Abstract
The recent development of a cell culture infection model for hepatitis C virus (HCV) permits the production of infectious particles in vitro. In this report, we demonstrate that infectious particles are present both within the infected cells and in the supernatant. Kinetic analysis indicates that intracellular particles constitute precursors of the secreted infectious virus. Ultracentrifugation analyses indicate that intracellular infectious viral particles are similar in size (∼65 to 70 nm) but different in buoyant density (∼1.15 to 1.20 g/ml) from extracellular particles (∼1.03 to 1.16 g/ml). These results indicate that infectious HCV particles are assembled intracellularly and that their biochemical composition is altered during viral egress.
Hepatitis C virus (HCV) is a major cause of chronic hepatitis worldwide. Approximately 3% of the human population is infected, and more than 80% of all HCV infections progress to chronicity, ultimately leading to fibrosis, cirrhosis, and hepatocellular carcinoma (24). There is no vaccine against HCV, and the most widely used therapy involves the administration of type I interferon (α2A) combined with ribavirin. However, this treatment strategy is toxic and has been shown to be ineffective in a significant proportion of the cases (41).
HCV is a member of the Flaviviridae family and the sole member of the genus Hepacivirus (34). HCV is an enveloped virus with a single-strand positive RNA genome that codes for a unique polyprotein of approximatively 3,000 amino acids (11, 12). A single open reading frame is flanked by 5′ and 3′ untranslated regions that contain RNA sequences essential for RNA translation and replication, respectively (17, 18, 23). The translation of the single open reading frame is driven by an internal ribosomal entry site sequence present within the 5′ untranslated region (23), and the resulting polyprotein is processed by cellular and viral proteases into its individual components (reviewed in reference 42). The E1, E2, and core structural proteins are assembled into particles (3, 4), but are not essential for viral RNA replication or translation. The NS2, NS3, NS4A, NS4B, NS5A, and NS5B nonstructural proteins constitute the viral components necessary for efficient viral RNA replication, although NS2 is dispensable for this function (5, 33). In the linear sequence of the polyprotein, the structural proteins are separated from the nonstructural proteins by a small hydrophobic protein, p7 (29), whose function remains unknown but that has the potential to form ion channels (19). Viral proteins are localized in the cytoplasm, and it is assumed, by analogy with other members of the Flaviviridae family, that the entire life cycle of the virus is exclusively cytoplasmic (32).
The expression of the viral polyprotein leads to the formation of virus-like particles in HeLa (37) and Huh-7 cells (21), although the overexpression of core, E1, and E2 is sufficient for the formation of virus-like structures in insect cells (2). None of these particles has been shown to be infectious, even when full-length genomes are used for protein expression (2, 21, 37). The current model for HCV morphogenesis proposes that core particles containing the genome acquire the viral envelope by budding through the endoplasmic reticulum (ER) membrane (44), where viral glycoproteins are inserted as a complex (14, 15, 45).
It has been suggested that, in chronically infected patients, HCV circulates as low-density lipoprotein virus particles (1, 40), with various density profiles depending on the stage of the infection at which the sample was obtained (9, 43). The differences in density and infectivity have been attributed to the presence of host lipoproteins and antibodies bound to the circulating viral particles (22, 43). HCV immune complexes purified by protein A affinity chromatography are rich in HCV RNA, HCV core protein, triglycerides, and apolipoprotein B (apoB) (1). Recently, it has been shown that HCV RNA can be immunoprecipitated with antibodies against the apolipoproteins apoB and apoE, components of very-low-density lipoproteins (VLDL), suggesting that these host proteins are components of circulating HCV particles (40). It is currently unknown whether the viral particles acquire their lipoprotein components in the extracellular milieu or in the infected cell.
In this study, we demonstrate that intracellular viral precursors are infectious and exhibit a higher density than does the secreted virus, suggesting that the low-density configuration of infectious HCV particles might be acquired during viral egress.
MATERIALS AND METHODS
Cells and viruses.
Huh-7 cells (39), which are designated Huh-7/scr cells in this report, were maintained in Dulbecco's modified Eagle's medium (DMEM) medium (Cellgro; Mediatech, Herndon, VA) supplemented with 10% fetal calf serum (FCS) (Invitrogen, Carlsbad, CA), 10 mM HEPES (Invitrogen, Carlsbad, CA), 100 units/ml penicillin, 100 mg/ml streptomycin, and 2 mM l-glutamine (Invitrogen, Carlsbad, CA) in 5% CO2 at 37°C. The plasmid containing the JFH-1 genome was kindly provided by T. Wakita (National Institute of Infectious Diseases, Tokyo, Japan). The original virus was obtained by JFH-1 RNA transfection as previously described (54). JFH-1 virus stocks were obtained by the infection of Huh-7/scr cells at a low multiplicity of infection (MOI) of 0.01 as previously described (54).
Cell lysate preparation.
At the indicated times postinfection, cells were washed once with phosphate buffered saline (PBS; 1.47 mM KH2PO4, 10 mM Na2HPO4, 2.7 mM KCl, 137 mM NaCl, pH 7.4) and incubated with trypsin-EDTA (Invitrogen, Carlsbad, CA) for 2 min at 37°C. Cells were resuspended in PBS and collected by centrifugation at 1,500 rpm for 3 min. The cell pellet was resuspended in DMEM-10% FCS, and cells were lysed by four freeze-thaw cycles in dry ice and a 37°C water bath, respectively. Cell debris was pelleted by centrifugation for 5 min at 4,000 rpm. The supernatant was collected and used for the infection of naive cells or stored at −80°C.
Sedimentation equilibrium gradients.
Gradients were formed by equal-volume (700 μl) steps of 20, 30, 40, 50, and 60% sucrose solutions in TNE buffer (10 mM Tris-HCl, pH 8, 150 mM NaCl, 2 mM EDTA). Iodixanol gradients were prepared identically by using solutions from 10 to 50% in TNE. The samples (250 μl) were overlaid on the gradients, and equilibrium was reached by ultracentrifugation for 16 h at 36,000 rpm (135,000 × g) in an SW60Ti rotor at 4°C in a Beckman L8-80 M preparative ultracentrifuge. Gradient fractions (15 × 250 μl) were collected from the top and titrated for virus infectivity and HCV RNA as described above. The density of the fractions was determined by measuring the mass of 100-μl aliquots of each sample.
Sedimentation velocity gradients.
Infected cell supernatants were subjected to ultracentrifugation in continuous sucrose gradients (11 ml) from 10 to 50% sucrose solutions in TNE. The gradients were overlaid with 250 to 500 μl of the purified virus and centrifuged for 1 h at 39,500 rpm (200,000 × g) in a SW40.Ti rotor at 4°C. Gradient fractions (650 μl) were collected from the top and titrated for virus infectivity and HCV RNA as described above. The migration distance was estimated based on the fraction volume and tube diameter.
Hepatitis B virus (HBV) core particles, purified from liver extracts of HBV transgenic mice as previously described (52), were used as sedimentation velocity markers. The approximate sedimentation coefficient was calculated using the tables previously described by C. R. McEwen (35), where an average density in sucrose of 1.1 g/ml was used for the supernatant HCV particles and the described density of 1.2 g/ml (27) was used for HBV core particles. The approximate sedimentation coefficient of HBV core particles was ∼100S, in good agreement with the expected coefficient (16). The Stokes radius calculated for HBV core particle is ∼36 nm, as observed previously (13). The approach used to calculate the approximate radius of HCV particles was the same as that described by Nielsen et al. (40). It is noteworthy that these results were obtained by using a preparative ultracentrifuge and that the presented values should be considered an approximation.
Infectivity titration.
The infectivity titer was determined on Huh-7/scr cells by end point dilution and immunofluorescence as previously described (54). Typically, 25 μl of each sample were serially diluted fivefold in DMEM-10% FCS and 100 μl was used to inoculate Huh-7/scr cells. Infection was examined 72 h postinoculation by immunofluorescence using a 1:1,000 dilution of a rabbit polyclonal anti-NS5A antibody (MS5 serum provided by Chiron, Emeryville, CA) or a recombinant monoclonal human immunoglobulin G (IgG) anti-E2 antibody (C1 antibody, a gift from Dennis Burton, the Scripps Research Institute) with the appropriate secondary Alexa 555-conjugated antibodies.
RNA extraction and HCV RNA quantitation.
The RNA was extracted from 50 to 100 μl of sample by using the guanidinium isothiocyanate extraction method (10) after adding 2 μg of yeast tRNA per sample as a carrier. HCV RNA was measured by real-time quantitative PCR (RT-qPCR) using primers specific for JFH-1 as described previously (26, 54).
Core protein quantitation.
Samples (50 μl) were diluted 1:1 in radioimmunoprecipitation assay buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8, 0.1% NP-40, 1% sodium dodecyl sulfate, 0.5% deoxycholate) and used to coat enzyme-linked immunosorbent assay plates (F96 MaxiSorp Immunoplate; Nunc, Rochester, NY) overnight at 4°C. The plates were washed twice with 200 μl of PBS before adding 300 μl of blocking solution (5% dry nonfat milk and 10% FCS in PBS). The plates were incubated for 1 h at room temperature. The blocking solution was discarded, and 50 μl of an anti-NP antiserum (MS3; Chiron) dilution (1:1,000) in washing solution (PBS-0.05% Tween 20) was added to the wells. After 1 h of incubation at room temperature, the wells were washed four times with 200 μl of washing solution. The bound primary antibody was detected by incubation with 50 μl of a peroxidase-conjugated goat anti-rabbit IgG antibody (1:10,000 in washing solution). After 1 h of incubation at room temperature, the wells were washed four times with 200 μl of washing solution. The assay was developed using 50 μl of the colorimetric tetramethyl benzidine liquid substrate system (Sigma, St Louis, MO). The reaction was stopped by adding 50 μl of 1N H2SO4, and the relative amount of core protein was determined by reading the optical density at the recommended wavelength (450 nm).
RNase treatment.
Infectious supernatants were treated with a concentrated (2 mg/ml) RNase A solution at a final concentration of 40 μg/ml for 1 h at 37°C. To test whether HCV RNA degradation could be monitored using RT-qPCR, 1 μg of total RNA isolated from HCV-infected cells was diluted in complete medium (DMEM-10% FCS) and exposed to RNase A in parallel with the supernatant samples. An internal control containing the green fluorescent protein (GFP) gene under the T7 promoter was transcribed in vitro using the MEGAscript T7 transcription kit (Ambion, Austin, TX). This control transcript (125 ng) was included in the supernatant-RNase A mixture. The RNA was extracted from the mixture as described above. HCV and GFP RNAs were quantitated by RT-qPCR using specific primers as described above.
Protease digestion of the cell-associated particles.
The infected cells were collected by trypsinization and washed once with PBS. Aliquots of infected cell suspension were prepared, and the cells were pelleted (4,000 rpm, 3 min) and either exposed directly to the protease cocktail or used to prepare a cell lysate by freeze-thaw cycles (as described above) and subsequently exposed to the protease cocktail.
A 10-mg/ml stock solution of the protease cocktail pronase (Fluka; Seelze, Germany) was prepared in PBS. This stock was diluted to a final concentration of 0.5 mg/ml in the infectious supernatant in the infected cell lysate, and the same dilution in DMEM-10% FCS was used to resuspend the infected cell pellet. The mixture was incubated for 10 min at 37°C. This treatment did not affect the integrity of the cells as assessed by trypan blue staining (data not shown). After the protease treatment, the infected cell suspension was washed once with PBS and used to prepare a cell lysate.
Supernatant and cell lysates were diluted in 2 ml of DMEM-10% FCS and partially purified through a 10% sucrose cushion (2 ml) in TNE (3 h at 280,000 × g). The pelleted material was resuspended in 200 μl of DMEM-10% FCS, and the infectivity titer of each sample was determined as described above.
Infectivity neutralization assay.
To determine whether the infectivity of cell-associated particles is dependent on the E2 protein, we preincubated infectious particles, partially purified from supernatants and cell lysates in sedimentation velocity gradients, with 50 μg/ml of a previously described anti-E2 IgG1 antibody (30, 54) and an isotype control anti-Dengue virus IgG1 antibody (DEN3, a gift from Dennis Burton, the Scripps Research Institute). After incubation, the mixtures were serially diluted in complete medium to determine the infectivity titer as described above.
RESULTS
Buoyant density and sedimentation velocity of secreted infectious HCV particles.
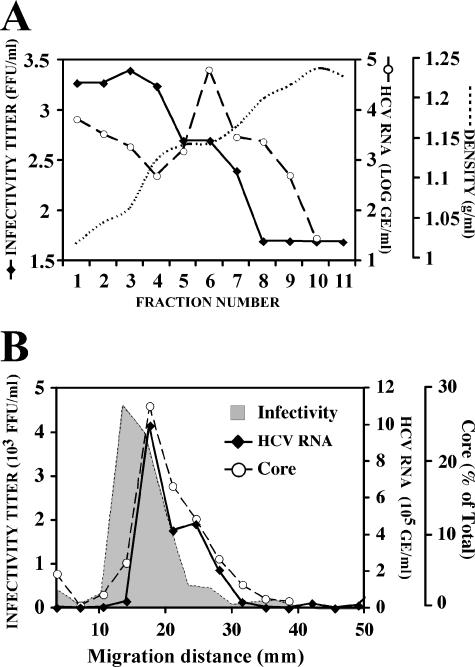
To determine the buoyant density profile of the infectious particles generated in cell culture, supernatants of infected cells were collected on days 8 to 10 postinfection and overlaid onto discontinuous sucrose gradients (20 to 60% in TNE) as described in Materials and Methods. After ultracentrifugation, the infectivity titer and HCV RNA content were determined in all the fractions. As shown in Fig. 1A, the infectivity was observed in numerous fractions corresponding to densities ranging from 1.03 to 1.16 g/ml. Similar results were obtained using gradients of 10 to 50% iodixanol, as shown in Fig. S1a in the supplemental material. As expected, HCV RNA was present in all fractions that contained infectious particles. However, the peak of HCV RNA coincided with the fractions of lowest infectivity, around 1.14 g/ml (Fig. 1A). It is noteworthy that the RNA content is in large excess relative to the number of infectious particles (100:1; 1,000:1) detected in the supernatant of HCV-infected Huh-7/scr cells. The foregoing results corroborate previous observations in similar in vitro HCV infections in human hepatoma cell lines (30, 31, 50, 54).
FIG. 1.
Biophysical properties of the infectious HCV particles in the supernatant. Infected cell supernatants were subjected to ultracentrifugation as described in Materials and Methods. (A) Representative buoyant density profile of viral infectivity (FFU/ml, solid line) and HCV RNA (genome equivalents [GE]/ml, dashed line) in step gradients of 20 to 60% sucrose. Dotted line represents the density (g/ml) measured in each fraction. (B) Representative sedimentation velocity profile of viral infectivity (FFU/ml, gray area), HCV RNA (GE/ml, solid line) and core protein (percent total, dashed line) in continuous gradients of 10 to 50% sucrose. FFU, focus-forming units.
In order to estimate the approximate size of the infectious particle, the sedimentation velocity of infectious HCV particles was determined by rate zonal ultracentrifugation in sucrose gradients as described in Materials and Methods. The supernatant of JFH-1-infected Huh-7/scr cells collected on days 8 to 10 postinfection was overlaid on continuous gradients of 10 to 50% sucrose. The gradients were subjected to ultracentrifugation for 1 h at 200,000 × g, and fractions were collected and analyzed for infectivity, core protein, and HCV RNA content. As shown in Fig. 1B, infectious particles were concentrated in a single peak migrating at an approximate velocity of 15 to 20 mm/h. By using the formulas described in Materials and Methods, we determined the sedimentation coefficient for the infectivity peak fractions to be ∼200S. A diameter of ∼70 nm was calculated for these particles by using this coefficient in the Stokes formula. HBV core particles were used as a reference to validate these approximations.
When the gradient fractions were analyzed for their HCV RNA content, a unique peak containing most of the viral RNA and core protein in the supernatant was observed in fractions corresponding to velocities around 20 mm/h (Fig. 1B). As shown in Fig. 1B and Fig. S1b in the supplemental material, a slight shift was consistently observed in the HCV RNA and core peak relative to the peak of infectivity. This small difference is likely due to the different density of the HCV RNA-containing particles that peak around 1.14 g/ml and the infectious particles whose average density is lower, as shown in Fig. 1A. These results suggest that most of the HCV RNA in the supernatant is encapsidated in noninfectious particles containing at least core protein.
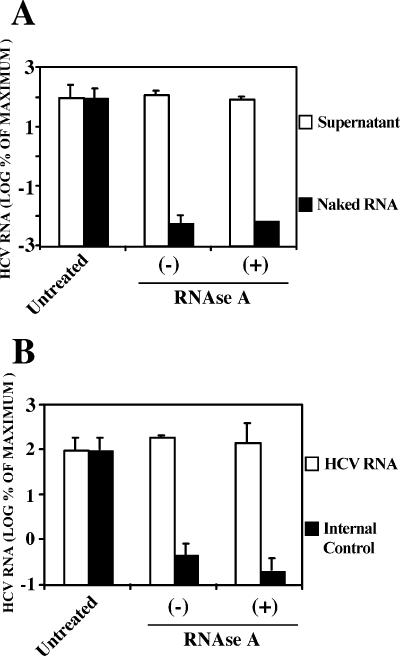
To test this hypothesis, the susceptibility of the HCV RNA to RNase digestion was examined. The cell supernatant was treated with 40 μg/ml of RNase A for 1 h at 37°C. As a control, total RNA purified from HCV-infected cells was resuspended in complete medium (DMEM-10% FCS) and incubated in parallel with the infectious supernatant. As shown in Fig. 2A, the levels of HCV RNA detected by RT-qPCR in the supernatants were comparable in the RNase A-treated samples and untreated samples, suggesting that the HCV RNA present in the infected cell supernatant was resistant to RNase A degradation (Fig. 2A). In contrast, the naked HCV RNA was readily degraded in the medium, even in the absence of added RNase A (Fig. 2A). To rule out the possibility that the RNase activity is inhibited in the infected cell supernatants, an in vitro-transcribed RNA containing the GFP gene was used as an internal control. This control RNA was added to the infectious supernatant before the RNase A treatment, and its stability was also monitored by RT-qPCR. This control was readily degraded after incubation at 37°C, even in the absence of added RNase (Fig. 2B). In contrast, HCV RNA remained intact after the RNase treatment (Fig. 2B). These results support the notion that the majority of the HCV RNA present in the supernatant of infected Huh-7 cells is encapsidated in viral particles.
FIG. 2.
Supernatant HCV RNA is resistant to RNase A degradation. Infected cell supernatants and controls were treated with an excess of RNase A (40 μg/ml) for 1 h at 37°C. The remaining RNA was quantitated by RT-qPCR by using untreated RNAs as a reference. Results are shown as means and standard deviations (error bars) (n = 3) of the percentage of the input RNA. (A) Supernatants (white bars) or naked HCV RNA (black bars) were treated in parallel and HCV RNA was quantitated by RT-qPCR in the different samples. (B) An internal naked RNA control was included in the supernatant-RNase A mixture and both HCV RNA (white bars) and control RNA (black bars) were quantitated by RT-qPCR in the same sample. −, absence of; +, presence of RNase A.
Infected cells contain infectious HCV particles.
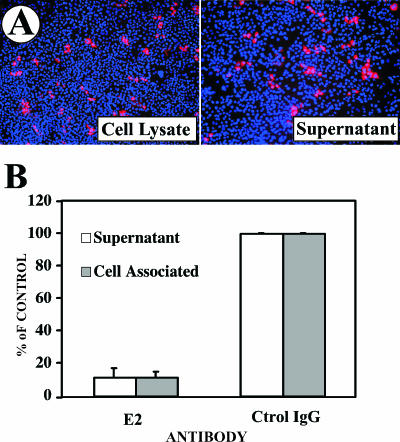
Since it is known that the infectious particles present in the supernatant of HCV-infected Huh-7/scr cells efficiently propagate the infection in vitro (54), we wanted to determine whether infectious particles are also physically associated with the infected cells. Crude cell lysates, prepared from extensively washed infected Huh-7/scr cells 7 days postinfection, were used to inoculate naive Huh-7/scr cells as described in Material and Methods. Infection foci were observed after 72 h in the cells infected with both the cell lysate and the supernatant (Fig. 3A). Importantly, the infectious particles partially purified from the cell lysate were also specifically neutralized by an antibody against the viral envelope glycoprotein E2 (Fig. 3B) as described previously for the particles in the supernatant (30, 54). This suggests that the infectivity of the intracellular particles reflected the presence of enveloped virus rather than the nonspecific uptake of viral RNA or protein from the lysate.
FIG. 3.
Infectious viral particles within infected Huh-7scr cells. Huh-7 cells were infected at a low multiplicity (MOI = 0.01) with JFH-1 virus stocks. Samples from the supernatant and the infected cells were collected at day 7 postinfection. Cell lysates were prepared by freeze-thaw cycles as described in Materials and Methods. The resulting lysates were serially diluted in complete medium and used to inoculate Huh-7 cells. After 72 h incubation, the cells were fixed and processed for immunofluorescence. (A) Infection foci were observed in cells infected with cell lysates and infectious supernatants. (B) In similar infection experiments, infectious virus was purified from the supernatants and cell lysates by ultracentrifugation in a continuous sucrose gradient. The fraction corresponding to the infectivity peak was preincubated for 1 h at 37°C with a neutralizing anti-E2 antibody (E2) or an isotype control antibody (Ctrol IgG) prior to infection. The results are shown as means and standard errors (error bars) of the percentage of the infection detected in the control in two independent infections.
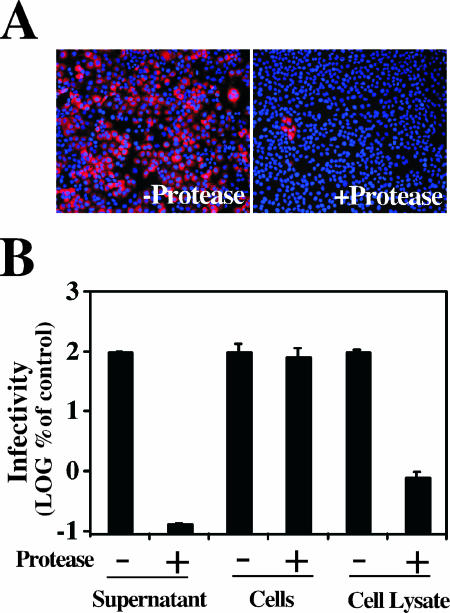
In order to determine whether the cell-associated infectivity was derived from intracellular particles or from extracellular particles that were bound to the cell surface, supernatants from infected cells, cell lysates, and single-cell suspensions were exposed to a protease cocktail pronase (Fluka; Seelze, Germany). The mixtures were incubated for 10 min at 37°C. After the treatment, the particles were partially purified as described in Materials and Methods to avoid any interference of the protease cocktail with the infectivity titration procedure. The infectivity present in the supernatant was very sensitive to the proteases (Fig. 4A) and reduced the infectious titer by more than three orders of magnitude (Fig. 4B). In contrast, the cell-associated infectivity was unaffected when the protease cocktail was added to the intact cell suspension (Fig. 4B), while it was accessible to proteolytic degradation when the protease was added after cell lysis (Fig. 4B), indicating that the cell-associated infectious particles are confined inside the infected cell.
FIG. 4.
Cell-associated particles are confined inside the infected cells. Infected cells and cell supernatants were treated with a protease cocktail as described in Materials and Methods. After the protease treatment, the infectivity titer of the partially purified material was determined in each sample. (A) Undiluted purified supernatants were used to infect naive cells. Seventy-two hours postinfection, the inoculated cells were fixed and processed for immunofluorescence using a monoclonal anti-E2 antibody. −protease, untreated supernatant; +protease, pronase-treated sample. (B) Infectivity titer of the supernatant (supernatant), cell suspension treated before lysis (cells), and cell lysate (cell lysate) in the protease cocktail-untreated (−) or −treated (+) samples. The results are shown as means and standard errors (error bars) (n = 2) of the percentage of the corresponding untreated sample. The infectivity titer in the purified supernatant was ∼105 focus-forming units (FFU)/ml and ∼104 FFU/ml in the purified cell lysates. −, absence of; +, presence of RNase A.
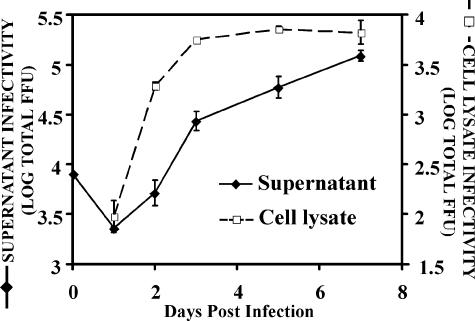
The kinetics of viral particle production was analyzed after the infection of Huh-7/scr cells at a low multiplicity (MOI = 0.01). As expected (54), progeny particles progressively accumulated in the supernatant of the infected cells (Fig. 5) and reached maximum titers after day 7 postinfection. In contrast, cell-associated infectious particles reached maximum titers 4 days postinfection, after which no significant increase in the total number of cell-associated infectious particles was observed (Fig. 5). The early accumulation of infectious cell-associated particles contrasts with the gradual release of infectious particles into the supernatant, suggesting that the production of infectious particles within the infected cell precedes their secretion.
FIG. 5.
Infectious viral particle production kinetics. Huh-7 cells were infected at a low multiplicity (MOI = 0.01) with JFH-1 virus stocks. Supernatants and infected cell lysates were collected at the indicated time points. Infected cells were subjected to lysis as described in Materials and Methods. The infectivity titer of both supernatant and cell lysate was determined by infection on naive cells. The total infectivity titers in the supernatant of 1.5 × 106 cells (black diamonds) and cell lysates (white squares) at different times postinfection are presented as the means and standard errors (error bars) of two independent infections. FFU, focus-forming units
The density profiles of cell-associated and secreted infectious particles are distinct.
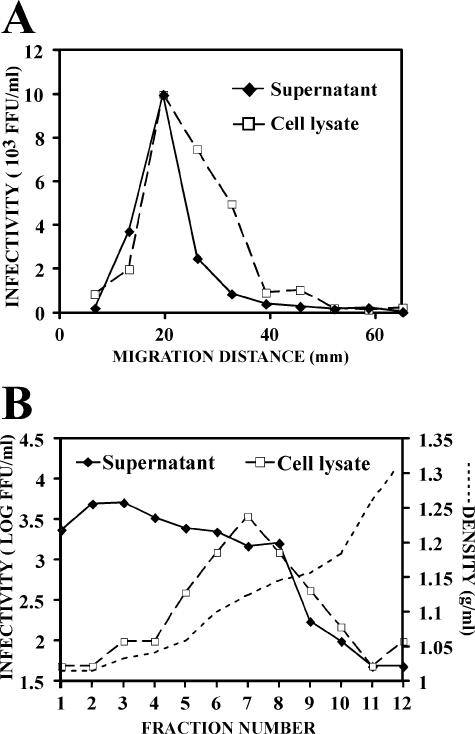
The sedimentation velocity and buoyant density profiles of secreted and intracellular infectious virus particles were analyzed in parallel to compare their biophysical properties. These samples were overlaid onto continuous sucrose gradients to determine their sedimentation velocities as described above. The total cell lysates displayed a single peak of infectivity in the fractions migrating at ∼20 mm/h (Fig. 6A). This sedimentation profile was very similar to that of the particles from the supernatant (Fig. 6A), suggesting that they share similar sizes and shapes.
FIG. 6.
Biophysical properties of the infectious particles present in the Huh-7 cell-lysates. Infected cell lysates were subjected to ultracentrifugation as described in Materials and Methods. (A) Representative sedimentation velocity profile of the infectious viral particles in the supernatant (focus-forming units [FFU]/ml, black diamonds) and the cell lysate (FFU/ml, white squares) in continuous gradients of 10 to 50% sucrose. (B) Representative buoyant density profile of viral infectivity in the supernatant (black diamonds) and in the cell lysate (open squares) in 10 to 50% iodixanol step gradients. The dotted line represents the density (g/ml) measured in each fraction.
The buoyant density of the cell-associated and supernatant-derived infectivity was also analyzed by equilibrium sedimentation ultracentrifugation in gradients of 10 to 50% iodixanol as described above. The broad distribution of the infectivity in the supernatant contrasted with a relatively narrow peak found in the cell lysate that displayed a mean density of ∼1.15 g/ml. Similar results were obtained in gradients of 20 to 60% sucrose (see Fig. S2a in the supplemental material). This density difference was not due to the exposure of the particles to enzymatic activities present in the cell lysate since infectious supernatants incubated with uninfected Huh-7/scr lysates showed a density profile identical to that of the original supernatant sample (see Fig. S2b in the supplemental material). The different buoyant density observed in the secreted and cell-associated infectious particles could reflect differences in their biochemical compositions.
DISCUSSION
The recent development of an in vitro cell culture infection system for HCV (30, 50, 54) provides the necessary tools to study viral entry, particle assembly, maturation, and egress, i.e., aspects of the viral life cycle that could not be studied in vivo or in previous in vitro models. Until recently, the only sources of infected cells were liver biopsies obtained from chronically infected individuals and infected livers from transplant recipients. The reconstitution of the entire life cycle of the virus in vitro uniquely enables the examination of events occurring during HCV infection before the viral particle is released to the extracellular milieu. In this study, we report for the first time the existence of infectious HCV particles in infected cells. These particles share a similar sedimentation velocity profile with the particles purified from the supernatant (Fig. 1B and 6A). However, the buoyant density profile of these particles is clearly different with a density range located between 1.15 to 1.20 g/ml in sucrose gradients, in contrast with the lower density and wider distribution (1.03 to 1.16 g/ml) observed for the particles in the supernatant (Fig. 6B). This different density distribution might account for a slight shift in the sedimentation velocity profile observed for the intracellular particles (Fig. 5A) and suggests that the biochemical composition of the intracellular infectious particles may differ from that of the infectious particles in the supernatant. The detection of infectious particles in densities above 1.15 g/ml in the cell lysates suggests that the low-density configuration of the particles found in the supernatant is not required for in vitro infectivity.
In the present study, we also provide evidence that the infectious particles generated in vitro using the consensus JFH-1 molecular HCV clone display sedimentation velocity and buoyant density profiles similar to those described for HCV particles isolated from the sera of HCV-infected patients (40). Our results suggest that the infectivity and the HCV RNA are associated with particles of similar size (Fig. 1B), although most of the RNA is encapsidated in noninfectious particles containing the core protein (Fig. 1B and 2). Thus, the sedimentation profile of the JFH-1 RNA present in the supernatant of the infected cells is similar to the RNA sedimentation profile of HCV virions previously described in samples from HCV-infected patients (28, 40). Based on this sedimentation coefficient, we calculated an approximative diameter of ∼65 to 70 nm for the JFH-1 infectious particles, a size that is compatible with that of the infectious particles found in infected patient sera as estimated by filtration and subsequent infection in chimpanzees (7, 20).
While the sedimentation coefficient calculated for HCV both in vivo and in vitro falls into the range described for other Flaviviridae (∼200S) (32), the broad density range of the particles in the supernatant appears to be a unique feature characteristic of HCV (1, 22, 25, 30, 36, 40, 43, 48, 49, 51, 54). This broad range of buoyant densities is thought to be partially dependent on the presence of host lipoprotein components, including apoB and apoE, in the viral particle (1, 40, 47, 48). The fact that this characteristic distribution was also observed in the in vitro-produced infectious HCV particles (Fig. 1A) (30) suggests that their density is also determined by the presence of host lipoprotein components, as are the HCV particles found in patients. Although it has been shown that recombinant envelope glycoproteins have the capacity to bind human lipoproteins in vitro (38), whether this interaction is responsible for the association with the host lipoproteins remains unknown. Interestingly, the density profile of a fully infectious mutant of JFH-1 virus bearing a point mutation in E2 is different than that of wild-type JFH-1 virus (55). These results suggest that viral components (53) (e.g., the E2 protein) as well as host factors (31) (e.g., lipoproteins) determine the density profiles of infectious HCV particles. Differential flotation ultracentrifugation analyses showed that populations of infectious particles with low, intermediate, and high densities could be isolated (data not shown). Indeed, the buoyant density profile illustrated in a previous report from our laboratory reflects a virus population that was preselected for higher density by ultracentrifugation in a low-density medium (54).
Based on the differential density associated with the infectious intracellular particles, we hypothesize that infectious HCV particles are assembled in intracellular compartments, as high-density (>1.15-g/ml) precursors, and that they acquire elements that confer a low-density (<1.14-g/ml) configuration along their egress, resulting in the secretion of low-density particles that can be found in the supernatant of the in vitro-infected cells. It is noteworthy that in normal hepatocytes, apoB-containing VLDL precursors are assembled in the rough ER lumen as high-density (>1.18-g/ml) particles (6, 46). These precursors are not secreted unless they undergo further lipidation steps occurring in undefined post-ER compartments where they acquire the final VLDL configuration (8).
The molecular mechanisms underlying the biophysical transformation that HCV particles undergo during their secretion process require further investigation. However, the detection of apoB associated with circulating particles (1, 40) could suggest that, like VLDL-precursors, high-density intracellular HCV particles incorporate apoB during their secretion process. This association could drive the virus precursors into the VLDL maturation and secretion pathway. The addition of low-density lipids (e.g., triglycerides) could be as essential for the secretion of the HCV particles as it is for the secretion of VLDL (8). The resulting secreted particles would contain apoB (40), would be rich in triglycerides (1), and would have a lower density (Fig. 1A).
Supplementary Material
Acknowledgments
We thank Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan) for kindly providing the infectious molecular clone JFH-1; Dennis Burton (Department of Immunology, the Scripps Research Institute, La Jolla, CA) for providing the recombinant human IgG anti-E2; Michael Houghton (Chiron, Emeryville, CA) for providing polyclonal antibodies against NS5A and core HCV proteins; and Holly Maier, Stefan Wieland, and Bruno Sainz in our laboratory and Malcolm R. Wood in the Core Microscopy Facility at the Scripps Research Institute for helpful comments and discussions. We also thank Christina Whitten, Angelina Eustaquio, and Brian Boyd for technical support.
This study was supported by grant R01-CA108304 from the National Institutes of Health. This is manuscript number 18238 from the Scripps Research Institute.
Footnotes
Published ahead of print on 6 September 2006.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Andre, P., F. Komurian-Pradel, S. Deforges, M. Perret, J. L. Berland, M. Sodoyer, S. Pol, C. Brechot, G. Paranhos-Baccala, and V. Lotteau. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 76:6919-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumert, T. F., S. Ito, D. T. Wong, and T. J. Liang. 1998. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 72:3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumert, T. F., J. Vergalla, J. Satoi, M. Thomson, M. Lechmann, D. Herion, H. B. Greenberg, S. Ito, and T. J. Liang. 1999. Hepatitis C virus-like particles synthesized in insect cells as a potential vaccine candidate. Gastroenterology 117:1397-1407. [DOI] [PubMed] [Google Scholar]
- 4.Blanchard, E., D. Brand, S. Trassard, A. Goudeau, and P. Roingeard. 2002. Hepatitis C virus-like particle morphogenesis. J. Virol. 76:4073-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1975. [DOI] [PubMed] [Google Scholar]
- 6.Boren, J., L. Graham, M. Wettesten, J. Scott, A. White, and S. O. Olofsson. 1992. The assembly and secretion of ApoB 100-containing lipoproteins in Hep G2 cells. ApoB 100 is cotranslationally integrated into lipoproteins. J. Biol. Chem. 267:9858-9867. [PubMed] [Google Scholar]
- 7.Bradley, D. W., K. A. McCaustland, E. H. Cook, C. A. Schable, J. W. Ebert, and J. E. Maynard. 1985. Posttransfusion non-A, non-B hepatitis in chimpanzees. Physicochemical evidence that the tubule-forming agent is a small, enveloped virus. Gastroenterology 88:773-779. [PubMed] [Google Scholar]
- 8.Brodsky, J. L., V. Gusarova, and E. A. Fisher. 2004. Vesicular trafficking of hepatic apolipoprotein B100 and its maturation to very low-density lipoprotein particles; studies from cells and cell-free systems. Trends Cardiovasc. Med. 14:127-132. [DOI] [PubMed] [Google Scholar]
- 9.Carabaich, A., M. Ruvoletto, E. Bernardinello, N. Tono, L. Cavalletto, L. Chemello, A. Gatta, and P. Pontisso. 2005. Profiles of HCV core protein and viremia in chronic hepatitis C: possible protective role of core antigen in liver damage. J. Med. Virol. 76:55-60. [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 11.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, P. J. Barr, and et al. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choukhi, A., S. Ung, C. Wychowski, and J. Dubuisson. 1998. Involvement of endoplasmic reticulum chaperones in the folding of hepatitis C virus glycoproteins. J. Virol. 72:3851-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crowther, R. A., N. A. Kiselev, B. Bottcher, J. A. Berriman, G. P. Borisova, V. Ose, and P. Pumpens. 1994. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell 77:943-950. [DOI] [PubMed] [Google Scholar]
- 14.Deleersnyder, V., A. Pillez, C. Wychowski, K. Blight, J. Xu, Y. S. Hahn, C. M. Rice, and J. Dubuisson. 1997. Formation of native hepatitis C virus glycoprotein complexes. J. Virol. 71:697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duvet, S., L. Cocquerel, A. Pillez, R. Cacan, A. Verbert, D. Moradpour, C. Wychowski, and J. Dubuisson. 1998. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J. Biol. Chem. 273:32088-32095. [DOI] [PubMed] [Google Scholar]
- 16.Fields, H. A., F. B. Hollinger, J. Desmyter, J. L. Melnick, and G. R. Dreesman. 1977. Biochemical and biophysical properties of hepatitis B core particles derived from Dane particles and infected hepatocytes. Intervirology 8:336-350. [DOI] [PubMed] [Google Scholar]
- 17.Friebe, P., J. Boudet, J. P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffin, S. D., L. P. Beales, D. S. Clarke, O. Worsfold, S. D. Evans, J. Jaeger, M. P. Harris, and D. J. Rowlands. 2003. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, amantadine. FEBS Lett. 535:34-38. [DOI] [PubMed] [Google Scholar]
- 20.He, L. F., D. Alling, T. Popkin, M. Shapiro, H. J. Alter, and R. H. Purcell. 1987. Determining the size of non-A, non-B hepatitis virus by filtration. J. Infect. Dis. 156:636-640. [DOI] [PubMed] [Google Scholar]
- 21.Heller, T., S. Saito, J. Auerbach, T. Williams, T. R. Moreen, A. Jazwinski, B. Cruz, N. Jeurkar, R. Sapp, G. Luo, and T. J. Liang. 2005. An in vitro model of hepatitis C virion production. Proc. Natl. Acad. Sci. USA 102:2579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hijikata, M., Y. K. Shimizu, H. Kato, A. Iwamoto, J. W. Shih, H. J. Alter, R. H. Purcell, and H. Yoshikura. 1993. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J. Virol. 67:1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honda, M., M. R. Beard, L. H. Ping, and S. M. Lemon. 1999. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 73:1165-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoofnagle, J. H. 2002. Course and outcome of hepatitis C. Hepatology 36:S21-S29. [DOI] [PubMed] [Google Scholar]
- 25.Kanto, T., N. Hayashi, T. Takehara, H. Hagiwara, E. Mita, M. Naito, A. Kasahara, H. Fusamoto, and T. Kamada. 1995. Density analysis of hepatitis C virus particle population in the circulation of infected hosts: implications for virus neutralization or persistence. J. Hepatol. 22:440-448. [DOI] [PubMed] [Google Scholar]
- 26.Kapadia, S. B., A. Brideau-Andersen, and F. V. Chisari. 2003. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc. Natl. Acad. Sci. USA 100:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kessler, H. A., J. Dixon, C. R. Howard, K. Tsiquaye, and A. J. Zuckerman. 1981. Effects of amphotericin B on hepatitis B virus. Antimicrob. Agents Chemother. 20:826-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein, K. C., S. J. Polyak, and J. R. Lingappa. 2004. Unique features of hepatitis C virus capsid formation revealed by de novo cell-free assembly. J. Virol. 78:9257-9269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, C., B. D. Lindenbach, B. M. Pragai, D. W. McCourt, and C. M. Rice. 1994. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 68:5063-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 31.Lindenbach, B. D., P. Meuleman, A. Ploss, T. Vanwolleghem, A. J. Syder, J. A. McKeating, R. E. Lanford, S. M. Feinstone, M. E. Major, G. Leroux-Roels, and C. M. Rice. 2006. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. USA 103:3805-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincot Williams & Wilkins, Phildelphia, Pa. [Google Scholar]
- 33.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 34.Maniloff, J. 1995. Identification and classification of viruses that have not been propagated. Arch. Virol. 140:1515-1520. [DOI] [PubMed] [Google Scholar]
- 35.McEwen, C. R. 1967. Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal. Biochem. 20:114-149. [DOI] [PubMed] [Google Scholar]
- 36.Miyamoto, H., H. Okamoto, K. Sato, T. Tanaka, and S. Mishiro. 1992. Extraordinarily low density of hepatitis C virus estimated by sucrose density gradient centrifugation and the polymerase chain reaction. J. Gen. Virol. 73:715-718. [DOI] [PubMed] [Google Scholar]
- 37.Mizuno, M., G. Yamada, T. Tanaka, K. Shimotohno, M. Takatani, and T. Tsuji. 1995. Virion-like structures in HeLa G cells transfected with the full-length sequence of the hepatitis C virus genome. Gastroenterology 109:1933-1940. [DOI] [PubMed] [Google Scholar]
- 38.Monazahian, M., S. Kippenberger, A. Muller, H. Seitz, I. Bohme, S. Grethe, and R. Thomssen. 2000. Binding of human lipoproteins (low, very low, high density lipoproteins) to recombinant envelope proteins of hepatitis C virus. Med. Microbiol. Immunol. 188:177-184. [DOI] [PubMed] [Google Scholar]
- 39.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 40.Nielsen, S. U., M. F. Bassendine, A. D. Burt, C. Martin, W. Pumeechockchai, and G. L. Toms. 2006. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J. Virol. 80:2418-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patel, K., and J. G. McHutchison. 2004. Initial treatment for chronic hepatitis C: current therapies and their optimal dosing and duration. Clevel. Clin. J. Med. 71(Suppl. 3):S8-12. [DOI] [PubMed] [Google Scholar]
- 42.Penin, F., J. Dubuisson, F. A. Rey, D. Moradpour, and J. M. Pawlotsky. 2004. Structural biology of hepatitis C virus. Hepatology 39:5-19. [DOI] [PubMed] [Google Scholar]
- 43.Pumeechockchai, W., D. Bevitt, K. Agarwal, T. Petropoulou, B. C. Langer, B. Belohradsky, M. F. Bassendine, and G. L. Toms. 2002. Hepatitis C virus particles of different density in the blood of chronically infected immunocompetent and immunodeficient patients: Implications for virus clearance by antibody. J. Med. Virol. 68:335-342. [DOI] [PubMed] [Google Scholar]
- 44.Roingeard, P., C. Hourioux, E. Blanchard, D. Brand, and M. Ait-Goughoulte. 2004. Hepatitis C virus ultrastructure and morphogenesis. Biol. Cell 96:103-108. [DOI] [PubMed] [Google Scholar]
- 45.Rouille, Y., F. Helle, D. Delgrange, P. Roingeard, C. Voisset, E. Blanchard, S. Belouzard, J. McKeating, A. H. Patel, G. Maertens, T. Wakita, C. Wychowski, and J. Dubuisson. 2006. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J. Virol. 80:2832-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rustaeus, S., K. Lindberg, P. Stillemark, C. Claesson, L. Asp, T. Larsson, J. Boren, and S. O. Olofsson. 1999. Assembly of very low density lipoprotein: a two-step process of apolipoprotein B core lipidation. J. Nutr. 129:463S-466S. [DOI] [PubMed] [Google Scholar]
- 47.Thomssen, R., S. Bonk, C. Propfe, K. H. Heermann, H. G. Kochel, and A. Uy. 1992. Association of hepatitis C virus in human sera with beta-lipoprotein. Med. Microbiol. Immunol. 181:293-300. [DOI] [PubMed] [Google Scholar]
- 48.Thomssen, R., S. Bonk, and A. Thiele. 1993. Density heterogeneities of hepatitis C virus in human sera due to the binding of beta-lipoproteins and immunoglobulins. Med. Microbiol. Immunol. 182:329-334. [DOI] [PubMed] [Google Scholar]
- 49.Trestard, A., Y. Bacq, L. Buzelay, F. Dubois, F. Barin, A. Goudeau, and P. Roingeard. 1998. Ultrastructural and physicochemical characterization of the hepatitis C virus recovered from the serum of an agammaglobulinemic patient. Arch. Virol. 143:2241-2245. [DOI] [PubMed] [Google Scholar]
- 50.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watson, J. P., D. J. Bevitt, G. P. Spickett, G. L. Toms, and M. F. Bassendine. 1996. Hepatitis C virus density heterogeneity and viral titre in acute and chronic infection: a comparison of immunodeficient and immunocompetent patients. J. Hepatol. 25:599-607. [DOI] [PubMed] [Google Scholar]
- 52.Wieland, S. F., A. Eustaquio, C. Whitten-Bauer, B. Boyd, and F. V. Chisari. 2005. Interferon prevents formation of replication-competent hepatitis B virus RNA-containing nucleocapsids. Proc. Natl. Acad. Sci. USA 102:9913-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong, J., P. Gastaminza, J. Chung, Z. Stamataki, M. Isogawa, G. Cheng, J. A. McKeating, and F. V. Chisari. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082-11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.