated and challenged animals were observed with respect to weight loss, respiratory distress, and nasal or ocular discharge. It should be noted that infection of 10-week-old pigs does not produce the clinical signs observed in 4-week-old pigs with the same SIV isolates. All pigs in the unvaccinated and unchallenged control group (group 1, mock infected) were free of macroscopic lung lesions. Typical SIV lesions, characterized by marked, plum-colored, consolidated areas on lung lobes, were observed in all pigs in the unvaccinated and challenged groups (groups 2 and 3) as well as the group vaccinated and challenged with the heterosubtypic H1N1 virus (group 6). Average lung lesion scores are given in Table 2. There was no significant difference among average lung scores of groups 2, 3, and 6. Lesions were most prominent in cardiac and apical lobes, while they were less severe in intermediate and diaphragmatic lobes. Average lung lesion scores of vaccinated and mock-challenged (group 4) or homologous virus-challenged (group 5) groups were not significantly different from that of the healthy-control, mock-challenged group (group 1) but were significantly lower than those of unvaccinated pigs challenged with H3N2 (homosubtypic) and H1N1 (heterosubtypic) control viruses (groups 2 and 3) and those of vaccinated pigs challenged with the heterosubtypic SIV (group 6).
TABLE 2.
Summary of each tested parameter for pathological findings and virus titersa
| Treatment group | Lung lesion score | Microscopic lung lesion scoreb | IHC scorec | Virus titerd
|
||
|---|---|---|---|---|---|---|
| BALF (5 days p.i.) | Nasal swab
|
|||||
| 2 days p.i. | 5 days p.i. | |||||
| Mock challenge alone | 0.0 ± 0.0 B (0/8) | 0.0 ± 0.0 C (0/8) | 0.0 ± 0.0 B (0/8) | 0.0 ± 0.0 B (0/8) | 0.0 ± 0.0 C (0/8) | 0.0 ± 0.0 C (0/8) |
| Mock with rTX98 challenge | 13.4 ± 6.9 A (8/8) | 1.9 ± 0.6 A (8/8) | 1.5 ± 0.5 A (8/8) | 5.6 ± 0.5 A (8/8) | 2.4 ± 1.7 A (8/8) | 4.5 ± 0.0 A (8/8) |
| Mock with MN99 challenge | 18.6 ± 6.8 A (8/8) | 2.7 ± 0.8 A (8/8) | 1.6 ± 0.8 A (8/8) | 5.1 ± 0.4 A (8/8) | 1.7 ± 1.0 AB (8/8) | 4.3 ± 0.4 A (8/8) |
| MLV with mock challenge | 0.2 ± 0.5 B (0/8) | 0.0 ± 0.0 C (0/8) | 0.0 ± 0.0 B (0/8) | 0.0 ± 0.0 B (0/8) | 0.0 ± 0.0 C (0/8) | 0.0 ± 0.0 C (0/8) |
| MLV with rTX98 challenge | 0.2 ± 0.4 B (0/8) | 0.0 ± 0.0 C (0/8) | 0.0 ± 0.0 B (0/8) | 0.0 ± 0.0 B (0/8) | 0.0 ± 0.0 C (0/8) | 0.0 ± 0.0 C (0/8) |
| MLV with MN99 challenge | 19.2 ± 9.3 A (9/9) | 0.9 ± 0.9 B (5/9) | 0.0 ± 0.0 B (0/9) | 0.4 ± 1.2 C (6/9) | 0.7 ± 1.2 BC (3/9) | 1.4 ± 1.7 B (4/9) |
Means ± standard deviations are shown, with numbers of positive samples/total numbers of samples in parentheses. Letters A, B, and C after each number represent groups of means in each parameter that are significantly different from each other, with a P value of <0.05. Means labeled with the same letter are not significantly different, while those with different letters are significantly different.
Only the right cardiac lung lobe was analyzed.
IHC was performed on the right cardiac lung lobe only.
Values have been transferred into a log10 scale for calculation and report.
Microscopic lung lesions and presence of SIV antigen in lung tissue of MLV-vaccinated pigs after homosubtypic and heterosubtypic challenge.
Histopathological changes in lungs and trachea of pigs in the mock-challenged group (group 1) as well as vaccinated and mock-challenged (group 4) or homosubtypic SIV-challenged (group 5) groups corresponded to the macroscopic lesions. In the group with mock challenge alone, there were no lesions in lungs or trachea suggestive of SIV infection in any of these pigs.
Unvaccinated pigs challenged with H3N2 TX98 (group 2) or H1N1 MN99 (group 3) produced the most severe histopathological changes, all typical for SIV infection. Lesions in the H3N2-challenged group were mild to moderate, while lesions in the H1N1-challenged group tended to be moderate to severe. Tracheal lesions were mild in both groups and were found in about half of the animals (four of eight animals in group 2 and five of eight animals in group 3). Average microscopic lesion scores of both groups were high, did not differ from each other (Table 2), and were significantly higher than those of the group with mock challenge alone (group 1) or the vaccinated groups mock challenged (group 4) or challenged with homo- and heterosubtypic SIVs (groups 5 and 6) (Table 2). Lesions consisted of mild and occasional lymphocytic cuffing of bronchioles, occasional accumulation of neutrophils in large airways, and multifocal atelectasis (Fig. 1C and E). None of the pigs in the unvaccinated or vaccinated and mock-challenged groups (groups 1 and 4) produced microscopic lesions in the lung (Fig. 1A and G). In one animal from group 1, mild lesions were found in the upper trachea, probably due to physical injury from intratracheal inoculation. Similarly, vaccinated pigs challenged with H3N2 (group 5) produced no lung lesions (Fig. 1I). Mild tracheal changes were noted in two pigs, but these were mild and also considered incidental due to intratracheal-challenge trauma. Average microscopic lesion scores of this group were similar to those of the mock-infected groups 1 and 4 (Table 2).
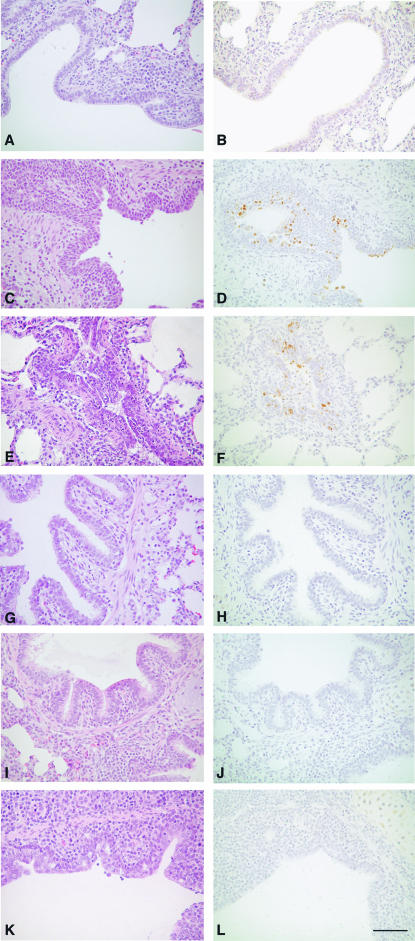
FIG. 1.
Histopathologic evaluation of lungs from vaccinated and unvaccinated pigs challenged with either H3N2 (TX98) or H1N1 or mock challenged. (A) Hematoxylin and eosin (H&E)-stained section of lung from an unvaccinated, mock-challenged pig with normal bronchiolar epithelium. (B) Immunohistochemical staining in lung from the pig represented in panel A, containing no positive signal. (C) H&E-stained section of lung from an unvaccinated pig challenged with H3N2 (TX98). The bronchiolar epithelium is attenuated in early stages of recovery. (D) Immunohistochemical staining of lung from the pig represented in panel C, identifying SIV antigen in the bronchiolar epithelium (brown staining). (E) H&E-stained section of lung from an unvaccinated pig challenged with H1N1 (MN99). The bronchiolar epithelium is attenuated, and the lumen contains necrotic cellular debris and exudates. (F) Immunohistochemical staining of lung from the pig represented in panel E, identifying SIV antigen in the bronchiolar epithelium (brown staining). (G) H&E-stained section of lung from a pig vaccinated with the modified live vaccine and mock challenged. The bronchiolar epithelium is unaffected. (H) Immunohistochemical staining of lung from the pig represented in panel G, containing no positive signal. (I) H&E-stained section of lung from a pig vaccinated with the modified live vaccine and challenged with H3N2 (TX98). The bronchiolar epithelium is unaffected. (J) Immunohistochemical staining of lung from the pig represented in panel I, containing no positive signal. (K) H&E-stained section of lung from a pig vaccinated with the modified live vaccine and challenged with H1N1. The bronchiolar epithelium is mildly attenuated and proliferative. (L) Immunohistochemical staining of lung from the pig represented in panel K, containing no positive signal. Magnification, ×225. Bar = 75 μm.
Vaccinated pigs challenged with the heterosubtypic H1N1 SIV (group 6) produced severe macroscopic lung lesions but only mild microscopic lesions at day 5 p.i. (Fig. 1K). Microscopic lesions were observed in the right cardiac lobes of five of nine pigs in group 6; however, the cardiac lobe lesion-negative pigs had few histopathological lesions in other lung lobes. In contrast, cardiac lobe lesions were present in all unvaccinated, H1N1-challenged pigs (group 3). Tracheal lesions were mild and found in four of the nine pigs in group 6. Microscopic lung lesion scores of group 6 (Table 2) were mild to moderate but significantly lower than those of the unvaccinated, H1N1-challenged group (group 3).
Detection of SIV antigen by IHC in lung cells revealed that all unvaccinated animals challenged with the homosubtypic and heterosubtypic SIVs were positive for SIV antigen by IHC at day 5 p.i. (Fig. 1D and F). In contrast, IHC influenza-positive cells were not found in lung tissues of the MLV-vaccinated pigs challenged with homosubtypic and heterosubtypic SIVs (Fig. 1J and L). Also, all pigs in the unvaccinated and vaccinated and mock-infected groups were negative for the presence of SIV antigen (Fig. 1B and H). IHC scores of both unvaccinated groups (group 2 and 3) were not different but were significantly higher than those of groups with mock challenge alone (group 1) and the three MLV-vaccinated groups (groups 4 to 6) (Table 2).
Vaccination reduces virus shedding.
Nasal swabs collected at 2 and 5 days p.i. and BALF collected at necropsy were 10-fold serially diluted from 100 to 10−6 and inoculated onto a monolayer of MDCK cells. The titer was determined as TCID50/ml. Mean virus titers from each group are given in Table 2. When present, virus titers of nasal swabs at 5 days p.i. were higher than at 2 days p.i., except for the titer of one pig in group 2. Virus titers of BALF from pigs in groups of nonvaccinated, challenged animals (groups 2 and 3) were higher than nasal swab titers from the corresponding pigs. Mean virus titers of nasal swabs as well as BALF from pigs unvaccinated and challenged with either H3N2 (group 2) or H1N1 (group 3) SIV were not significantly different from each other.
No virus was detected in nasal swabs and BALF of pigs vaccinated and challenged with the homosubtypic H3N2 virus (group 5), indicating that the MLV vaccine induced complete protection against infection with the homologous virus. Virus was detected in nasal secretion collected at 2 days p.i. from three of nine pigs vaccinated and challenged with heterosubtypic H1N1 SIV (group 6), with titers ranging from 101 to 103.7 TCID50/ml. Nasal secretions obtained at 5 days p.i. of four of nine pigs in this group had virus titers of more than 102.7 TCID50/ml, while those of the remaining pigs were negative. BALF was positive from six of nine pigs in this group, with titers ranging from 102.5 to 104.7 TCID50/ml. Importantly, mean virus titers in nasal secretions and BALF 5 days p.i. from pigs in this group were significantly lower than those of the unvaccinated H1N1 challenge control group (Table 2).
Antibodies specific to SIV. (i) HI.
Antibodies specific for H3 were determined in sera collected prior to the first vaccination (−31 days p.i.) from pigs in all groups. All sera were negative by HI (titers of less than 1:10). Similarly, sera from all pigs in unvaccinated groups (groups 1 to 3) collected at necropsy (5 days p.i.) had HI titers less than 1:10. Sera of pigs in vaccinated groups (groups 4 to 6) collected prior to the second vaccination (day −10 p.i.), at challenge (day 0 p.i.), and at necropsy (day 5 p.i.) were also tested in HI assays to H3, and results are listed in Table 3. The differences in HI titers before and after boost within individual treatment groups are not significant. There was also no significant difference between mean HI titers to H3 at 5 days p.i. from pigs in groups 1 to 3 or groups 4 to 6 (Table 4). However, there were significant differences in mean HI titers to H3 between vaccinated and unvaccinated groups independent of their challenge treatment (Table 4). The HI titers of sera at 5 days p.i. obtained from pigs challenged with MN99 H1N1 SIV (groups 3 and 6) were also determined using H1 antigen. The HI titers to H1 in sera of these pigs were less than 1:10.
TABLE 3.
HI titers to TX98 H3N2 antigens of sera collected prior to second vaccination and challenge as well as at necropsy from pigs in vaccinated groups
| Treatment group | HI titer (mean± SD [range]) at:
|
||
|---|---|---|---|
| −10 days p.i. | 0 days p.i. | 5 days p.i. | |
| MLV with mock challenge | 70 ± 41.4 (40-160) | 50 ± 26.19 (20-80) | 27.50 ± 10.35 (20-40) |
| MLV with TX98 challenge | 111.27 ± 93.42 (10-320) | 66.25 ± 45.65 (10-160) | 53.75 ± 47.49 (10-160) |
| MLV with MN99 challenge | 75.56 ± 37.12 (40-160) | 57.78 ± 21.08 (40-80) | 42.22 ± 15.63 (20-80) |
TABLE 4.
Summary of serum and mucosal antibodies tested by HI and ELISA
| Treatment group |
|
OD ELISA values (mean ± SD)a
|
||||
|---|---|---|---|---|---|---|
| TX98 HI serum | H3 ELISA serum | TX98 mucosal IgG | TX98 mucosal IgA | MN99 mucosal IgG | MN99 mucosal IgA | |
| Mock challenge alone | 0.0 ± 0.0 B | −0.002 ± 0.02 C | 0.3 ± 0.1 C | 0.6 ± 0.1 B | 0.2 ± 0.1 B | 0.1 ± 0.3 B |
| Mock with rTX98 challenge | 0.8 ± 1.5 B | 0.02 ± 0.03 C | 0.3 ± 0.1 C | 0.7 ± 0.1 B | 0.2 ± 0.1 B | 0.1 ± 0.1 B |
| Mock with MN99 challenge | 0.0 ± 0.0 B | 0.01 ± 0.03 C | 0.3 ± 0.1 C | 0.7 ± 0.1 B | 0.2 ± 0.1 B | 0.1 ± 0.1 B |
| MLV with mock challenge | 4.7 ± 0.5 A | 0.9 ± 0.3 B | 1.6 ± 0.2 B | 1.7 ± 0.2 A | 0.4 ± 0.1 B | 0.3 ± 0.2 AB |
| MLV with rTX98 challenge | 5.7 ± 1.2 A | 1.0 ± 0.3 AB | 1.6 ± 0.1 B | 1.8 ± 0.1 A | 0.4 ± 0.1 B | 0.3 ± 0.2 AB |
| MLV with MN99 challenge | 5.3 ± 0.5 A | 1.2 ± 0.2 A | 1.8 ± 0.03 A | 1.7 ± 0.1 A | 1.0 ± 0.2 A | 0.5 ± 0.3 A |
Letters A, B, and C after each value represent groups of means in each parameter that are significantly different from each other, with a P value of <0.05. Means labeled with the same letter are not significantly different, while those with different letters are significantly different. TX98 HI serum values have been transferred into a log2 scale for calculation and report. H3 ELISA serum values represent S/P ratio (mean ± standard deviation). All other values represent values for antibodies in sera or BALF collected at 5 days postchallenge.
(ii) Serum antibodies to SIV measured by ELISA.
Antibodies specific to either subtype H1 or subtype H3 of SIV from pigs in every group obtained at necropsy (5 days p.i.) were determined using ELISA. Means and standard deviations of S/P ratios are presented in Table 4. Results from an H3 subtype-specific ELISA showed that sera from unvaccinated groups (groups 1 to 3) were negative, while those from vaccinated groups (groups 4 to 6) were positive. In addition, mean S/P ratios of sera from pigs in vaccinated animals were similar (Table 4). When tested with an H1 subtype-specific ELISA, sera from pigs in all groups were negative, with mean S/P ratios of less than 0.02, including for MN99-challenged groups (groups 3 and 6).
(iii) Mucosal antibodies to SIV measured by ELISA.
BALF collected at necropsy (5 days p.i.) from pigs in all groups was tested for the presence of IgG and IgA isotype antibodies specific to either TX98 or MN99 antigen. In the TX98 mucosal ELISA, mean corrected ODs of groups with mock challenge alone and unvaccinated groups (groups 1 to 3) were not significantly different for IgG or IgA (Table 4). Levels of TX98-specific IgG were highest in the heterosubtypic-challenge group (group 6). IgG levels for groups 4 and 5 were significantly higher than those for unvaccinated pigs but were not different from one another. IgA levels in BALF from pigs in vaccinated groups (groups 4 to 6) were similar to, but significantly different from, those from unvaccinated pigs (groups 1 to 3) (Table 4). The mean OD using the MN99 H1N1 mucosal IgG ELISA for BALF was significantly higher in group 6 than in all other groups, including the unvaccinated, MN99-challenged group (group 3). Levels of IgA specific to MN99 from pigs in all groups were low; however, they demonstrated a pattern similar to that of the the IgG mucosal antibody by ELISA (Table 4).
Antibodies specific to NS1 protein of SIV.
An NS1 ELISA was performed, as described in Materials and Methods, with sera obtained from pigs in vaccinated groups (groups 4 to 6) collected at day of challenge (day 0 p.i.). Antibodies to the NS1 protein of SIV were detected in the serum of 1 pig out of 24 animals, with a rather low OD of 0.26 (cutoff of 0.15). The remaining pigs had corrected ODs ranging from −0.014 to 0.1 and a mean OD of 0.03. The average corrected OD values of sera from pigs infected with wild-type TX98 (n = 19) were 0.63, 0.76, and 1.1 at 7, 14, and 21 or 28 days p.i., respectively.
DISCUSSION
As presented in this study, TX98 NS1▴126 MLV, a live attenuated SIV in 4-week-old piglets (46), confers the ability to induce a strong protective immune response against a homosubtypic H3N2 challenge and a partial protective immune response against a heterosubtypic H1N1 SIV challenge. TX98 NS1▴126 MLV therefore has great potential as an MLV vaccine for SIV in swine. Although several studies previously conducted with mice showed the vaccine properties of NS1-truncated mutants (8, 10, 48), this is the first demonstration of vaccine efficacy of an NS1-modified live attenuated influenza virus in a natural host. The results of both macroscopic and microscopic lesions in the respiratory tract as well as virus titers in BALF and nasal secretions of vaccinated pigs challenged with rTX98 indicate that TX98 NS1▴126 MLV completely protected these animals from infection with the virulent parental virus. Tracheal lesions in a few pigs of this group as well as in the nonvaccinated, mock-challenged group were most likely caused by the intratracheal inoculation procedure. Since no virus was detected in BALF and nasal secretions of these pigs, MLV vaccination effectively prevents virus shedding from the respiratory tract. Consequently, MLV vaccination and infection with wild-type live virus confer the same level of protection against a homologous challenge in pigs (15, 50, 52).
Attenuation characteristics of the TX98 NS1▴126 virus are due to the mutation of the NS1 protein. Influenza viruses isolated from different host species, such as human (A/Puerto Rico/8/34 and B/Yamagata/1/73/ AWBY-234) and equine (A/equine/Kentucky/5/02) isolates harboring C-terminal NS1 truncations or entire NS1 deletions, were shown to be highly attenuated (6, 8, 10, 13, 39, 48). Reduction in virulence of these NS1 mutants is linked to their sensitivity to the IFN system, since the PR8 virus with a truncated or deleted NS1 open reading frame could not grow efficiently in cells expressing type I IFN (6, 14). NS1 contains two major functional domains: the RNA binding domain spans the N terminus (first 73 residues), and the effector domain is located in the C-terminal half of the protein (21). The effector domain appears to be involved in general inhibition of host cell pre-mRNA processing by binding to the cellular mRNA processing factors CPSF and PABII (28, 44). In addition to its effector functions, the C-terminal domain of NS1 plays a structural role in NS1 function by stabilizing the functional NS1 dimer (55). The N-terminal region of NS1, containing a double-stranded RNA binding domain, attenuates the induction of transcription factors involved in IFN synthesis, therefore reducing the levels of IFN secretion by infected cells (13). This region also inhibits the activation of the IFN-inducible antiviral proteins PKR and OAS (1, 25, 27). Other functions associated with the NS1 protein include inhibition of mRNA splicing (29), enhancement of viral protein translation (2), and modulation of RNA replication and transcription (9). In our studies with pigs, we have used a mutant TX98 virus expressing a truncated NS1 protein that contains the RNA binding domain but lacks the effector domain. This truncated NS1 protein is poorly expressed, resulting in high levels of IFN induction by the mutant virus and in attenuation (46). In addition, the NS1 mutant lacks the CPSF binding domain that has been implicated in regulation of cellular gene expression (28, 32). Recently, a motif at the very-C-terminal portion of the NS1 has been identified as a PDZ ligand domain (33). Although this domain has been proposed to modulate virulence, many naturally occurring strains of influenza virus, including the wild-type swine TX98 virus used in our experiments, lack this domain.
In the MLV (TX98 NS1▴126)-vaccinated and H1N1-challenged group, significant lung damage was macroscopically observed at day 5 p.i., although SIV antigens could not be detected by IHC at this time. Whether the lung damage associated with the heterosubtypic SIV challenge in this study is due mainly to the host immune response or to virus-mediated cell damage is not clear. Previously, DNA vaccination of pigs with a swine influenza M2-NP fusion protein prior to inoculation with SIV H1N1 produced more-severe clinical signs than control pigs (17). Antibodies to both proteins, in contrast to anti-HA, are not neutralizing but possibly promote death of infected cells via antibody-mediated mechanisms. On the other hand, it has been demonstrated previously that timing of a virus-specific immune response relative to viral dissemination is critical in determining the balance between immunoprotection and immunopathology (34, 41).
The results from our present study showed that swine vaccinated with MLV H3N2 NS1▴126 were also partially protected from subsequent inoculation with a heterosubtypic H1N1 SIV, although there is no HI cross-reactivity between the subtypes. The mucosal immune response elicited by vaccinated pigs challenged with the heterosubtypic virus was significantly higher than that of unvaccinated and H1N1-challenged pigs. Rapid production of heterosubtypic mucosal antibodies in the MLV plus H1N1-challenged group suggests the presence of a cross-immune response prior to challenge. Cross-protective immunity between different subtypes of influenza A virus is called heterosubtypic immunity. Het-I in swine was first described when pigs were intranasally infected with H1N1 SIV followed by a challenge with H3N2 SIV; the animals neither produced clinical symptoms nor transmitted the virus to unprimed sentinel pigs in the same pen (15).
In mice, this cross-protection was found to be based on a specific immune response to common epitopes of influenza A virus proteins, since it could not protect against a distantly related pathogen, such as influenza B virus (7, 24). B cells and CD4+ cells have been shown to be required for a full memory response against influenza virus, especially in heterosubtypic immunity (7, 31). It is assumed that memory T helper cells, specific for conserved internal proteins such as NP and M, are able to rapidly stimulate B cells to produce antibodies with specificity for the HA protein (43). Antibodies specific to the external domain of the M2 protein (15) and/or the conserved stem region of HA (45) may also contribute to heterosubtypic protection. These early M2- or HA-specific antibodies may not be neutralizing but are likely to interfere with and be capable of reducing virus load and spread from cell to cell within the respiratory tract. In addition to the role of CD4+ T cells in Het-I, CD8+ T cells were observed in respiratory tracts of mice challenged with H3N2 virus after H1N1 priming as a result of activation of memory CD8+ T cells, suggesting a role of these cells in Het-I (11, 15, 30). Internal antigens, such as NP and M proteins, were thought to be responsible for Het-I induction via cell-mediated immune responses since they contained common epitopes among influenza A viruses (57). In fact, a high proliferation of CD8+ T cells recognizing the shared immunodominant epitope of NP was observed after challenge of mice with H1N1 virus primed with H3N2 virus (11, 12).
Vaccination with TX98 NS1▴126 MLV decreased virus titers in BALF of animals challenged with the heterosubtypic H1N1 virus by several orders of magnitude compared with results for the nonvaccinated, H1N1-challenged group. The significant reduction of BALF virus in this group (in some animals, negative; in others, by 4 orders of magnitude) most likely reflects an early strong immune response to the heterosubtypic SIV antigens mediated by infiltrating immune cells that remain present in the lung for a period of time even after the virus is cleared. Het-I protection has also been reported for European SIV. HA genes from European H1N1 and H1N2 SIVs are not related since the HA of the H1N1 virus originated from avian lineage while the HA of the H1N2 virus is of human origin (26). Despite heterologous HA genes, pigs previously infected intranasally with live H1N1 SIV followed by live H3N2 SIV were clinically protected from a subsequent H1N2 challenge and virus replication was markedly reduced (50, 51). In contrast, postinfection immunity elicited by infection with H1N1 or H3N2 alone was not able to protect against disease after H1N2 challenge. However, intramuscular vaccination with inactivated bivalent H1N1 and H3N2 commercial vaccines did not protect pigs from H1N2 challenge (50), again suggesting a role for cell-mediated immune responses in Het-I. It might be possible that priming and boosting with live viruses representing two subtypes or one subtype, as done in our study, provide cross-protection. This would suggest that a booster with viruses containing conserved internal proteins of SIV may enhance the Het-I response. However, it remains to be seen whether single immunization with MLV H3N2 NS1▴126 results in homosubtypic and heterosubtypic protection.
Inactivated vaccines induce mainly humoral immune responses, whereas live viruses infect cells and induce CD4+ and CD8+ T-cell responses important for Het-I protection. Although recent studies with inactivated viruses given intranasally demonstrated the induction of a broad range of Het-I mediated by mucosal immunity (47, 49), further studies employing both inactivated and MLV vaccines side by side are warranted to compare the efficacies of these vaccination approaches. These data indicate that the type of immunogen and route of administration also play a role in Het-I stimulation (16). It is also possible that our results could be explained, at least in part, by the induction of innate immune responses (type I IFN) by the MLV H3N2 NS1▴126 that are still present at day 10 postimmunization and that might contribute to inhibition of replication of the heterosubtypic H1N1 virus.
It is well known that vaccination can complicate serological surveillance activities that follow eradication if the antibody response induced by vaccination is indistinguishable from that which follows infection. This disadvantage can be overcome by the use of DIVA vaccines and their companion diagnostic tests. The term DIVA is now generally used as an acronym for “differentiating infected from vaccinated animals.” The term was originally applied to the use of marker vaccines, which are based on deletion mutants of wild-type microbes, in conjunction with a differentiating diagnostic test. The DIVA strategy has been extended to include subunit and killed-whole-virus vaccines. This system makes possible the mass vaccination of a susceptible animal population without compromising the serological identification of convalescent individuals. The DIVA approach has been applied successfully to pseudorabies and avian influenza eradication and has been proposed for use in foot-and-mouth disease and classical swine fever eradication campaigns (38). Based on an NS1 differentiating diagnostic test, the modified live TX98 NS1▴126 virus has the potential to be used as a DIVA vaccine.
We demonstrated that inoculation of pigs twice with the modified live TX98 NS1▴126 virus followed by H1N1 or H3N2 SIV challenge induced no or a very low level of antibodies to NS1 protein by use of our NS1 ELISA. These results suggest a potential use of this vaccine and the NS1-based ELISA to differentiate infected from vaccinated animals. This DIVA system could be developed further and used as a crucial epidemiological tool for swine influenza control. In summary, our results suggest that NS1-modified SIV vaccines could represent a new vaccine generation against swine influenza. This will be the case if NS1-modified SIV-based vaccines prove to be more advantageous in the field than the traditional inactivated vaccines currently in use. In this report, we used the intratracheal route of inoculation to test the protective efficacy of our MLV vaccine in order to assure a consistent infection of the pigs with the MLV. Future studies are planned to test the efficacy of the MLV vaccine after intranasal inoculation. Similar vaccination approaches based on NS1-modified influenza viruses might be developed for other hosts, such as horses (39), dogs, birds, or humans.
Acknowledgments
These studies were partially supported by NIH grant R01AI46954 and by grants from NIH-funded centers NBC (North Biodefense Center, U54AI57158) and CIVIA (Center for Investigating Virus Interferon Antagonism, U19AI62623) to A.G.-S.
We thank Chalermpol Lekcharoensuk for statistical analysis as well as Richard Cádagan, Deborah Clauser, and Kevin Hassall for excellent technical assistance. We appreciate Andrew Gibson for animal care.
Mount Sinai School of Medicine owns patent positions for reverse genetics of influenza viruses.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Footnotes
Published ahead of print on 30 August 2006.
REFERENCES
- 1.Bergmann, M., A. García-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts the PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgui, I., T. Aragon, J. Ortin, and A. Nieto. 2003. PABP1 and eIF4GI associate with influenza virus NS1 protein in viral mRNA translation initiation complexes. J. Gen. Virol. 84:3263-3274. [DOI] [PubMed] [Google Scholar]
- 3.Choi, Y. K., S. M. Goyal, M. W. Farnham, and H. S. Joo. 2002. Phylogenetic analysis of H1N2 isolates of influenza A virus from pigs in the United States. Virus Res. 87:173-179. [DOI] [PubMed] [Google Scholar]
- 4.Choi, Y. K., S. M. Goyal, and H. S. Joo. 2002. Prevalence of swine influenza virus subtypes on swine farms in the United States. Arch. Virol. 147:1209-1220. [DOI] [PubMed] [Google Scholar]
- 5.de Jong, J. C., P. P. Heinen, W. L. Loeffen, A. P. van Nieuwstadt, E. C. Claas, T. M. Bestebroer, K. Bijlsma, C. Verweij, A. D. Osterhaus, G. F. Rimmelzwaan, R. A. Fouchier, and T. G. Kimman. 2001. Antigenic and molecular heterogeneity in recent swine influenza A(H1N1) virus isolates with possible implications for vaccination policy. Vaccine 19:4452-4464. [DOI] [PubMed] [Google Scholar]
- 6.Egorov, A., S. Brandt, S. Sereinig, J. Romanova, B. Ferko, D. Katinger, A. Grassauer, G. Alexandrova, H. Katinger, and T. Muster. 1998. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol. 72:6437-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epstein, S. L., C. Y. Lo, J. A. Misplon, C. M. Lawson, B. A. Hendrickson, E. E. Max, and K. Subbarao. 1997. Mechanisms of heterosubtypic immunity to lethal influenza A virus infection in fully immunocompetent, T cell-depleted, beta2-microglobulin-deficient, and J chain-deficient mice. J. Immunol. 158:1222-1230. [PubMed] [Google Scholar]
- 8.Falcon, A. M., A. Fernandez-Sesma, Y. Nakaya, T. M. Moran, J. Ortin, and A. García-Sastre. 2005. Attenuation and immunogenicity in mice of temperature-sensitive influenza viruses expressing truncated NS1 proteins. J. Gen. Virol. 86:2817-2821. [DOI] [PubMed] [Google Scholar]
- 9.Falcon, A. M., R. M. Marion, T. Zurcher, P. Gomez, A. Portela, A. Nieto, and J. Ortin. 2004. Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein. J. Virol. 78:3880-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferko, B., J. Stasakova, J. Romanova, C. Kittel, S. Sereinig, H. Katinger, and A. Egorov. 2004. Immunogenicity and protection efficacy of replication-deficient influenza A viruses with altered NS1 genes. J. Virol. 78:13037-13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flynn, K. J., G. T. Belz, J. D. Altman, R. Ahmed, D. L. Woodland, and P. C. Doherty. 1998. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity 8:683-691. [DOI] [PubMed] [Google Scholar]
- 12.Flynn, K. J., J. M. Riberdy, J. P. Christensen, J. D. Altman, and P. C. Doherty. 1999. In vivo proliferation of naive and memory influenza-specific CD8+ T cells. Proc. Natl. Acad. Sci. USA 96:8597-8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Sastre, A. 2002. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes Infect. 4:647-655. [DOI] [PubMed] [Google Scholar]
- 14.García-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 15.Heinen, P. P., E. A. de Boer-Luijtze, and A. T. Bianchi. 2001. Respiratory and systemic humoral and cellular immune responses of pigs to a heterosubtypic influenza A virus infection. J. Gen. Virol. 82:2697-2707. [DOI] [PubMed] [Google Scholar]
- 16.Heinen, P. P., A. P. van Nieuwstadt, E. A. de Boer-Luijtze, and A. T. Bianchi. 2001. Analysis of the quality of protection induced by a porcine influenza A vaccine to challenge with an H3N2 virus. Vet. Immunol. Immunopathol. 82:39-56. [DOI] [PubMed] [Google Scholar]
- 17.Heinen, P. P., F. A. Rijsewijk, E. A. de Boer-Luijtze, and A. T. Bianchi. 2002. Vaccination of pigs with a DNA construct expressing an influenza virus M2-nucleoprotein fusion protein exacerbates disease after challenge with influenza A virus. J. Gen. Virol. 83:1851-1859. [DOI] [PubMed] [Google Scholar]
- 18.Janke, B. H. 1998. Classical swine influenza. Large Anim. Pract. 19:24-29. [Google Scholar]
- 19.Karasin, A. I., M. M. Schutten, L. A. Cooper, C. B. Smith, K. Subbarao, G. A. Anderson, S. Carman, and C. W. Olsen. 2000. Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977-1999: evidence for wholly human and reassortant virus genotypes. Virus Res. 68:71-85. [DOI] [PubMed] [Google Scholar]
- 20.Karasin, A. I., J. Landgraf, S. Swenson, G. Erickson, S. Goyal, M. Woodruff, G. Scherba, G. Anderson, and C. W. Olsen. 2002. Genetic characterization of H1N2 influenza A viruses isolated from pigs throughout the United States. J. Clin. Microbiol. 40:1073-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krug, R. M., W. Yuan, D. L. Noah, and A. G. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309:181-189. [DOI] [PubMed] [Google Scholar]
- 22.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1531. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 23.Larsen, D. L., A. Karasin, F. Zuckermann, and C. W. Olsen. 2000. Systemic and mucosal immune responses to H1N1 influenza virus infection in pigs. Vet. Microbiol. 74:117-131. [DOI] [PubMed] [Google Scholar]
- 24.Liang, S., K. Mozdzanowska, G. Palladino, and W. Gerhard. 1994. Heterosubtypic immunity to influenza type A virus in mice. Effector mechanisms and their longevity. J. Immunol. 152:1653-1661. [PubMed] [Google Scholar]
- 25.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214:222-228. [DOI] [PubMed] [Google Scholar]
- 26.Marozin, S., V. Gregory, K. Cameron, M. Bennett, M. Valette, M. Aymard, E. Foni, G. Barigazzi, Y. Lin, and A. Hay. 2002. Antigenic and genetic diversity among swine influenza A H1N1 and H1N2 viruses in Europe. J. Gen. Virol. 83:735-745. [DOI] [PubMed] [Google Scholar]
- 27.Min, J. Y., and R. M. Krug. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. USA 103:7100-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 29.Nemeroff, M. E., U. Utans, A. Krämer, and R. M. Krug. 1992. Identification of cis-acting intron and exon regions in influenza virus NS1 mRNA that inhibit splicing and cause the formation of aberrantly sedimenting presplicing complexes. Mol. Cell. Biol. 12:962-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen, H. H., Z. Moldoveanu, M. J. Novak, F. W. van Ginkel, E. Ban, H. Kiyono, J. R. McGhee, and J. Mestecky. 1999. Heterosubtypic immunity to lethal influenza A virus infection is associated with virus-specific CD8+ cytotoxic T lymphocyte responses induced in mucosa-associated tissues. Virology 254:50-60. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen, H. H., F. W. van Ginkel, H. L. Vu, J. R. McGhee, and J. Mestecky. 2001. Heterosubtypic immunity to influenza A virus infection requires B cells but not CD8+ cytotoxic T lymphocytes. J. Infect. Dis. 183:368-376. [DOI] [PubMed] [Google Scholar]
- 32.Noah, D. L., K. Y. Twu, and R. M. Krug. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAS. Virology 307:386-395. [DOI] [PubMed] [Google Scholar]
- 33.Obenauer, J. C., J. Denson, P. K. Mehta, X. Su, S. Mukatira, D. B. Finkelstein, X. Xu, J. Wang, J. Ma, Y. Fan, K. M. Rakestraw, R. G. Webster, E. Hoffmann, S. Krauss, J. Zheng, Z. Zhang, and C. W. Naeve. 2006. Large-scale sequence analysis of avian influenza isolates. Science 311:1576-1580. [DOI] [PubMed] [Google Scholar]
- 34.Oehen, S., H. Hengartner, and R. M. Zinkernagel. 1991. Vaccination for disease. Science 251:195-198. [DOI] [PubMed] [Google Scholar]
- 35.Olsen, C. W., S. Carey, L. Hinshaw, and A. I. Karasin. 2000. Virologic and serologic surveillance for human, swine and avian influenza virus infections among pigs in the north-central United States. Arch. Virol. 145:1399-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsen, C. W. 2002. The emergence of novel swine influenza viruses in North America. Virus Res. 85:199-210. [DOI] [PubMed] [Google Scholar]
- 37.Palmer, D. F., M. T. Coleman, W. R. Dowdle, and G. C. Schild. 1975. Advanced laboratory techniques for influenza diagnosis, p. 51-52. Immunology series no. 6. U.S. Department of Health, Education, and Welfare, Washington, D.C.
- 38.Pasick, J. 2004. Application of DIVA vaccines and their companion diagnostic tests to foreign animal disease eradication. Anim. Health Res. Rev. 5:257-262. [DOI] [PubMed] [Google Scholar]
- 39.Quinlivan, M., D. Zamarin, A. García-Sastre, A. Cullinane, T. Chambers, and P. Palese. 2005. Attenuation of equine influenza viruses through truncations of the NS1 protein. J. Virol. 79:8431-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 41.Richt, J. A., A. Schmeel, K. Frese, K. M. Carbone, O. Narayan, and R. Rott. 1994. Borna disease virus-specific T cells protect against or cause immunopathological Borna disease. J. Exp. Med. 179:1467-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richt, J. A., K. M. Lager, B. H. Janke, R. D. Woods, R. G. Webster, and R. J. Webby. 2003. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J. Clin. Microbiol. 41:3198-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scherle, P. A., and W. Gerhard. 1986. Functional analysis of influenza-specific helper T cell clones in vivo. T cells specific for internal viral proteins provide cognate help for B cell responses to hemagglutinin. J. Exp. Med. 164:1114-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimizu, K., A. Iguchi, R. Gomyou, and Y. Ono. 1999. Influenza virus inhibits cleavage of the HSP70 pre-mRNAs at the polyadenylation site. Virology 254:213-219. [DOI] [PubMed] [Google Scholar]
- 45.Smirnov, Y. A., A. S. Lipatov, A. K. Gitelman, Y. Okuno, R. Van Beek, A. D. Osterhaus, and E. C. Claas. 1999. An epitope shared by the hemagglutinins of H1, H2, H5, and H6 subtypes of influenza A virus. Acta Virol. 43:237-244. [PubMed] [Google Scholar]
- 46.Solórzano, A., R. J. Webby, K. M. Lager, B. H. Janke, A. García-Sastre, and J. A. Richt. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 79:7535-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takada, A., S. Matsushita, A. Ninomiya, Y. Kawaoka, and H. Kida. 2003. Intranasal immunization with formalin-inactivated virus vaccine induces a broad spectrum of heterosubtypic immunity against influenza A virus infection in mice. Vaccine 21:3212-3218. [DOI] [PubMed] [Google Scholar]
- 48.Talon, J., M. Salvatore, R. E. O'Neill, Y. Nakaya, H. Zheng, T. Muster, A. García-Sastre, and P. Palese. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. USA 97:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tumpey, T. M., M. Renshaw, J. D. Clements, and J. M. Katz. 2001. Mucosal delivery of inactivated influenza vaccine induces B-cell-dependent heterosubtypic cross-protection against lethal influenza A H5N1 virus infection. J. Virol. 75:5141-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Reeth, K., I. Brown, S. Essen, and M. Pensaert. 2004. Genetic relationships, serological cross-reaction and cross-protection between H1N2 and other influenza A virus subtypes endemic in European pigs. Virus Res. 103:115-124. [DOI] [PubMed] [Google Scholar]
- 51.Van Reeth, K., V. Gregory, A. Hay, and M. Pensaert. 2003. Protection against a European H1N2 swine influenza virus in pigs previously infected with H1N1 and/or H3N2 subtypes. Vaccine 21:1375-1381. [DOI] [PubMed] [Google Scholar]
- 52.Van Reeth, K., G. Labarque, S. De Clercq, and M. Pensaert. 2001. Efficacy of vaccination of pigs with different H1N1 swine influenza viruses using a recent challenge strain and different parameters of protection. Vaccine 19:4479-4486. [DOI] [PubMed] [Google Scholar]
- 53.Vincent, A., K. M. Lager, W. Ma, P. Lekcharoensuk, M. R. Gramer, C. M. Loiacono, and J. A. Richt. Evaluation of hemagglutinin subtype 1 swine influenza viruses from the United States. J. Vet. Microbiol., in press. [DOI] [PubMed]
- 54.Vincent, L. L., B. H. Janke, P. S. Paul, and P. G. Halbur. 1997. A monoclonal-antibody-based immunohistochemical method for the detection of swine influenza virus in formalin-fixed, paraffin-embedded tissues. J. Vet. Diagn. Investig. 9:191-195. [DOI] [PubMed] [Google Scholar]
- 55.Wang, X., C. F. Basler, B. R. Williams, R. H. Silverman, P. Palese, and A. García-Sastre. 2002. Functional replacement of the carboxy-terminal two-thirds of the influenza A virus NS1 protein with short heterologous dimerization domains. J. Virol. 76:12951-12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Webby, R. J., S. L. Swenson, S. L. Krauss, P. J. Gerrish, S. M. Goyal, and R. G. Webster. 2000. Evolution of swine H3N2 influenza viruses in the United States. J. Virol. 74:8243-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woodland, D. L., R. J. Hogan, and W. Zhong. 2001. Cellular immunity and memory to respiratory virus infections. Immunol. Res. 24:53-67. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, N. N., D. A. Senne, J. S. Landgraf, S. L. Swenson, G. Erickson, K. Rossow, L. Liu, K. Yoon, S. Krauss, and R. G. Webster. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J. Virol. 73:8851-8856. [DOI] [PMC free article] [PubMed] [Google Scholar]