Abstract
Papillomaviruses are small DNA viruses which establish persistent infection in the epithelial tissue of various animal species. Three papillomavirus proteins encoded by the bovine papillomavirus type 1 E2 open reading frame have a common C-terminal DNA binding and dimerization domain and function as dimeric proteins in the regulation of viral gene expression, genome replication, and maintenance. The full-length E2 protein, expressed usually at the lowest level of the three, is an activator, while shorter forms of E2, lacking the transactivation domain, serve as repressors of replication and transcription. In virally infected cells, the full-length E2 protein forms heterodimers with repressor forms of the E2 protein and the biological activities of such heterodimers are poorly known. In order to study the functionality of E2 heterodimers, we joined the full-length E2 protein and E2 repressor by a flexible polypeptide hinge so that they formed a single-chain intramolecular dimer. The single-chain E2 heterodimers folded correctly to form genuine pseudodimers capable of binding to the specific E2 protein binding site with high affinity. Characterization of the activities of this protein in transcription showed that it functions as an effective transcriptional activator, which is comparable to what was found for the full-length E2 protein. The single-chain heterodimer is dependent to some extent on Brd4 protein and is able to support papillomavirus origin replication; however, it does not support the partitioning of the multimeric E2 binding site containing plasmids in dividing cells. Our results suggest that E2 heterodimers serve as activators of transcription and replication during the viral life cycle.
The proteins encoded by the papillomavirus E2 open reading frame (ORF) play a crucial role in the viral life cycle and serve as major viral regulators of the transcription, replication, and segregation of the viral genome in the infected cells. The E2 proteins are composed of three domains: the N-terminal transactivation domain, the C-terminal dimerization and sequence-specific DNA binding domain (DBD), and a flexible hinge region separating these two functional and structural domains. The carboxy-terminal DBD binds as a dimer to consensus sequence ACCN6GGT (3), and the N-terminal activation domain is required for the replication, transactivation, and segregation functions of the protein (1, 6, 19, 26).
Bovine papillomavirus type 1 (BPV1) and human papillomavirus (HPV) types 11, 16, and 31 have been shown to encode mRNAs coding for short forms of the E2 protein (8-10, 14, 24). For BPV1, mRNA for E2C is transcribed from a promoter internal to the open reading frame, and the other protein, E8/E2, is created by splicing E8 ORF sequences to an acceptor located within the E2 ORF. These proteins maintain the DBDs of the E2 protein; however, they lack the activation domain and are not able to activate transcription and replication from the E2 binding site-dependent promoters. In addition, the E2 protein and E2C are distributed differently within the cell nucleus (13). The putative repressor proteins encoded by HPVs are similar to the BPV1 E8/E2 protein, since they contain a small conserved E8 ORF (HPV 31 and 11) or a fragment of E1 ORF (HPV 11) fused to the C terminus of E2. Transient overexpression assays have suggested that repressor proteins act as negative regulators of E2 (8, 18, 24). However, despite being a negative regulator, HPV 31 and at least one of the BPV1 repressors are required for the long-term maintenance of viral episomes (15, 24). In another work, Lim et al. have shown that heterodimers composed of BPV1 E2 and E2C can activate DNA replication in a cell-free system (18).
The full-length E2 protein and short forms of E2 are able to form heterodimers through the common C-terminal DBD (19). The full-length E2 protein is an activator, and the short forms of E2 lacking the transactivation domain serve as repressors, but the functions of heterodimers containing only one activation domain are still not clear. In order to study the biological activities of E2 heterodimers, we used a tethering strategy and joined the full-length E2 protein and E2 repressor by a flexible polypeptide so that they formed a single-chain protein capable of forming an intramolecular dimer. In this study, we show that such single-chain heterodimers are fully functional in sequence-specific binding to the DNA. Additionally, we studied the functions of these single-chain E2 heterodimers in transcription, replication, and segregation assays. Our data show that the E2 heterodimer with one activation domain is a transactivator that is able to support papillomavirus DNA replication and efficiently activate the E2-dependent promoters but is inactive in partitioning the E2 binding site containing plasmids in dividing cells.
MATERIALS AND METHODS
Plasmids.
Bovine papillomavirus E2 expression vectors pCGE2, pCGE2C, pRetE2, and pRetE2E39A; E1 expression vector pCGEag; and replication reporter pUCAlu have been described previously (1, 26). pCGE2-DBD contains the DNA binding domain of the BPV1 E2 protein (amino acids [aa] 310 to 410) in frame with the N-terminal epitope tag E2Tag SSTSSDFRDR derived from BPV1 E2 (aa 199 to 208). To construct the E2 single-chain heterodimers, we first deleted the stop codon of E2 and created additional restriction sites for SalI and BamHI by PCR (oligonucleotide 5′ CGACTAGTGGATCCGTCGACGAAGTCCAAGCTGGC). The BamHI site was used to clone PCR-generated fragments coding E2 aa 284 to 410, 310 to 410, and 325 to 410 and the SalI site to clone the double-stranded oligonucleotide linker encoding the polypeptide tether (5′ TCGACGGGGGATCAGGCGGAGGTGGAGGATCCGGTGGCGGTGGCTCTC) (see Fig. 2A). Constructed coding regions were verified by sequencing. pRet vectors expressing E2 single-chain heterodimers were constructed by replacing the EheI-SfiI fragment in the wild-type E2 coding region with the corresponding fragment from the respective pCG plasmid.
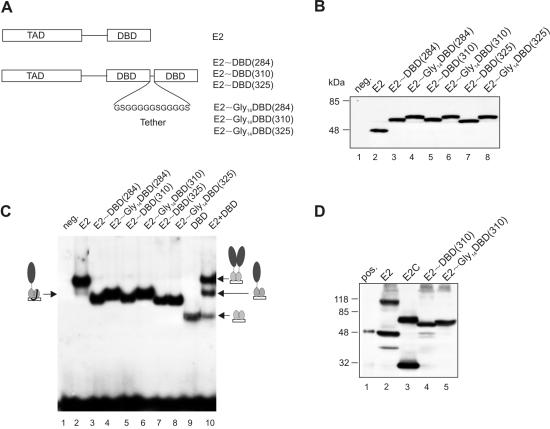
FIG. 2.
(A) Schematic representations of designed E2 single-chain heterodimers. A tilde indicates a tethered dimer, and the numbers in parentheses show the starting amino acid positions for the DBD; Gly14 indicates the glycine-rich tether. TAD, transactivation domain. (B) Immunoblot analysis of single-chain E2 heterodimers. The lysates of COS-7 cells transfected with E2 expression plasmids were subjected to SDS-PAGE and immunoblotted with E2-specific antibodies. neg., no E2. (C) DNA binding assay with single-chain E2 heterodimers. The sequence-specific binding of E2 to one E2 binding site was determined by a gel retardation assay. The electrophoretic mobility shift assay was performed with equal amounts of cell extracts of COS-7 cells transfected with expression plasmids for E2, E2DBD, E2 and E2DBD together, single-chain E2 heterodimers, and radiolabeled E2BS. Protein-DNA complexes were resolved on 6% PAGE gels in 0.25× Tris-borate-EDTA buffer. (D) Immunoblot analysis of UV-cross-linked COS-7 cells transfected with E2 expression plasmids. pos., 5 ng of bacterially purified E2 protein. Membrane was incubated with E2-specific antibody 5E11.
pCG-Brd4-CTD contains N-terminal epitope tag E2Tag, the nuclear localization signal from the simian virus 40 large T antigen, and the last 315 amino acids of human Brd4 in the pCG vector (11).
Reporter plasmid 3E2BS-Luc contains three E2 binding sites, three 21-bp GC-rich repeats, and an enhancerless simian virus 40 early promoter in front of the firefly luciferase gene. URR-Luc contains the BPV1 upstream regulatory region (URR; nucleotides 7476 to 7494 from the BPV1 genome) (11). pRL-TK was purchased from Promega.
Cells and transfections.
Chinese hamster ovary (CHO) cells were maintained in Ham's F-12 medium supplemented with 10% fetal calf serum, and COS-7, Jurkat, and C127- and BPV1-transformed C127 cells were grown in Iscove's modified Dulbecco's medium containing 10% fetal bovine serum.
Transfections into CHO, Jurkat, and COS-7 cells were performed as described previously (1, 2, 27). For the replication assay, CHO cells were electroporated with 100 ng of pUCAlu DNA, 500 ng of pCGEag, and 250 ng of pCGE2 or its derivatives. Low-molecular-weight DNA was isolated and analyzed by a DpnI sensitivity assay and Southern blotting as described in reference 27.
For the transcription assay, CHO cells were transfected with 25 ng of pRL-TK, 100 ng of E2-responsive reporter plasmid, and increasing concentrations of pCGE2 or pCG E2∼Gly14DBD(310). To test the influence of the Brd4 C-terminal domain (CTD) on E2 transcriptional activation, CHO cells were transfected with 25 ng of pRL-TK, 100 ng of E2-responsive reporter plasmid 3E2BS-Luc, 200 ng of pCGE2 or pCG E2∼Gly14DBD(310), and increasing concentrations of pCGBrd4-CTD. Thirty hours later, cells were harvested and lysed by freezing-thawing, and luciferase activity was analyzed by a TD-20/20 luminometer (Turner Designs), following the Dual-Luciferase reporter assay (Promega) manuals.
For the plasmid partitioning assay, Jurkat cells (5 × 106) were electroporated with 1 μg of the pRetE2 or pRet plasmid encoding single-chain E2 heterodimers. During 1 week, equal aliquots of the cell suspension were collected for flow cytometry analysis (FACSCalibur; Becton-Dickinson Biosciences) and the remaining cells were diluted with fresh medium, keeping the cell concentration between 0.3 × 106 and 1 × 106 cells per ml. The number of enhanced green fluorescent protein (EGFP)-positive cells was calculated as described in reference 1.
For the DNA binding assay, 106 COS-7 cells were electroporated with 250 ng of pCGE2 or its derivatives. Twenty-four hours later, the cells were collected and lysed in 50 μl of lysis buffer (20 mM Tris-HCl, pH 8.0, 30 mM KCl, 400 mM NaCl, 0.1 mM EDTA, 0.5% Triton X-100, 10 mM dithiothreitol, 5% glycerol, and protease inhibitors) on ice for 30 min. The cell debris were removed by centrifugation, and the supernatant was used for the DNA binding assay as described in reference 2. For Brd4-E2 coimmunoprecipitation analysis, COS-7 cells were electroporated with 200 ng of pCG-Brd4-CTD, 100 ng of pCGE2, and 200 ng of pCGE2C.
Immunoprecipitation and immunoblotting analysis.
For immunoprecipitation, 1 × 107 C127 and C127-BPV cells were lysed in 200 μl of radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1 mM dithiothreitol, protease inhibitors) containing 0.5% sodium dodecyl sulfate (SDS) and then diluted five times with RIPA buffer without SDS. Cells were disrupted by sonication on ice (twice, 5 s each). Cell extracts were clarified by centrifugation, and the E2 proteins were immunoprecipitated from supernatant with 3 μg of E2-specific rabbit polyclonal antibody for 1 h at 4°C. Immunocomplexes were collected on protein G-Sepharose (Amersham Biosciences, Uppsala, Sweden), washed twice with RIPA buffer containing 0.5 M NaCl and twice with RIPA buffer, resuspended in SDS loading buffer, and subjected to immunoblotting analysis with E2-specific antibodies 1E4 and 1H10 (12). UV treatment of cells was done on phosphate-buffered saline-washed cell culture plates in a UV Stratalinker 1800 (Stratagene) at 150 mJ. Brd4-E2 coimmunoprecipitation analysis was performed with extracts of COS-7 cells as depicted in reference 30, with 3 μg of rabbit polyclonal antibody against the Brd4 C terminus (Abgent).
For immunoblotting analysis, proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred by a semidry blotting method to a polyvinylidene difluoride membrane (Millipore Corp.). Membranes were incubated with anti-E2 antibodies 5E11 and 1E4 (12) and with a secondary horseradish peroxidase (HRP)-conjugated antibody (LabAs Ltd., Tartu, Estonia) according to the manufacturer's recommendations. Detection was performed using an ECL detection kit (Amersham Pharmacia Biotech), following the manufacturer's manual.
RESULTS
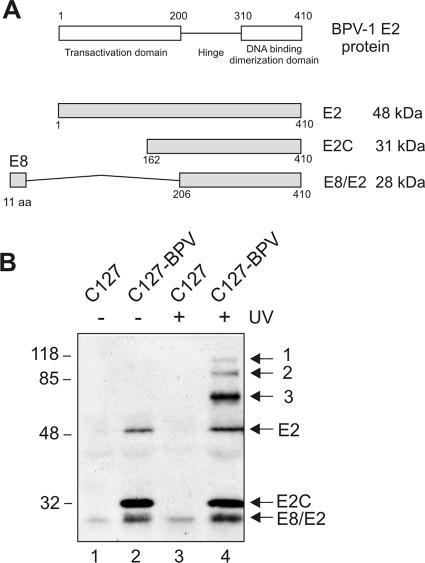
The BPV1 genome encodes three different E2 proteins (Fig. 1A), which share a common C-terminal DBD (20), and the three E2 proteins are able to form homo- and heterodimers through the DBD (19). To confirm the formation of E2 dimers in BPV1-transformed C127 cells, we irradiated the cells with UV light, lysed the cells, and immunoprecipitated the E2 proteins from cell lysates with E2-specific antibodies. UV light produces an interchain cross-link between Trp360 molecules of E2 within a dimer, providing a simple assay for E2 dimerization (21). As shown in Fig. 1B, lane 4, full-length E2 homodimers (96 kDa) (band 1), truncated E2C-E2C (62-kDa) and E8/E2-E8/E2 (56-kDa) homo- and E2C-E8/E2 (59-kDa) heterodimers (band 3), and E2-E2C (79-kDa) and E2-E8/E2 (76-kDa) (band 2) heterodimers formed after UV irradiation of BPV1-transformed C127 cells. As a control, parallel immunoprecipitations of extracts of the parental C127 cells (Fig. 1B, lanes 1 and 3) and C127-BPV cells without the UV cross-link (Fig. 1B, lane 2) were also carried out. The relative ratio of E2 proteins in the BPV1-transformed cells was 1:5:1.5 for E2-E2C-E8/E2; however, it has been shown to change during the cell cycle (29). Despite the low abundance of E2 proteins in transformed cells, we tried to estimate the ratios for different forms within the cells. The relative ratio of E2 dimers was 1:4:24 for bands 1, 2, and 3, but we suggest that the ratio of E2 homo- and heterodimers is also changing during the cell cycle. Regardless of that, it is clear that the amounts of E2-E2C and E2-E8/E2 heterodimers within the cells are always larger than or at least equal to the amount of full-length E2 homodimers, suggesting therefore that E2 heterodimers are the preferential form for E2 protein in infected cells.
FIG. 1.
E2 proteins in BPV1-transformed cells. (A) Schematic representation of E2 proteins. (B) Western blot analysis of proteins immunoprecipitated by E2-specific polyclonal antibody from C127 and BPV1-transformed C127 cells. UV indicates cells that were cross-linked at 150 mJ before immunoprecipitation. Locations of the three E2 proteins and the full-length E2 homodimers (band 1); the truncated E2C-E2C, E8/E2-E8/E2 homo-, and E2C-E8/E2 heterodimers (band 3); and the E2-E2C and E2-E8/E2 heterodimers (band 2) are indicated on the right. Proteins were subjected to SDS-PAGE and immunoblotted with E2-specific antibodies 1E4 and 1H10.
Construction of tethered E2 heterodimers.
In order to study the functions of E2 heterodimers containing only one activation domain, we constructed single-chain heterodimers as described in references 4 and 23. We fused the coding regions of the C-terminal DBD of E2 starting from amino acids 284, 310, and 325 in frame to the open reading frame of the full-length E2 protein (Fig. 2A). As a linker, we added a synthetic oligonucleotide encoding a glycine-rich polypeptide tether of 14 amino acids to E2 fusion proteins to guarantee the formation of correctly folded dimers. Glycine residues were used to maximize flexibility and serine residues to increase hydrophilicity. Recent studies have shown that intramolecular folding could be achieved as soon as the linker was of sufficient length and flexibility (4, 23). Theoretically, the local concentration of two proximal DBDs is always higher than the concentration of DBDs for different polypeptides, and we suggest that the formation of the intramolecular dimer is strongly preferred compared to that of the intermolecular dimer. To test whether the resulting molecules formed single-chain E2 dimers by forced dimerization, pCG plasmids coding the sequences of E2, E2∼DBD(284), E2∼DBD(310), and E2∼DBD(325) (a tilde indicates a tethered dimer, and the numbers in parenthesis show the starting amino acid positions for the DBD) as well as of E2∼Gly14DBD(284), E2∼Gly14DBD(310), and E2∼Gly14DBD(325) (Gly14 indicates the tether) were transfected into the COS-7 cells and analyzed by immunoblotting with E2-specific antibodies. As shown in Fig. 2B, all expressed proteins migrated at the expected sizes. UV irradiation of transfected COS-7 cells showed the formation of dimers in the case of full-length E2 (Fig. 2D, lane 2) and E2C (Fig. 2D, lane 3) proteins but not in the case of E2∼DBD(310) and E2∼Gly14DBD(310) (Fig. 2D, lanes 4 and 5) proteins. These data suggest that in the case of single-chain E2∼DBD(310) and E2∼Gly14DBD(310) proteins, only the intramolecular dimer is formed.
Next, we performed a gel shift assay (electrophoretic mobility shift assay) with the lysates of transfected COS-7 cells by using the E2 binding site E2BS9 as a probe (2). The mobilities of the full-length E2 protein and the DBD of E2 are shown in Fig. 2C, lanes 2 and 9, respectively. E2 single-chain heterodimers E2∼Gly14DBD(284) and E2∼Gly14DBD(310) bound E2BS with a mobility shift similar to that of a mixture of the full-length E2 protein and the DBD of E2 (Fig. 2C, lanes 4, 6, and 10). However, fusion proteins E2∼Gly14DBD(325), E2∼DBD(284), E2∼DBD(310), and E2∼DBD(325) showed mobilities slightly different from those of the mixture of separate proteins (Fig. 2C, compare lanes 3, 5, 7, and 8 with lane 10). In fact, all single-chain E2 heterodimers bind DNA sequence specifically, demonstrating the maintenance of specific DNA recognition after intramolecular dimerization. These results strongly suggest that the single-chain E2 heterodimers folded correctly to form genuine pseudodimers functional in DNA binding.
The activity of single-chain E2 heterodimers in the replication function.
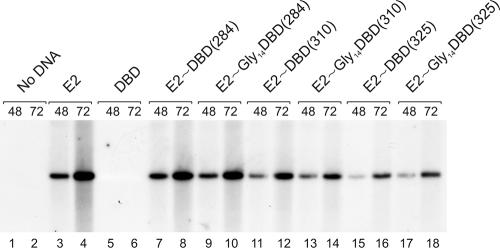
The E2 protein is able to support papillomavirus DNA replication, and first, we tested the ability of single-chain E2 heterodimers to support the replication of BPV1 origin-containing plasmids in a transient replication assay. The expression plasmid for the single-chain E2 protein was cotransfected with the origin-containing plasmid pUCAlu and E1 expression vector pCGEag into CHO cells by electroporation. Episomal DNA was extracted, purified, and digested with linearizing enzyme and DpnI, which digests only unreplicated input DNA, and analyzed by Southern blotting as described earlier (26). As shown in Fig. 3, the single-chain E2 heterodimers were able to support the replication of BPV1 origin-containing plasmid pUCAlu. However, the replication signal was dependent on the length of the added sequence; E2 single-chain heterodimers E2∼DBD(284) and E2∼Gly14DBD(284) were able to support the DNA replication on the wild-type level, but E2∼DBD(325) and E2∼Gly14DBD(325) were less efficient (Fig. 3, compare lanes 7 to 10 with lanes 15 to 18). Single-chain heterodimers E2∼DBD(310) and E2∼Gly14DBD(310) gave an intermediate result (Fig. 3, lanes 11 to 14). Despite these variations, we conclude that single-chain E2 heterodimers are activators of papillomavirus DNA replication in vivo.
FIG. 3.
The abilities of single-chain E2 proteins to support DNA replication of BPV1 origin-containing plasmid pUCAlu in CHO cells. Cells were transfected with 100 ng of pUCAlu, 500 ng of pCGEag, and 250 ng of pCG vector expressing E2, the DBD, or single-chain E2 heterodimers and harvested either 48 or 72 h after electroporation. Episomal DNA was isolated, digested with DpnI and linearizing enzyme, and analyzed by Southern blotting using the radiolabeled probe specific to the ori reporter plasmid.
The activity of the single-chain E2 heterodimer in the transcription function.
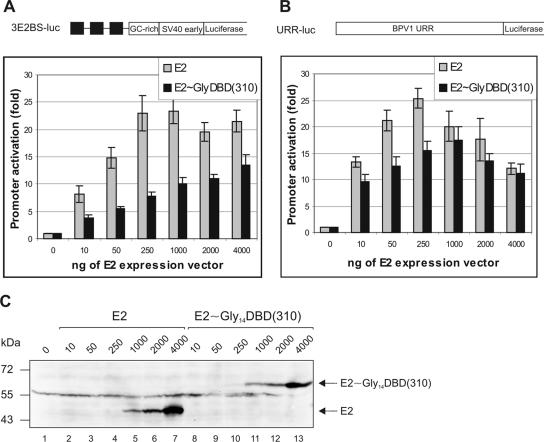
The E2 protein is a transactivator, and next, we analyzed the ability of the E2 heterodimer with one activation domain to activate transcription. In this assay, we used the E2 single-chain heterodimer E2∼Gly14DBD(310), because the DNA binding domain consisting of 101 residues (aa 310 to 410) is more stable than the DBD of 85 residues (aa 326 to 410) and the additional 26 residues of the hinge do not add any stability to the DBD (28). The transactivation function of the single-chain E2 heterodimer was tested by the Dual-Luciferase reporter assay system (Promega) using two different E2-dependent reporter plasmids (Fig. 4). The E2-specific reporters were cotransfected with pCG plasmid encoding E2 or its derivative and with pRL-TK, an E2-nonspecific reporter, into CHO cells, and the luciferase activities were measured. The data for E2-responsive firefly luciferase expression were normalized to the data for E2-nonresponsive Renilla luciferase expression, and the results are presented relative to basal activity levels (no E2 protein). First, CHO cells were transfected with E2-responsive reporter plasmid 3E2BS-Luc, which contains three E2 binding sites, and increasing amounts of E2 expression vectors (Fig. 4A). The activation of the promoter by E2 as well as by the E2 heterodimer with one activation domain was dependent on the amount of the input of E2 expression vectors. At the optimal E2 plasmid concentrations (250 ng of E2 expression plasmid), E2 enhanced levels of luciferase expression from the E2-responsive reporter plasmid 3E2BS-Luc 23-fold and E2∼Gly14DBD(310) 7.5-fold. However, the ability of the E2 single-chain heterodimer to activate the E2-responsive reporter plasmid increased at higher concentrations. Figure 4C shows the results for the Western blot analysis of the expression levels of E2 proteins in parallel with those for the luciferase assay. To address whether the E2 heterodimer with one activation domain is also able to activate the papillomavirus natural promoter, the BPV1 URR containing reporter plasmid URR-Luc was transfected with increasing concentrations of E2 expression plasmids into CHO cells. Using 250 ng of E2 expression plasmids, E2 enhanced luciferase expression levels 25-fold and E2∼Gly14DBD(310) 15-fold above basal activity levels (Fig. 4B). At higher E2 concentrations, the abilities of both E2 proteins to activate transcription from reporter plasmid URR-Luc were comparable and both proteins caused a reduction in transactivation. E2 single-chain heterodimer E2∼Gly14DBD(284) gave similar results, and E2 repressor protein E2C as well as E2DBD was not able to activate E2-responsive promoters (data not shown). From these results, we conclude that the E2 heterodimer with one activation domain functions as a transcriptional activator; however, it is a weaker transactivator than the E2 dimer with two activation domains. At optimal E2 concentrations, the E2 heterodimer with one activation domain is able to activate transcription from a synthetic promoter, which contains three E2 binding sites, to one-third of the wild-type protein activity and from the authentic BPV1 promoter to 60% of the E2 activity.
FIG. 4.
Transcription activation assay. CHO cells were transiently transfected with expression vectors for E2 or E2 single-chain heterodimer E2∼Gly14DBD(310) and the E2-responsive reporter plasmid 3E2BS-Luc (A) or BPV1-native promoter URR-luc (B) and were analyzed for luciferase activity. Luciferase activity levels were normalized to those for Renilla luciferase expression from the non-E2-responsive thymidine kinase promoter of cotransfected plasmid pRL-TK. The data are presented as promoter activities relative to those for the reporter alone and are averages of results from three independent transfections. The schematic diagrams of E2-responsive reporter plasmids used are indicated above the diagram. Solid boxes, E2 binding sites. (C) Western blot analysis of transfected cells. CHO cells were transfected with increasing concentrations of E2 expression plasmids and the E2-responsive reporter plasmid 3E2BS-Luc. Equal amounts of the total protein were subjected to immunoblotting with E2-specific antibodies. Locations of E2 and the single-chain E2 heterodimer are indicated on the right; the middle band (∼55 kDa) corresponds to nonspecific binding.
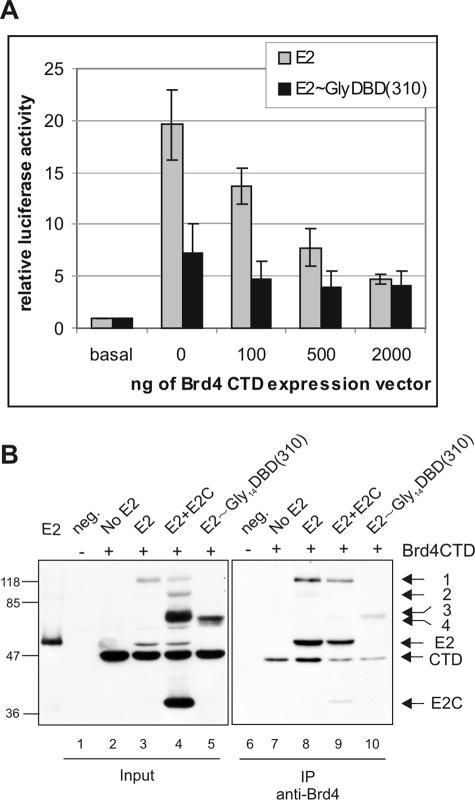
The Brd4 CTD inhibits E2 and single-chain E2 heterodimer transcriptional activation.
The cellular bromodomain protein Brd4 is one of the many cellular proteins involved in E2 transcriptional activation, and the E2-activated transcription is efficiently downregulated by the CTD of Brd4 (11, 22). We next tested whether the Brd4 CTD could also affect single-chain E2 heterodimer transcriptional activation. For this, CHO cells were transfected with E2-responsive reporter plasmid 3E2BS-Luc, E2-nonresponsive reporter pRL-TK, the E2 or E2∼Gly14DBD(310) expression plasmids, and increasing amounts of a Brd4 CTD expression plasmid. The data for E2-responsive firefly luciferase expression were normalized to the data for E2-nonresponsive Renilla luciferase expression, and the results are presented relative to those for the reporter alone. As shown in Fig. 5A, the expression of the Brd4 CTD inhibited the transcriptional activation function of E2 in a dose-dependent manner, resulting in a decrease in E2-activated transcription levels from 20-fold to 4.5-fold. E2 single-chain heterodimer-activated transcription levels decreased from sevenfold to fourfold, and this decrease was achieved already at the lowest Brd4 CTD concentration used. These results suggest that the E2 homodimer is more sensitive to Brd4 CTD overexpression than the E2 single-chain heterodimer because the Brd4 CTD was able to compete Brd4 out from the heterodimer complex at lower concentrations, compared to what was found for the E2 homodimer. Although E2 retained 20% of its initial activity and the single-chain E2 heterodimer retained 60% independently of Brd4 CTD expression level, both proteins were still able to activate transcription at levels fourfold above the basal level.
FIG. 5.
(A) Role of Brd4 in E2 transcriptional activation. CHO cells were transiently transfected with 200 ng of expression vectors for E2 or E2 single-chain heterodimer E2∼Gly14DBD(310) and the E2-responsive reporter plasmid 3E2BS-Luc, pRL-TK, and increasing concentrations of Brd4 CTD expression vector. Luciferase activity levels were measured and normalized to those for Renilla luciferase expression, and the data are presented as promoter activity levels relative to those for the reporter alone. The data represent the averages for three experiments. (B) Brd4 interaction with E2 dimers. Brd4 CTD and E2 proteins were expressed in COS-7 cells, and the Brd4 CTD was immunoprecipitated from UV-cross-linked cell extracts with anti-Brd4 antibody. Shown is an autoradiograph of SDS-PAGE analysis of cell extracts (lanes 1 to 5) and the precipitated proteins (lanes 6 to 10). Locations of E2 proteins, the Brd4 CTD, and full-length E2 homodimers (band 1), E2-E2C heterodimers (band 2), E2C homodimers (band 3), and single-chain E2 heterodimer E2∼Gly14DBD(310) (band 4) are indicated on the right. Membrane was immunoblotted with HRP-conjugated E2-specific antibody 5E11, which recognizes also the E2Tag in front of the Brd4 CTD. neg., empty vector.
The Brd4 CTD is able to interact with the full-length E2 protein but not with E2C, which is missing the transactivation domain (30). In order to test whether Brd4 could also interact with the E2 heterodimer with one activation domain, we cotransfected the COS-7 cells with expression plasmids for the Brd4 CTD and E2 proteins, immunoprecipitated E2 proteins from UV-cross-linked COS-7 cells with anti-Brd4 antibody, and analyzed by immunoblotting. We could follow the expression of the Brd4 CTD together with that of the E2 proteins, as the Brd4 CTD was N-terminally epitope tagged with E2Tag from BPV1 E2, and the HRP-conjugated 5E11 antibody used in this assay recognized all the expressed proteins. As shown in Fig. 5B, the Brd4 CTD efficiently immunoprecipitated the full-length E2 protein as well as the E2 homodimer (Fig. 5B, lane 8) from UV-treated cells. Furthermore, the Brd4 CTD was able to preferentially immunoprecipitate the full-length E2 homodimer (band 1) but not the E2C homodimer (band 3) or the E2-E2C heterodimer (band 2) from the mixture of E2 dimers formed after coexpression of full-length E2 and E2C proteins in COS-7 cells (Fig. 5B, lane 9). We also expressed the Brd4 CTD together with single-chain E2 heterodimer E2∼Gly14DBD(310) in COS-7 cells; antibodies against Brd4 immunoprecipitated this protein very poorly (Fig. 5B, lane 10). These data show that Brd4 interacts efficiently with the E2 homodimer and with low affinity with the E2 heterodimer with one activation domain.
Taken together, these data suggest that the Brd4 CTD efficiently downregulates the E2 heterodimer-activated transcription because Brd4 interacts poorly with the E2 heterodimer with one activation domain. The fact that the Brd4 CTD suppresses the E2 homodimer- and single-chain heterodimer-activated transcriptions to the same level (fourfold above the basal level) suggests that the E2 protein can form two transcription complexes. In one complex, strong activation of transcription is achieved as a result of Brd4, and a second complex lacks Brd4, giving activation levels fourfold above the basal level. This allows us to conclude that by use of the reporter plasmid with three E2 binding sites, approximately 80% of the E2 transcriptional activity is mediated directly by Brd4 and the remaining 20% is Brd4-independent transcription, which is mediated by other cellular transcription factors.
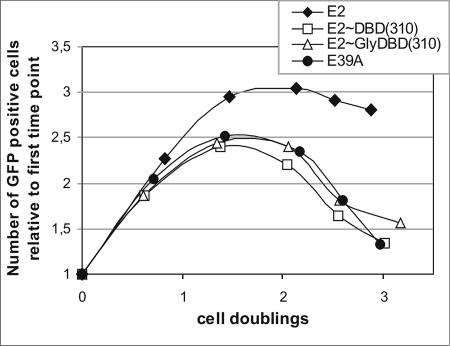
The activity of single-chain E2 heterodimers in the plasmid partitioning function.
We also analyzed the partitioning function of E2 heterodimers in a functional-partitioning assay developed in our laboratory (1). The Jurkat cells were transfected with pRet plasmid containing the destabilized EGFP, 10 E2 binding sites, and the encoding sequence for the full-length E2 protein, E2 mutant E39A, or single-chain E2 heterodimer E2∼DBD(310) or E2∼Gly14DBD(310). The number of EGFP-positive cells was determined by fluorescence-activated cell sorter analysis during 1 week after transfection and plotted against the number of cell doublings. As shown in Fig. 6, the full-length E2 protein supported the partitioning of pRet plasmid containing the E2 binding sites, but E2 mutant E39A did not, as described previously (1). The single-chain E2 heterodimers E2∼DBD(310) and E2∼Gly14DBD(310) were not able to support the partitioning of the E2 binding site containing pRet plasmid (Fig. 6). From these data, we suggest that the E2 heterodimers are crippled in the segregation function. The partitioning of the BPV1 genome is dependent on the effective chromatin attachment mediated by E2 and multiple E2 binding sites (1). The cellular protein Brd4 is also the tethering factor involved in the mitotic association of BPV1 genomes (30), and Baxter et al. have shown that the ability of E2 to bind mitotic chromosomes correlates with the interaction with Brd4 (7). However, Brd4 is not able to efficiently interact with the E2 heterodimer with one activation domain (Fig. 5B), suggesting that the defect of single-chained E2 heterodimers in the plasmid partitioning function is probably caused by weakening of the Brd4-E2 interaction.
FIG. 6.
Plasmid partitioning assay. Jurkat cells were transfected with pRetE2 plasmids containing E2 binding sites and expression cassettes for E2 or its derivatives and destabilized EGFP. The total number of cells and the number of EGFP-positive cells per milliliter were determined by fluorescence-activated cell sorter analysis at different time points. From these data, the numbers of cell doublings and of EGFP-positive cells relative to those for the first time point (20 h posttransfection) were calculated.
DISCUSSION
In this study, we show that E2 heterodimers are functional in the activation of E2-dependent transcription and replication; however, they are inactive in supporting the partitioning of the E2 binding site containing plasmids in dividing cells.
Three E2 proteins encoded by the BPV1 genome are able to form homo- and heterodimers through their common DBD (10, 19), resulting in the formation of the mixture of E2 homo- and heterodimers, which are all capable of DNA binding and, furthermore, compete for binding to the viral DNA (18). All E2 activities are dependent upon E2 binding sites in BPV or vector DNA, and competition for DNA binding sites between the short and long forms of E2 dimers underlies repression in BPV1 DNA replication and transcription control (5, 18). To study the functions of E2 dimers composed of full-length E2 and E2 repressors, we have to eliminate the competitive binding of other E2 dimers to DNA in order to investigate directly the biological functions of E2 heterodimers with only one activation domain. To overcome these difficulties, we have used a tethering strategy. Inspired by successful examples in the literature using other dimeric molecules (4, 23), we joined the full-length E2 protein and E2 DBD by a flexible polypeptide so that they formed an intramolecular dimer. The DNA binding properties of single-chain E2 proteins suggest that tethered proteins fold into active pseudodimers and that such linked molecules behave like natural dimers in sequence-specific DNA binding. It is possible that two tethered molecules might associate with each other to generate a heterodimer with two dimeric DBDs, or alternatively, linker proteolysis could produce a functional E2 homodimer from the tethered construct. However, we could not detect such proteins in any assay used, suggesting that tethered constructs fold into the heterodimer with one activation domain rather than forming an intermolecular dimer with two dimeric DBDs or dimers of proteolytic products.
The E2 heterodimers are able to support the replication of BPV1 origin-containing plasmids (Fig. 3). These data are consistent with observations by Lim et al. (18), who showed that heterodimers composed of BPV1 E2 and E2C were able to load helicase E1 to its cognate site and to activate DNA replication in a cell-free system. Our data show that E2 heterodimers are activators of replication in vivo, which suggests that E2 heterodimers are able to work on chromatin templates.
In this study, we show that the E2 heterodimer with one activation domain is a transactivator in vivo. However, it is a weaker transactivator than the full-length E2 homodimer. The E2 heterodimer with one activation domain is able to activate transcription from the E2-responsive promoter to one-third of the wild-type protein activity level and from the native BPV1 promoter to two-thirds of the E2 activity level. Previous studies have suggested that E2-E2C heterodimers could be repressors of transcription, as cotransfection of a vector encoding E2C with the full-length E2 protein resulted in up to 98% repression of E2-dependent reporter gene expression (5). However, cotransfection of E2 and E2C results in the formation of E2- E2 homodimers, E2-E2C heterodimers, and E2C-E2C homodimers which all are capable of DNA binding, and the fourfold excess of the E2C plasmid compared to the full-length E2 plasmid used in the study of Barsoum et al. (5) probably resulted in the formation of a large excess of E2C-E2C homodimers, which are the repressors of transcription. So, the competition for DNA binding between the E2C repressor protein and E2 heterodimers could hide the transactivation seen in our study by using the single-chain E2 heterodimer.
Our data show that the E2 heterodimer with one activation domain activates transcription from the E2-responsive promoter by a different mechanism than the E2 homodimer. E2-activated transcription is mediated mostly by cellular bromodomain protein Brd4 and is sensitive to Brd4 CTD overexpression (11, 22). At the same time, E2 heterodimer-activated transcription is downregulated by the Brd4 CTD to the same level as the E2 homodimer-activated transcription (Fig. 5). This is consistent with coimmunoprecipitation studies indicating that the E2 heterodimer with one activation domain interacts weakly with Brd4 and was therefore efficiently competed out at low Brd4 CTD concentrations. The E2 heterodimer has lost one-third of its initial activity after coexpression with the Brd4 CTD. One possibility is that the E2 heterodimer is still able to interact with Brd4, but this interaction is much weaker and therefore more unstable than the E2-Brd4 interaction. The other explanation is that in addition to the direct participation of Brd4 in E2-activated transcription, Brd4 is also involved indirectly, through interactions with other cellular transcription factors participating in E2 transactivation. In this case, Brd4 mediates E2 transcriptional activation by two different mechanisms, directly, through direct interaction with E2, and indirectly, through interactions with other cellular proteins interacting with E2.
E2 heterodimers with one activation domain are able to bind one (Fig. 2C) and two (data not shown) E2 binding sites as efficiently as full-length E2. In transcription, the E2 binding sites function synergistically in vivo; a very weak activation of transcription has been observed with a single binding site for E2, while increasing the number of binding sites to two or three enhances the transactivation (25). The differences in transcriptional activation occur after DNA binding; the single-chain E2 homodimer is either not able to modify the chromatin or not able to recruit the transcription initiation complex (16). This may also suggest that two transactivation domains are required in each dimer for interactions with some components of transcription machinery. For example, cellular protein Brd4 interacts preferentially with the full-length E2 homodimer, suggesting that this interaction requires two activation domains in each dimer. Recent crystallographic data support these findings by showing that a tetrameric structure consisting of a twofold symmetric dimer from an E2 dimer/Brd4 monomer complex is formed within the unit cell (E. A. Abbate and M. R. Botchan, personal communication).
Genetic studies have shown that the E2 repressor of HPV 31 and at least one of the BPV1 repressors are required for the long-term maintenance of viral episomes (15, 17, 24), demonstrating the important role of E2 repressors in the viral life cycle. In the steady state, there is an excess of repressor forms of the E2 activator protein throughout the cell cycle. The E2 repressor forms act at two levels; first, they compete with full-length E2 for binding to the viral DNA, and second, they form heterodimers. Our data show that the E2 heterodimer can initiate DNA replication but is crippled in the segregation of viral genomes. In addition, the E2 heterodimer is able to activate transcription from BPV1-native promoters; it activates transcription from E2-responsive promoters by a different mechanism than the E2 homodimer. Thus, there are two different E2 transactivator proteins in the virus-transformed cells, whose capacities for activation of BPV1-native promoters are comparable if we take into account the amounts of E2 homo- and heterodimers in the cells (ratio, 1:3). We suggest that the formation of heterodimers gives E2 additional opportunities for regulation of viral gene expression. These data also show that the E2 heterodimer is an integral part of the regulatory network of E2 activities. For all E2 activities, the full-length E2 homodimer is essential only for segregation of viral genomes; the replication and transcription activities are mediated also by the E2 heterodimer. Furthermore, our data suggest that E2 functions as a heterodimer and the E2 heterodimer is the main transactivator of BPV1. In addition, it will be easy to regulate the level of E2 dimers through the ratio of full-length E2 and E2 repressors and the formation of heterodimers during the cell cycle as well as during differentiation of epithelial cells.
Acknowledgments
We thank Marc Piechaczyk for the initial idea to use forced dimerization.
This study was supported in part by grant ETF5994 to R.K. and ETF6903 to A.A. from the Estonian Science Foundation and by long-term research program SF0182566 from the Estonian Ministry of Education and Science and grant INTNL 55000339 from the Howard Hughes Medical Institute to M.U.
Footnotes
Published ahead of print on 30 August 2006.
REFERENCES
- 1.Abroi, A., I. Ilves, S. Kivi, and M. Ustav. 2004. Analysis of chromatin attachment and partitioning functions of the bovine papillomavirus type 1 E2 protein. J. Virol. 78:2100-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abroi, A., R. Kurg, and M. Ustav. 1996. Transcriptional and replicational activation functions in the bovine papillomavirus type 1 E2 protein are encoded by different structural determinants. J. Virol. 70:6169-6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Androphy, E., D. Lowy, and J. Schiller. 1987. Bovine papillomavirus E2 trans-activating gene product binds to specific sites in papillomavirus DNA. Nature 325:70-73. [DOI] [PubMed] [Google Scholar]
- 4.Bakiri, L., K. Matsuo, M. Wisniewska, E. F. Wagner, and M. Yaniv. 2002. Promoter specificity and biological activity of tethered AP-1 dimers. Mol. Cell. Biol. 22:4952-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barsoum, J., S. S. Prakash, P. Han, and E. J. Androphy. 1992. Mechanism of action of the papillomavirus E2 repressor: repression in the absence of DNA binding. J. Virol. 66:3941-3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastien, N., and A. McBride. 2000. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology 270:124-134. [DOI] [PubMed] [Google Scholar]
- 7.Baxter, M. K., M. G. McPhillips, K. Ozato, and A. A. McBride. 2005. The mitotic chromosome binding activity of the papillomavirus E2 protein correlates with interaction with the cellular chromosomal protein, Brd4. J. Virol. 79:4806-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang, C., T. Broker, and L. Chow. 1991. An E1M-E2C fusion protein encoded by human papillomavirus type 11 is a sequence-specific transcription repressor. J. Virol. 65:3317-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doorbar, J., A. Parton, K. Hartley, L. Banks, T. Crook, M. Stanley, and L. Crawford. 1990. Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology 178:254-262. [DOI] [PubMed] [Google Scholar]
- 10.Hubbert, N., J. Schiller, D. Lowy, and E. Androphy. 1988. Bovine papilloma virus-transformed cells contain multiple E2 proteins. Proc. Natl. Acad. Sci. USA 85:5864-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ilves, I., K. Maemets, T. Silla, K. Janikson, and M. Ustav. 2006. Brd4 is involved in multiple processes of the bovine papillomavirus type 1 life cycle. J. Virol. 80:3660-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurg, R., J. Parik, E. Juronen, T. Sedman, A. Abroi, I. Liiv, U. Langel, and M. Ustav. 1999. Effect of bovine papillomavirus E2 protein-specific monoclonal antibodies on papillomavirus DNA replication. J. Virol. 73:4670-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurg, R., K. Sild, A. Ilves, M. Sepp, and M. Ustav. 2005. Association of bovine papillomavirus E2 protein with nuclear structures in vivo. J. Virol. 79:10528-10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambert, P., N. Hubbert, P. Howley, and J. Schiller. 1989. Genetic assignment of multiple E2 gene products in bovine papillomavirus-transformed cells. J. Virol. 63:3151-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambert, P., B. Monk, and P. Howley. 1990. Phenotypic analysis of bovine papillomavirus type 1 E2 repressor mutants. J. Virol. 64:950-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lefebvre, O., G. Steger, and M. Yaniv. 1997. Synergistic transcriptional-activation by the papillomavirus E2 protein occurs after DNA binding and correlates with a change in chromatin structure. J. Mol. Biol. 266:465-478. [DOI] [PubMed] [Google Scholar]
- 17.Lehman, C., D. King, and M. Botchan. 1997. A papillomavirus E2 phosphorylation mutant exhibits normal transient replication and transcription but is defective in transformation and plasmid retention. J. Virol. 71:3652-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim, D., M. Gossen, C. Lehman, and M. Botchan. 1998. Competition for DNA binding sites between the short and long forms of E2 dimers underlies repression in bovine papillomavirus type 1 DNA replication control. J. Virol. 72:1931-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBride, A., J. Byrne, and P. Howley. 1989. E2 polypeptides encoded by bovine papillomavirus type 1 form dimers through the common carboxyl-terminal domain: transactivation is mediated by the conserved amino-terminal domain. Proc. Natl. Acad. Sci. USA 86:510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McBride, A., R. Schlegel, and P. Howley. 1988. The carboxy-terminal domain shared by the bovine papillomavirus E2 transactivator and repressor proteins contains a specific DNA binding activity. EMBO J. 7:533-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prakash, S. S., S. R. Grossman, R. B. Pepinsky, L. A. Laimins, and E. J. Androphy. 1992. Amino acids necessary for DNA contact and dimerization imply novel motifs in the papillomavirus E2 trans-activator. Genes Dev. 6:105-116. [DOI] [PubMed] [Google Scholar]
- 22.Schweiger, M. R., J. You, and P. M. Howley. 2006. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J. Virol. 80:4276-4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sieber, M., and R. K. Allemann. 2000. Thermodynamics of DNA binding of MM17, a ‘single chain dimer’ of transcription factor MASH-1. Nucleic Acids Res. 28:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stubenrauch, F., M. Hummel, T. Iftner, and L. A. Laimins. 2000. The E8E2C protein, a negative regulator of viral transcription and replication, is required for extrachromosomal maintenance of human papillomavirus type 31 in keratinocytes. J. Virol. 74:1178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thierry, F., N. Dostatni, F. Arnos, and M. Yaniv. 1990. Cooperative activation of transcription by bovine papillomavirus type 1 E2 can occur over a large distance. Mol. Cell. Biol. 10:4431-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ustav, M., E. Ustav, P. Szymanski, and A. Stenlund. 1991. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 10:4321-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veeraraghavan, S., C. C. Mello, E. J. Androphy, and J. D. Baleja. 1999. Structural correlates for enhanced stability in the E2 DNA-binding domain from bovine papillomavirus. Biochemistry 38:16115-16124. [DOI] [PubMed] [Google Scholar]
- 29.Yang, L., I. Mohr, R. Li, T. Nottoli, S. Sun, and M. Botchan. 1991. Transcription factor E2 regulates BPV-1 DNA replication in vitro by direct protein-protein interaction. Cold Spring Harbor Symp. Quant. Biol. 56:335-346. [DOI] [PubMed] [Google Scholar]
- 30.You, J., J. Croyle, A. Nishimura, K. Ozato, and P. Howley. 2004. Interaction of the bovine papillomavirus E2 protein with brd4 tethers the viral DNA to host mitotic chromosomes. Cell 117:349-360. [DOI] [PubMed] [Google Scholar]