Abstract
Primary and laboratory-adapted variants of human immunodeficiency virus type 1 (HIV-1) exhibit a wide range of sensitivities to neutralization by antibodies directed against the viral envelope glycoproteins. An antibody directed against an artificial FLAG epitope inserted into the envelope glycoproteins of three HIV-1 isolates with vastly different neutralization sensitivities inhibited all three viruses equivalently. Thus, naturally occurring HIV-1 isolates that are neutralization resistant are not necessarily more impervious to the inhibitory consequences of bound antibody. Moreover, the binding affinity of the anti-FLAG antibody correlated with neutralizing potency, underscoring the dominant impact on neutralization of antibody binding to the envelope glycoproteins.
Human immunodeficiency virus type 1 (HIV-1) is the etiological agent of AIDS. HIV-1 establishes persistent infections in humans and has evolved to be relatively resistant to antibodies generated during natural infection (1, 2, 5, 8, 13, 26, 27, 34, 39, 42). Primary (clinical) HIV-1 strains exhibit a range of sensitivities to antibody-mediated neutralization, but they are generally more resistant than the T-cell line-adapted isolates that have been cultured extensively in vitro (9, 20, 24, 30, 44).
The viral targets of neutralizing antibodies are the gp120 exterior and gp41 transmembrane envelope glycoproteins (Envs), which are assembled into trimers on the virion surface (40). During virus entry, gp120 binds host CD4 and chemokine receptors, whereas gp41 mediates the fusion of the viral and target cell membranes. The binding of a single antibody molecule to the HIV-1 envelope glycoprotein trimer is sufficient to inactivate its function, independent of the HIV-1 strain from which the Envs are derived or the particular gp120 or gp41 epitope recognized by the monoclonal antibody (MAb) (36, 41). Even an unrelated antibody, the M2 anti-FLAG antibody, can effectively neutralize HIV-1 virions that carry an exogenous FLAG epitope in the gp120 V4 variable region (33). The V4 region has no known structural or functional roles in viral entry, consistent with the large amount of sequence diversity in this region for different HIV-1 isolates (14, 19, 21, 22, 40). These results suggest the hypothesis that the binding of an antibody anywhere on the HIV-1 envelope glycoprotein spike leads to neutralization and that the infectious trimers on the surface of primary HIV-1 virions resist such antibody binding.
Despite the appeal of the above model, the establishment of antibody-binding assays that reliably predict the neutralization sensitivity of a given HIV-1 isolate has proven to be elusive. To date, no antibody-binding assay using recombinant HIV-1 glycoproteins as binding targets perfectly predicts the HIV-1-neutralizing activity of an antibody, probably due to the failure of the recombinant forms to perfectly imitate the Env spikes on HIV-1 virions (29). Virion-binding assays, in which the ability of an anti-HIV-1-Env antibody to bind virus particles in vitro is examined, are not exact prognostic indicators for neutralization potency either (7, 16, 32, 42). Potential reasons for such difficulty include (i) the existence, often in vastly overwhelming proportions, of nonfunctional (including uncleaved) HIV-1 Env trimers in viral stocks (16, 32); (ii) the replication defectiveness of the vast majority (greater than 99.9%) of HIV-1 virions (4, 23); (iii) the small and varying number of intact Env trimers per HIV-1 virion (10, 15, 23, 43); and (iv) spontaneous and/or ligand-induced dissociation (“shedding”) of gp120 from the Env spikes (28, 31, 35). Thus, the precise measurement of MAb binding to functionally relevant HIV-1 Env spikes remains an elusive goal. Consequently, our understanding of the mechanistic basis of HIV-1 resistance to neutralization by antibodies is still incomplete. Here, we study antibody-mediated neutralization in a controlled context by introducing FLAG artificial epitopes into the gp120 V4 region of HIV-1 viruses with dramatically different sensitivities to neutralization; our approach overcomes some of the above difficulties by focusing on the functional portion of HIV-1 Env spikes on virions.
HIV-1YU2 is a primary isolate that is extremely resistant to neutralization (24, 25). HIV-1JR-FL is another primary HIV-1 isolate, but it exhibits intermediate sensitivity to neutralization (12). Both HIV-1YU2 and HIV-1JR-FL use CCR5 as a second coreceptor. HIV-1HXBc2 is a T-cell line-adapted HIV-1 that uses CXCR4 as a coreceptor and is very susceptible to neutralizing antibodies (44). Recombinant HIV-1 encoding firefly luciferase was pseudotyped with the wild-type Envs of HIV-1YU2, HIV-1JR-FL, and HIV-1HXBc2. Viruses produced by transfection of 293T cells were used to measure infectivity and neutralization sensitivity, as described previously (33, 38). The infectivity of recombinant viruses with HIV-1YU2 and HIV-1JR-FL Envs was measured by incubating the viruses with Cf2Th-CD4/CCR5 cells, and the infectivity of viruses with HIV-1HXBc2 Envs was measured by using Cf2Th-CD4/CXCR4 target cells. For neutralization assays, viruses were incubated with antibodies and 1 μM of Polybrene at 37°C for 4 h prior to exposure to the target cells.
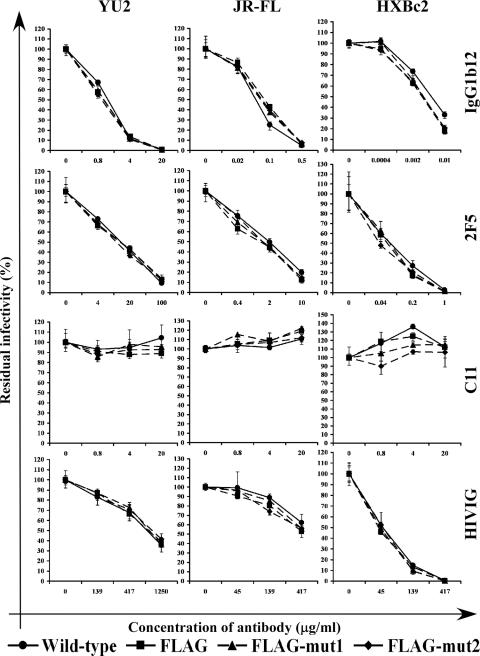
The sensitivity of the viruses with wild-type HIV-1YU2, HIV-1JR-FL, and HIV-1HXBc2 Envs to neutralization by three relatively potent human MAbs (IgG1b12, 2G12, and 2F5) was assessed. Viruses with wild-type HIV-1JR-FL Envs were neutralized by IgG1b12, 2F5, and 2G12 MAbs at 10- to 100-fold-lower concentrations than those required to neutralize the HIV-1YU2 viruses (Fig. 1 and data not shown). Viruses with the HIV-1HXBc2 Envs were neutralized by IgG1b12 and 2F5 MAbs at concentrations 100- to 1,000-fold lower than the HIV-1YU2 viruses (Fig. 1). Sensitivity to neutralization by a polyclonal immunoglobulin preparation (HIVIG) from HIV-1-infected individuals exhibited the following order: HXBc2 > JR-FL > YU2 (Fig. 1). Thus, the Envs from these three HIV-1 strains specify vastly different sensitivities to neutralization by natural anti-HIV-1 Env MAbs.
FIG. 1.
Envelope glycoproteins from distinct HIV-1 strains exhibit very different sensitivities to neutralization by anti-HIV-1-Env antibodies. Recombinant luciferase-expressing reporter viruses containing the indicated HIV-1 Envs were produced as described previously (17). Viruses with Envs originating from one HIV-1 strain were produced and analyzed for neutralization sensitivity as a set. Within each set, the same amounts of viruses were incubated for 4 h at 37°C with the indicated concentrations of a given antibody in growth medium containing 1 μM Polybrene. Residual infectivities were measured in a single-round entry assay using appropriate target cells (Cf2Th-CD4/CCR5 for HIV-1YU2 and HIV-1JR-FL; Cf2Th-CD4/CXCR4 for HIV-1HXBc2). The infectivities after virus/antibody incubation were normalized to that of the same virus without antibody incubation, which was set at 100%. The means and ranges of variation from three parallel measurements are shown. All experiments were repeated at least once, and the results of a typical experiment are shown.
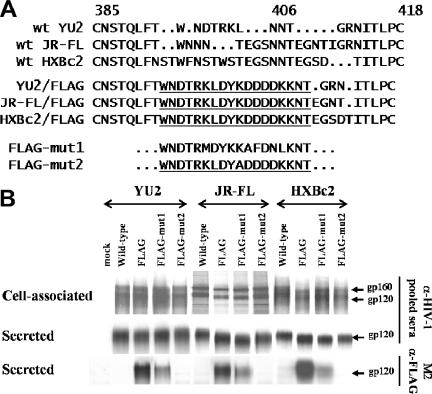
HIV-1 isolates that differ in neutralization sensitivity levels could hypothetically differ in the binding levels of antibodies to the Envs or in the consequences of antibody binding with respect to virus infectivity. To examine these possibilities, we inserted an artificial epitope, the FLAG tag or its variants, into the V4 region of the HIV-1YU2, HIV-1JR-FL, and HIV-1HXBc2 gp120 glycoproteins (Fig. 2A). The FLAG-mut1 and FLAG-mut2 variants contain sequence changes designed to decrease the affinity of the M2 anti-FLAG MAb for the epitope. Insertion of the artificial epitopes had no significant effect on the expression levels, proteolytic maturation, or subunit association of the Envs (Fig. 2B). Precipitation of the secreted gp120 glycoproteins by the M2 MAb revealed that the FLAG-mut1 gp120 glycoproteins bound M2 at reduced levels compared to the FLAG gp120 glycoproteins; the FLAG-mut2 gp120 glycoproteins did not detectably bind M2 under these conditions. The lower affinities of the FLAG-mut1 and FLAG-mut2 gp120 glycoproteins of the HIV-1YU2 strain for the M2 MAb, compared with the HIV-1YU2 FLAG gp120, were confirmed by surface plasmon resonance analysis (data not shown). Thus, we have created epitope-tagged HIV-1 gp120 variants that bind the M2 MAb in the following order: FLAG > FLAG-mut1 > FLAG-mut2.
FIG. 2.
Design and expression of HIV-1 Envs carrying FLAG artificial epitopes. (A) The amino acid sequences of the wild-type (wt) V4 region from the three studied HIV-1 gp120 Envs are aligned in the top panel. The amino acid numbering corresponds to that of HIV-1HXBc2, per current convention (18). The YU2/FLAG construct contains an insertion of the FLAG epitope tag “DYKDDDDK” and an N406K change to remove a glycosylation site in the HIV-1YU2 gp120 V4 region. The underlined YU2/FLAG sequences were used to replace the corresponding sequences in the HIV-1JR-FL and HIV-1HXBc2 Envs to create the JR-FL/FLAG and HXBc2/FLAG Envs, respectively. To generate different FLAG derivatives with reduced affinities for the M2 anti-FLAG antibody, the wild-type FLAG sequence was replaced with “AMDYKAFDNL” and “DYADDDDK” to generate the FLAG-mut1 and FLAG-mut2 constructs, respectively. The FLAG-mut1 and FLAG-mut2 constructs were generated by using the Envs from the three studied HIV-1 isolates. (B) HIV-1 Envs were transiently expressed in 293T cells and labeled with [35S]methionine/cysteine. The cell-associated and supernatant Envs were precipitated by pooled sera from HIV-1-infected individuals or by the M2 anti-FLAG antibody. The precipitated proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The duration of exposure of the autoradiographs differs for the Envs derived from each strain of HIV-1.
The infectivity of recombinant luciferase reporter viruses carrying these HIV-1 Envs was tested by a standard single-round entry assay (33). Viruses carrying the artificial epitope tags entered the appropriate target cells at levels within threefold of those of the wild-type glycoproteins (data not shown). Therefore, the insertion of these FLAG variants into the gp120 V4 region exerted little detrimental effect on the basal ability of HIV-1 Envs to support virus entry. We used a standard neutralization assay to evaluate the effect of the inserted sequences on the general neutralization sensitivity of viruses with the modified Envs. Viruses bearing Envs with the inserted FLAG epitopes were indistinguishable from the viruses bearing the parental glycoproteins in terms of sensitivity to neutralization by neutralizing MAbs IgG1b12 and 2F5 as well as by a nonneutralizing MAb, C11 (Fig. 1, top three panels). Additionally, viruses with the FLAG-tagged and wild-type Envs were neutralized equivalently by HIVIG (Fig. 1, bottom panel). In summary, insertion of the artificial epitope tags had no deleterious effect on the general properties of the HIV-1YU2, HIV-1JR-FL, and HIV-1HXBc2 Envs with respect to protein expression, processing, trimer stability, entry function, and general neutralization sensitivity.
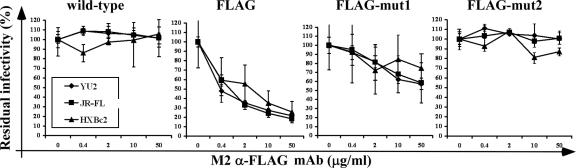
Next, we compared the neutralization efficiency of the M2 MAb against the viruses. Incubation with 50 μg/ml or less of the M2 MAb did not significantly inhibit infection by viruses carrying the wild-type glycoproteins of HIV-1YU2, HIV-1JR-FL, and HIV-1HXBc2 (Fig. 3, left panel). In the same concentration range, the M2 MAb equivalently neutralized viruses with the FLAG-tagged HIV-1YU2, HIV-1JR-FL, and HIV-1HXBc2 Envs (Fig. 3, second panel from left). Therefore, even though the HIV-1YU2, HIV-1JR-FL, and HIV-1HXBc2 Envs specify large differences in sensitivity to neutralization by many antibodies, the consequences of antibody binding with respect to inactivation of infectivity are similar for these Env variants.
FIG. 3.
Sensitivity of recombinant viruses to neutralization by the M2 anti-FLAG antibody. Recombinant luciferase-expressing reporter viruses were produced and tested for sensitivity to neutralization by the M2 anti-FLAG antibody (Sigma). The same amount of viruses in each set (i.e., with Envs originating from one HIV-1 strain) was incubated for 4 h at 37°C with the indicated concentrations of the M2 antibody and 1 μM Polybrene. The residual infectivities after incubation with the M2 antibody were normalized to the infectivity observed in the absence of antibody treatment, which was set at 100%. For viruses with wild-type and FLAG-mut2 HIV-1 Envs, the means and ranges of variation from three parallel measurements from a typical experiment are shown. For viruses with Envs carrying the FLAG and FLAG-mut1 artificial epitopes, data from 10 parallel measurements made in three independent experiments were pooled, and the means and standard deviations are shown.
For all three HIV-1 strains, the M2 MAb neutralized the viruses with the FLAG epitope more efficiently than those with the FLAG-mut1 epitope (compare the middle two panels of Fig. 3). The M2 MAb did not appreciably neutralize the viruses with the FLAG-mut2 epitope (Fig. 3, right panel). Thus, the sensitivities of the viruses with FLAG epitope variants to neutralization by the M2 MAb directly reflected the M2-binding affinities of the viral Envs.
In summary, our results demonstrate that the Envs of neutralization-resistant, primary HIV-1 are not intrinsically resistant to the inactivating consequences of antibody binding. Furthermore, we were able to demonstrate a clear relationship between neutralization potency and the affinity of the antibody for its cognate epitope. Our results are consistent with a large body of evidence suggesting that antibody binding to the HIV-1 Env trimer is necessary and sufficient for at least some level of neutralization (6). Thus, limiting the access of antibodies to epitopes, particularly those that are well-conserved among HIV-1 strains, on the functional Env trimer is essential for the ability of HIV-1 to resist neutralization by antibodies. Multiple determinants in the gp120 variable loops and gp41 ectodomain can contribute to the high level of neutralization resistance of certain HIV-1 strains (3, 11, 37, 38). An understanding of the structure of the HIV-1 Env trimers will reveal the mechanistic basis of these complex interactions and thereby guide interventional strategies.
Acknowledgments
We thank Yvette McLaughlin, Elizabeth Carpelan, and Lisa Bradbury for manuscript preparation.
This work was supported by NIH grants (AI064009 to X.Y. and AI24755, AI39420, and AI40895 to J.S.), by a Center for HIV/AIDS Vaccine Immunology grant (AI67854), by a Center for AIDS Research grant (AI42848), by an unrestricted research grant from the Bristol-Myers Squibb Foundation, by a gift from the late William F. McCarty-Cooper, and by funds from the International AIDS Vaccine Initiative.
Footnotes
Published ahead of print on 6 September 2006.
REFERENCES
- 1.Albert, J., B. Abrahamsson, K. Nagy, E. Aurelius, H. Gaines, G. Nystrom, and E. M. Fenyo. 1990. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS 4:107-112. [DOI] [PubMed] [Google Scholar]
- 2.Arendrup, M., A. Sonnerborg, B. Svennerholm, L. Akerblom, C. Nielsen, H. Clausen, S. Olofsson, J. O. Nielsen, and J. E. Hansen. 1993. Neutralizing antibody response during human immunodeficiency virus type 1 infection: type and group specificity and viral escape. J. Gen. Virol. 74:855-863. [DOI] [PubMed] [Google Scholar]
- 3.Bouma, P., M. Leavitt, P. F. Zhang, I. A. Sidorov, D. S. Dimitrov, and G. V. Quinnan, Jr. 2003. Multiple interactions across the surface of the gp120 core structure determine the global neutralization resistance phenotype of human immunodeficiency virus type 1. J. Virol. 77:8061-8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourinbaiar, A. S. 1994. The ratio of defective HIV-1 particles to replication-competent infectious virions. Acta Virol. 38:59-61. [PubMed] [Google Scholar]
- 5.Bradney, A. P., S. Scheer, J. M. Crawford, S. P. Buchbinder, and D. C. Montefiori. 1999. Neutralization escape in human immunodeficiency virus type 1-infected long-term nonprogressors. J. Infect. Dis. 179:1264-1267. [DOI] [PubMed] [Google Scholar]
- 6.Burton, D. R., R. A. Williamson, and P. W. Parren. 2000. Antibody and virus: binding and neutralization. Virology 270:1-3. [DOI] [PubMed] [Google Scholar]
- 7.Cavacini, L., and M. Posner. 2004. Native HIV type 1 virion surface structures: relationships between antibody binding and neutralization or lessons from the viral capture assay. AIDS Res. Hum. Retrovir. 20:435-441. [DOI] [PubMed] [Google Scholar]
- 8.Cecilia, D., V. N. KewalRamani, J. O'Leary, B. Volsky, P. Nyambi, S. Burda, S. Xu, D. Littman, and S. Zolla-Pazner. 1998. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J. Virol. 72:6988-6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng-Mayer, C., and J. A. Levy. 1988. Distinct biological and serological properties of human immunodeficiency viruses from the brain. Ann. Neurol. 23(Suppl.):S58-S61. [DOI] [PubMed] [Google Scholar]
- 10.Chertova, E., J. W. Bess, Jr., B. J. Crise, I. R. Sowder, T. M. Schaden, J. M. Hilburn, J. A. Hoxie, R. E. Benveniste, J. D. Lifson, L. E. Henderson, and L. O. Arthur. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 76:5315-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Etemad-Moghadam, B., Y. Sun, E. K. Nicholson, G. B. Karlsson, D. Schenten, and J. Sodroski. 1999. Determinants of neutralization resistance in the envelope glycoproteins of a simian-human immunodeficiency virus passaged in vivo. J. Virol. 73:8873-8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frost, S. D., T. Wrin, D. M. Smith, S. L. Kosakovsky Pond, Y. Liu, E. Paxinos, C. Chappey, J. Galovich, J. Beauchaine, C. J. Petropoulos, S. J. Little, and D. D. Richman. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. USA 102:18514-18519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaschen, B., C. Kuiken, B. Korber, and B. Foley. 2001. Retrieval and on-the-fly alignment of sequence fragments from the HIV database. Bioinformatics 17:415-418. [DOI] [PubMed] [Google Scholar]
- 15.Hart, T. K., A. M. Klinkner, J. Ventre, and P. J. Bugelski. 1993. Morphometric analysis of envelope glycoprotein gp120 distribution on HIV-1 virions. J. Histochem. Cytochem. 41:265-271. [DOI] [PubMed] [Google Scholar]
- 16.Herrera, C., C. Spenlehauer, M. S. Fung, D. R. Burton, S. Beddows, and J. P. Moore. 2003. Nonneutralizing antibodies to the CD4-binding site on the gp120 subunit of human immunodeficiency virus type 1 do not interfere with the activity of a neutralizing antibody against the same site. J. Virol. 77:1084-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann, W., D. Schubert, J. LaBonte, L. Munson, S. Gibson, J. Scammell, P. Ferrigno, and J. Sodroski. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73:10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korber, B., B. Foley, C. Kuiken, S. Pillai, and J. Sodroksi. 1998. Numbering positions in HIV relative to HXBc2, p. III-102-IV-103. In B. Korber, C. L. Kuiken, B. Foley, B. Hahn, F. McCutchan, J. W. Mellors, and J. Sodroski (ed.), Human retroviruses and AIDS. Los Alamos National Laboratories, Los Alamos, N.Mex.
- 19.Kowalski, M., J. Potz, L. Basiripour, T. Dorfman, W. C. Goh, E. Terwilliger, A. Dayton, C. Rosen, W. Haseltine, and J. Sodroski. 1987. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 237:1351-1355. [DOI] [PubMed] [Google Scholar]
- 20.Koyanagi, Y., S. Miles, R. T. Mitsuyasu, J. E. Merrill, H. V. Vinters, and I. S. Chen. 1987. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236:819-822. [DOI] [PubMed] [Google Scholar]
- 21.Kuiken, C., B. Korber, and R. W. Shafer. 2003. HIV sequence databases. AIDS Rev. 5:52-61. [PMC free article] [PubMed] [Google Scholar]
- 22.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Layne, S. P., M. J. Merges, M. Dembo, J. L. Spouge, S. R. Conley, J. P. Moore, J. L. Raina, H. Renz, H. R. Gelderblom, and P. L. Nara. 1992. Factors underlying spontaneous inactivation and susceptibility to neutralization of human immunodeficiency virus. Virology 189:695-714. [DOI] [PubMed] [Google Scholar]
- 24.Li, Y., H. Hui, C. J. Burgess, R. W. Price, P. M. Sharp, B. H. Hahn, and G. M. Shaw. 1992. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J. Virol. 66:6587-6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Y., J. C. Kappes, J. A. Conway, R. W. Price, G. M. Shaw, and B. H. Hahn. 1991. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J. Virol. 65:3973-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montefiori, D. C., I. Y. Zhou, B. Barnes, D. Lake, E. M. Hersh, Y. Masuho, and L. B. Lefkowitz, Jr. 1991. Homotypic antibody responses to fresh clinical isolates of human immunodeficiency virus. Virology 182:635-643. [DOI] [PubMed] [Google Scholar]
- 27.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas III, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore, J. P., J. A. McKeating, R. A. Weiss, and Q. J. Sattentau. 1990. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250:1139-1142. [DOI] [PubMed] [Google Scholar]
- 29.Nara, P. L., and G. Lin. 2005. HIV-1: the confounding variables of virus neutralization. Curr. Drug Targets Infect. Disord. 5:157-170. [DOI] [PubMed] [Google Scholar]
- 30.Parren, P. W., M. Wang, A. Trkola, J. M. Binley, M. Purtscher, H. Katinger, J. P. Moore, and D. R. Burton. 1998. Antibody neutralization-resistant primary isolates of human immunodeficiency virus type 1. J. Virol. 72:10270-10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poignard, P., T. Fouts, D. Naniche, J. P. Moore, and Q. J. Sattentau. 1996. Neutralizing antibodies to human immunodeficiency virus type-1 gp120 induce envelope glycoprotein subunit dissociation. J. Exp. Med. 183:473-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poignard, P., M. Moulard, E. Golez, V. Vivona, M. Franti, S. Venturini, M. Wang, P. W. Parren, and D. R. Burton. 2003. Heterogeneity of envelope molecules expressed on primary human immunodeficiency virus type 1 particles as probed by the binding of neutralizing and nonneutralizing antibodies. J. Virol. 77:353-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren, X., J. Sodroski, and X. Yang. 2005. An unrelated monoclonal antibody neutralizes human immunodeficiency virus type 1 by binding to an artificial epitope engineered in a functionally neutral region of the viral envelope glycoproteins. J. Virol. 79:5616-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider, J., O. Kaaden, T. D. Copeland, S. Oroszlan, and G. Hunsmann. 1986. Shedding and interspecies type sero-reactivity of the envelope glycopolypeptide gp120 of the human immunodeficiency virus. J. Gen. Virol. 67:2533-2538. [DOI] [PubMed] [Google Scholar]
- 36.Schonning, K., O. Lund, O. S. Lund, and J. E. Hansen. 1999. Stoichiometry of monoclonal antibody neutralization of T-cell line-adapted human immunodeficiency virus type 1. J. Virol. 73:8364-8370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Si, Z., M. Cayabyab, and J. Sodroski. 2001. Envelope glycoprotein determinants of neutralization resistance in a simian-human immunodeficiency virus (SHIV-HXBc2P 3.2) derived by passage in monkeys. J. Virol. 75:4208-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sullivan, N., Y. Sun, J. Li, W. Hofmann, and J. Sodroski. 1995. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J. Virol. 69:4413-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 40.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 41.Yang, X., S. Kurteva, S. Lee, and J. Sodroski. 2005. Stoichiometry of antibody neutralization of human immunodeficiency virus type 1. J. Virol. 79:3500-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.York, J., K. E. Follis, M. Trahey, P. N. Nyambi, S. Zolla-Pazner, and J. H. Nunberg. 2001. Antibody binding and neutralization of primary and T-cell line-adapted isolates of human immunodeficiency virus type 1. J. Virol. 75:2741-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuste, E., J. D. Reeves, R. W. Doms, and R. C. Desrosiers. 2004. Modulation of Env content in virions of simian immunodeficiency virus: correlation with cell surface expression and virion infectivity. J. Virol. 78:6775-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, Y. J., R. Fredriksson, J. A. McKeating, and E. M. Fenyo. 1997. Passage of HIV-1 molecular clones into different cell lines confers differential sensitivity to neutralization. Virology 238:254-264. [DOI] [PubMed] [Google Scholar]