Abstract
Hepatitis C virus (HCV) entry is dependent on CD81. To investigate whether the CD81 sequence is a determinant of HCV host range, we expressed a panel of diverse CD81 proteins and tested their ability to interact with HCV. CD81 large extracellular loop (LEL) sequences were expressed as recombinant proteins; the human and, to a low level, the African green monkey sequences bound soluble HCV E2 (sE2) and inhibited infection by retrovirus pseudotype particles bearing HCV glycoproteins (HCVpp). In contrast, mouse or rat CD81 proteins failed to bind sE2 or to inhibit HCVpp infection. However, CD81 proteins from all species, when expressed in HepG2 cells, conferred susceptibility to infection by HCVpp and cell culture-grown HCV to various levels, with the rat sequence being the least efficient. Recombinant human CD81 LEL inhibited HCVpp infectivity only if present during the virus-cell incubation, consistent with a role for CD81 after virus attachment. Amino acid changes that abrogate sE2 binding (I182F, N184Y, and F186S, alone or in combination) were introduced into human CD81. All three amino acid changes in human CD81 resulted in a molecule that still supported HCVpp infection, albeit with reduced efficiency. In summary, there is a remarkable plasticity in the range of CD81 sequences that can support HCV entry, suggesting that CD81 polymorphism may contribute to, but alone does not define, the HCV susceptibility of a species. In addition, the capacity to support viral entry is only partially reflected by assays measuring sE2 interaction with recombinant or full-length CD81 proteins.
Hepatitis C virus (HCV) is an enveloped, positive-strand RNA virus and is the sole member of the Hepacivirus genus of the family Flaviviridae. An estimated 170 million individuals worldwide are infected with HCV and are at risk for the development of chronic liver disease and hepatocellular carcinoma. Animal models that support HCV infection are limited to the chimpanzee, an endangered species available in limited numbers, and immunodeficient mice with transplanted human hepatocytes.
HCV encodes two envelope glycoproteins, E1 and E2, which confer liver cell specificity to retroviral pseudotype particles (HCVpp) (3, 9, 18). The availability of HCVpp enabled functional studies of the glycoproteins (4, 10, 12, 18, 33), measurement of HCV-specific neutralizing antibodies (2, 26, 31, 47), and elucidation of cellular receptors required for viral entry (3, 4, 7, 29, 49). The recent discovery that the JFH strain of HCV is able to replicate and release infectious virus particles in cell culture (HCVcc) allows comparative studies on the entry processes of HCVcc and HCVpp (25, 46, 50).
Soluble, truncated forms of E2 (sE2) bind to human cells and were used to identify interactions with several cell surface molecules, including CD81 (35), scavenger receptor class B type I (SR-BI) (40), and dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN) (14, 27, 37).
CD81 is a member of the tetraspanin superfamily, possessing short intracellular N and C termini, four transmembrane domains, a small extracellular loop (SEL), and a large extracellular loop (LEL) (23, 24). The critical role of CD81 in HCV entry was demonstrated by experiments showing that HCVpp infection of primary human hepatocytes or hepatoma cell lines was inhibited by CD81-specific monoclonal antibodies (MAbs) or recombinant CD81 protein (3, 18), that infection of the Huh-7 hepatoma cell line was inhibited when CD81 expression was silenced by specific small interfering RNAs, and that CD81-negative HepG2 cells supported HCVpp infection when transduced to express CD81 (4, 49). Similar approaches confirmed that HCVcc infection is also dependent on CD81 expression (25, 46, 50). The LEL of CD81 was shown to bind sE2 (13, 35), and characterization of chimeric CD9/CD81 molecules confirmed that the LEL sequence is a determinant of HCVpp entry (49). We previously reported that sE2 binds to human but not to mouse, rat, or African green monkey (AGM) CD81 proteins, suggesting that CD81 may be a determinant of the species restriction of HCV infection (13, 15). However, CD81 cannot be the sole determinant, as transgenic mice expressing human CD81 fail to support HCV infection (28). In addition, tamarin CD81 has been reported to bind sE2, but tamarins are not susceptible to infection by HCV (1, 30).
SR-BI is a member of the scavenger receptor family and binds sE2 (40). Silencing of SR-BI expression and treatment of target cells with anti-SR-BI antibodies inhibited HCVpp infection, suggesting a role for SR-BI during HCV infection (4, 22). However, multiple cell lines expressing both SR-BI and CD81 fail to support HCVpp infection, indicating that additional factors are required for HCV entry (4, 18).
The recently reported HCVpp and HCVcc systems permit studies of the complete HCV entry mechanism, as opposed to the binding assays with recombinant proteins that were previously possible. Thus, the objectives of our study were to use HCVpp and HCVcc to (i) determine to what extent CD81 defines species restriction of HCV infection; (ii) assess whether assays using recombinant, truncated sE2 or CD81 LEL proteins predict the capacity of a CD81 sequence to support viral infection; and (iii) identify residues within CD81 that are critical for its receptor function. We expressed and characterized a panel of CD81 proteins, both as recombinant LEL fusion proteins and as full-length molecules in HepG2 cells, for their interaction with HCV. Our data show that CD81 derived from a broad range of species can support HCVpp and HCVcc infection and suggest that recombinant proteins are poor mimics for interactions between cellular CD81 and HCV. These findings have implications for the development of small animal models supporting HCV infection and for the design of inhibitors targeting CD81-dependent HCV entry.
MATERIALS AND METHODS
Cells and antibodies.
293T, Hep3B, BHK, and CV-1 cells were from the American Type Culture Collection (ATCC) and were propagated according to ATCC recommendations. HepG2 cells (a gift of Yoshiharu Matsuura, Osaka University, Osaka, Japan) were propagated in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). HepG2 cells were propagated on collagen type 1-coated tissue culture vessels. The mouse liver cell line Hepa1-6 was also obtained from the ATCC and propagated according to ATCC recommendations. All cells were grown at 37°C in 5% CO2.
Rat anti-E2 MAbs 6/1a, 7/59, and 7/16 have been described previously (13). Anti-glutathione S-transferase (GST) antibody was purchased from Sigma (St. Louis, MO) and anti-His6 antibody from Invitrogen (Carlsbad, CA). The MAbs 1.3.3.22 (Santa Cruz Biotechnology, Santa Cruz, CA), 1D6 (Serotec, Raleigh, NC) and JS-81 (BD Biosciences, San Jose, CA) against human CD81, and EAT-2 (Santa Cruz Biotechnology) against mouse CD81 were employed in this study. Horseradish peroxidase (HRP)-conjugated anti-mouse antibody was from Pierce (Rockford, IL). Anti-rat and anti-Armenian hamster antibodies were from Jackson ImmunoResearch (West Grove, PA). Alexa 488-conjugated antibodies were from Molecular Probes (Eugene, OR).
Cloning of CD81 cDNAs from different species.
The cDNAs for CD81 from several species were amplified by reverse transcription (RT)-PCR as described previously for the human CD81 sequence (49). RNA from tamarin and chimpanzee peripheral blood mononuclear cells was kindly provided by Robert Lanford (Southwest National Primate Research Center, San Antonio, TX). Rat, hamster, mouse, and AGM RNAs were isolated from the Rat-2, BHK, Hepa1-6, and CV-1 cell lines, respectively. RNA was extracted from between 3 × 106 and 7 × 106 of the appropriate cells by using the RNeasy minikit (QIAGEN, Valencia, CA), and RT was performed using SuperScript II (Invitrogen) according to the manufacturer's instructions with RU-O-4109 (5′-GCG CTC GAG TCA GTA CAC GGA GCT GTT CC-3′; the CD81 stop codon is underlined). PCR products were amplified with oligonucleotide primers RU-O-4104 (5′-CGC GGA TCC GCC ACC ATG GGA GTG GAG GGC TGC ACC-3′; the CD81 start codon is underlined) and RU-O-4109 and the Advantage 2 PCR kit (Clontech, Palo Alto, CA). The resulting PCR products were digested with BamHI and XhoI and were ligated with the similarly digested human immunodeficiency virus (HIV) vector pTRIP-EGFP (48). For expression of GST-CD81 LEL fusion proteins in Escherichia coli, cDNAs encoding residues 112 to 202 of human, AGM, rat, and mouse CD81 were amplified by PCR, using the appropriate pTRIP clones as templates. A C-terminal His6 tag was introduced during PCR through inclusion in the antisense primer. The resulting PCR products were ligated with pGEX-6P (Novagen, Madison, WI) by standard techniques.
Cloning of human-rat and rat-human chimeric CD81 proteins.
Small and large extracellular loop exchanges were made by amplifying the SEL or LEL fragments from 25 ng of the human or rat CD81 expression plasmids, using a proofreading polymerase (Roche Expand). Human or rat SEL was amplified using the primers RU-O-4104 and CD81SEL/LELPst1− (CCA GAT GCC TGC AGC TAC CTC ACA GGC; the silent PstI site is underlined), and the human or rat LEL was amplified using the primers CD81SEL/LELPst1+ (GCC TGT GAG GTA GCT GCA GGC ATC TGG; the silent PstI site is underlined) and RU-O-4109. PCR products were gel purified (QIAGEN Minelute kit), digested with BamHI and PstI (SEL) or PstI and XhoI (LEL), and ligated with gel-purified BamHI/XhoI-digested pTRIP-EGFP. Clones containing the correct fragments were verified by sequencing.
Expression and purification of GST-CD81 LEL fusion proteins.
A single colony of Rosetta-gami Escherichia coli (Novagen), freshly transformed with the appropriate GST-CD81 LEL expression plasmid, or the empty vector was used to seed a starter culture of Luria broth (LB) supplemented with 15 μg/ml kanamycin, 12.5 μg/ml tetracycline, 34 μg/ml chloramphenicol, and 100 μg/ml ampicillin This culture was incubated in an orbital shaker at 250 rpm overnight at 37°C and then diluted 1 to 100 into fresh LB (supplemented with antibiotics as previously) and incubated at 30°C and 250 rpm until an optical density at 600 nm of 0.6 to 0.8 was reached. Expression was induced by the addition of isopropyl-1-thio-β-d-galactopyranoside to a final concentration of 100 μM and the culture incubated overnight at 18°C and 250 rpm. Cells were collected by centrifugation at 6,000 × g for 15 min at 4°C.
For purification of GST-CD81 LEL fusion proteins, the cell pellets were resuspended in lysis buffer (1× phosphate-buffered saline [PBS], 10% glycerol) and lysed by three passages through an Avestin air emulsifier at 15,000 lb/in2. Lysates were clarified at 25,000 × g for 30 min at 4°C, and the clarified extracts were applied to GSTrap FF affinity columns according to the manufacturer's instructions (Amersham Biosciences, Piscataway, NJ). Columns were washed with 5 column volumes of lysis buffer, and GST fusion proteins were eluted with 5 column volumes of elution buffer (100 mM Tris-HCl, pH 8.0; 150 mM KCl; 15 mM reduced glutathione). Fractions of the eluate were collected and analyzed for protein content by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. Peak fractions were pooled, aliquoted, and stored at −80°C. Concentrations of partially purified GST-CD81 LEL fusion proteins were determined using the Bio-Rad protein assay (Bio-Rad, Hercules, CA).
EIA for E2 binding to GST-CD81 LEL.
The enzyme-linked immunoassay (EIA) was described previously (15). Briefly, GST-CD81 LEL fusion proteins were used to coat 96-well plates at 5 μg/ml in PBS overnight at 4°C. Plates were blocked for 1 h at room temperature with PBS supplemented with 5% bovine serum albumin. Supernatant from a culture of 293T cells transfected to express the sE2 protein was used as a source of E2 protein truncated at residue 661, as previously described (13). Serial dilutions of sE2 were allowed to bind at 4°C overnight in antibody diluent (1× PBS, 5% bovine serum albumin, 20% sheep serum, 0.05% Tween 20). After washing, bound sE2 was detected using rat anti-E2 antibodies 6/1a, 7/59, and 7/16, each diluted 1:10 in antibody diluent, for 1 h at room temperature, followed by HRP-conjugated anti-rat immunoglobulin and tetramethylbenzidine substrate. Absorbance was measured at 450 nm.
Generation of retroviral pseudotype particles and infection assay.
HIV pseudotype viruses were generated as previously described (18). Briefly, 293T cells were cotransfected with an HCV E1E2 expression plasmid and pNL4.3.Luc.R−E−, using Lipofectamine 2000 (Invitrogen). Lipid-DNA complexes were removed 4 to 6 h later and replaced with DMEM supplemented with 3% FBS. At 48 h, or 72 h for HCVpp-J6, posttransfection the culture medium was collected, clarified by low-speed centrifugation for 20 min, and then aliquoted and stored at −80°C before use. The particulate p24 content of pseudotype virus preparations was assessed as described previously (12). HCVpp were pelleted through a sucrose cushion, resuspended in PBS supplemented with 1% Empigen, and heat inactivated. The p24 content was quantified by EIA.
HCVpp infectivity was measured as previously described (18). Target cells were seeded at 8 × 103 cells per well of a 96-well plate the day prior to infection. For infection, medium was removed and pseudotype virus, diluted in DMEM supplemented with 3% FBS, was added. After overnight incubation, the inoculum was removed and replaced with DMEM supplemented with 3% FBS. At 72 h postinfection, the medium was removed and the cells lysed with 40 μl of cell lysis buffer (Promega, Madison, WI) per well. Luciferase activity was assayed by the addition of 35 μl of lysate to 50 μl of luciferase substrate and measured for 5 s in a luminometer (Lumat LB 9507). To study inhibition by soluble CD81 LEL, HCVpp was incubated with GST-CD81 LEL fusion protein for 1 h at 37°C prior to addition to the target cells. Alternatively, GST-CD81 LEL was incubated with Huh-7.5 target cells for 1 h at 37°C, and the cells were washed prior to addition of HCVpp.
Transduction of cells to express CD81.
HepG2 cells were transduced to express CD81 or CD9 as previously described (49). Briefly, packaged lentiviruses to express CD81 were generated by cotransfection of 293T cells with a plasmid encoding vesicular stomatitis virus G protein, a plasmid encoding HIV Gag-Pol, and pTRIP-CD81 (1:3:3 ratio). HepG2 cells were seeded at 8 × 105 cells per well of a six-well plate and infected 24 h later with the packaged lentivirus in DMEM supplemented with 3% FBS and 4 μg/ml Polybrene. After 12 h, cells were washed, trypsinized, and seeded into appropriate plates either for pseudotype virus infection or for flow cytometry.
Flow cytometry.
Cell surface CD81 expression was monitored by live cell staining with CD81-specific antibodies and irrelevant species- and isotype-matched antibodies. To detect human, AGM, chimpanzee, and tamarin CD81 sequences, MAbs 1.3.3.22 and JS-81 were used, while MAb EAT-2 was used to detect the mouse and rat CD81 proteins. Binding of sE2 to cells was assayed as previously described (13). Analyses were performed using a FACScalibur flow cytometer (BD Biosciences) and FlowJo software (Tree Star, San Carlos, CA).
HCVcc generation and infection assays.
Infectious J6/JFH particles were generated as previously described (25). Briefly, RNA was transcribed in vitro from a chimeric full-length J6/JFH genome by using the Megascript T7 kit (Ambion, Austin, TX) and electroporated into Huh-7.5 cells. Alternatively, a J6/JFH genome with a Renilla luciferase reporter upstream of core (fl-J6/JFH-5′C19Rluc2AUbi) was used (referred to here as J6/JFH(Rluc2Aubi); this construct is described elsewhere [45]). At 48 h, 96 h, and 8 days after electroporation, supernatants were collected, filtered, pooled, and stored at 4°C. One day before infection, target cells were seeded at 5 × 104 cells per well in 6-well plates for infection with J6/JFH or in 12-well plates for infection with J6/JFH(Rluc2AUbi). For infection, the pooled HCVcc-containing supernatant with HEPES (20 mM) and Polybrene (4 μg/ml) was applied undiluted. at 24 h after infection, the supernatants were replaced with regular growth medium. At 72 h postinoculation, infection was detected by methanol fixation and staining for NS5A antigen, using the anti-NS5A MAb 9E10 as a primary antibody and HRP-conjugated anti-mouse immunoglobulin G (Vector Laboratories, Burlingame, CA) as a secondary antibody. Alternatively, infection with J6/JFH(Rluc.2A.Ubi) was detected 48 or 72 h postinfection by measuring luciferase activity after lysis with 250 μl of Renilla luciferase lysis buffer (Promega) and addition of 100 μl Renilla luciferase substrate (Promega) to 100 μl of lysate.
RESULTS
Interaction of recombinant CD81 proteins with sE2 and HCVpp.
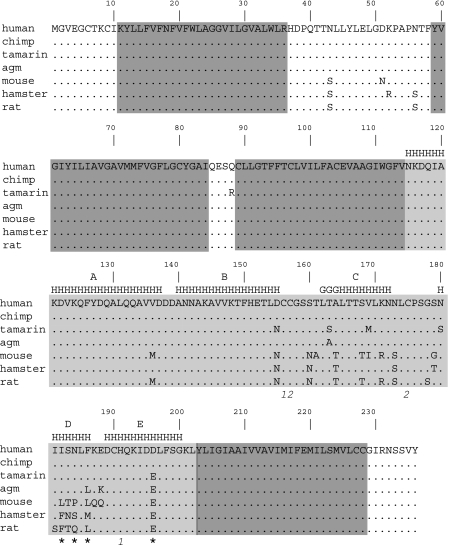
To assess CD81 sequence polymorphism, RNA was extracted from human, chimpanzee, tamarin, AGM, mouse, rat, and hamster cells and CD81 cDNA amplified by RT-PCR. The open reading frames were cloned into the pTRIP lentiviral vector, and the clones were sequenced and aligned (Fig. 1). Among the seven species tested, 26 residues (11% of 236) were polymorphic, and the majority of these residues were located within the LEL (21/89; 24%).
FIG. 1.
Alignment of the inferred CD81 amino acid sequences used in this study. Amino acids that differ from the human sequence are shown. Residues predicted to be included in the four transmembrane domains are indicated by dark shading: TMD1 residues 11 to 36, TMD2 residues 59 to 64, TMD3 residues 89 to 114, and TMD4 residues 203 to 228 (41). The LEL (residues 115 to 202) is indicated by light shading. Letters indicating the secondary structure of the LEL are above the sequences (20) (H, α-helix; G, 310 helix). Cysteine residues connected by disulfide bonds are indicated by numbers below the sequence (1, Cys156-Cys190; 2, Cys157-Cys175). Asterisks below the sequence indicate amino acid positions that, in the context of the complete human CD81 molecule, have previously been shown to be important for the interaction with a truncated E2: I182, N184, F186, and D196. Note that the human and chimpanzee amino acid sequences are identical.
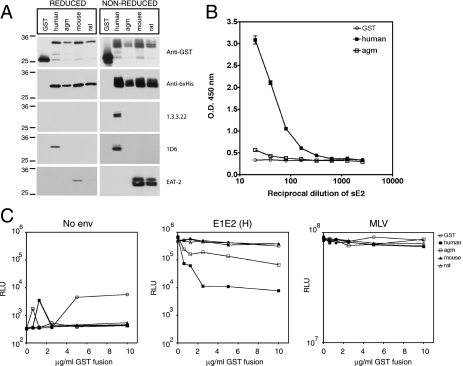
To examine the effect of CD81 polymorphism on sE2 protein binding, several species were selected and expressed as recombinant LEL fusion proteins. The observation that conserved LEL cysteine residues are important for the interaction with sE2 (15, 20, 34) led us to optimize our protocol for expressing CD81 fusion proteins. To enhance native protein folding and conformation, CD81 amino acid residues 112 to 202 were expressed as GST- and His6-tagged proteins in an E. coli strain that favors disulfide bond formation. To assess disulfide bond formation, the proteins were analyzed by reducing and nonreducing SDS-PAGE and detection by anti-CD81 MAbs whose reactivity is dependent upon disulfide bond integrity (Fig. 2A). The proteins were also characterized for their ability to bind antibodies whose recognition is independent of oxidation state (anti-GST, anti-His6, and anti-human CD81 MAb 1D6). After separation under reducing conditions, both anti-GST and anti-His6 antibodies recognized proteins of the expected molecular weights. However, under nonreducing conditions, these antibodies detected a diffusely migrating species containing at least two distinct bands. The differences in migration were most likely due to the formation of disulfide bonds within GST, as a similar pattern was observed when GST alone was expressed. As anticipated, MAb 1D6 recognized GST-human CD81 LEL under both reducing and nonreducing conditions, whereas MAb 1.3.3.22 detected GST- human CD81 LEL only after nonreducing SDS-PAGE. The EAT-2 MAb recognized both mouse and rat LEL proteins, and recognition was decreased following reduction (detection of the GST-rat CD81 LEL protein under reducing conditions was more apparent after longer exposures). These data suggest that correct disulfide bonds were formed in the GST-human, mouse, and rat CD81 LEL fusion proteins, allowing the formation of the 1.3.3.22 and EAT-2 epitopes. Due to the lack of an antibody reactive against the SDS-denatured protein, we were unable to confirm disulfide bond formation in the GST-AGM CD81 LEL fusion protein.
FIG. 2.
Characterization of GST-CD81 LEL fusion proteins and inhibition of HCVpp infectivity. (A) Formation of disulfide bonds in GST-CD81 LEL fusion proteins. Fusion proteins with the human, AGM, mouse, or rat LEL sequence, or GST alone, were isolated on glutathione-Sepharose and separated by reducing or nonreducing SDS-PAGE (12% polyacrylamide gels). Immunoblotting was performed with anti-GST, anti-His6, or anti-CD81 (1.3.3.22, 1D6, or EAT-2) antibodies. The migration of molecular mass markers is indicated (in kilodaltons). The expected molecular mass of the reduced GST-CD81 LEL proteins was ∼33 kDa. (B) Binding of truncated strain H sE2 protein to GST-CD81 LEL fusion proteins. A serial dilution of sE2 protein was allowed to bind to EIA plates coated with GST-CD81 LEL proteins. After washing, bound sE2 was detected using anti-E2 specific MAbs and anti-rat-HRP. Each dilution was tested in triplicate, and the mean absorbance is shown with standard deviation bars (not visible for many points). Absorbance readings obtained with GST, GST-mouse CD81 LEL, and GST-rat CD81 LEL were similar (coefficient of variation, <11%) hence only GST is shown for clarity. O.D., optical density. (C) Inhibition of HCVpp infection by GST-CD81 LEL fusion proteins. Pseudoparticles were generated and the particulate p24 content of the preparations was determined as described in Materials and Methods. Prior to infection of Hep3B cells, 1 ng of particulate p24 of each pseudoparticle preparation (no env, E1E2, or MLV) was incubated at 37°C for 1 h with GST-CD81 LEL fusion proteins. Three days after infection, cells were lysed and luciferase activity determined. All infections were performed in quadruplicate, and the mean luciferase activity is shown. Depicted are results from a representative of three separate experiments. RLU, relative light units.
The CD81 fusion proteins were tested for their ability to bind strain H sE2 by EIA. sE2 bound readily to the human sequence and failed to interact with GST alone or the mouse and rat LEL sequences (Fig. 2B and data not shown). The GST-AGM CD81 LEL bound low levels of sE2, contrary to our previous report (15), which may reflect our modified protocol for expression of fusion proteins with native LEL conformation. To study GST-CD81 LEL interaction with functional E1E2 glycoproteins, the GST fusion proteins were tested for their ability to neutralize HCVpp infectivity. Infection with pseudotypes bearing no viral glycoproteins (no Env; negative control) or the murine leukemia virus (MLV) glycoprotein (MLVpp; positive control) was unaffected by the proteins. GST-human CD81 LEL inhibited HCVpp infectivity, while addition of GST alone or the GST-mouse or rat LEL proteins had no effect (Fig. 2C). Consistent with the EIA data, GST-AGM CD81 LEL partially inhibited HCVpp infectivity (Fig. 2C).
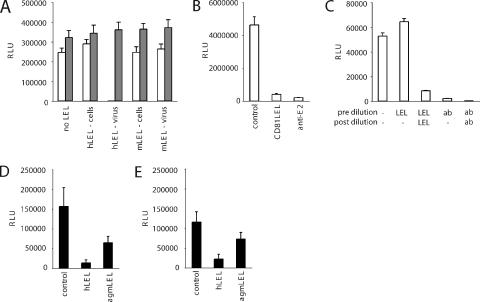
Recombinant forms of tetraspanin LELs have been reported to affect cellular functions such as sperm-egg fertilization, leukocyte adhesion and monocyte fusion (16, 44). More recently, a range of recombinant tetraspanin LELs was reported to inhibit HIV infection of macrophages by interacting with the target cell and associating with endogenous tetraspanins (17). To assess whether human CD81 LEL inhibits HCVpp infectivity by interacting with the target cell, recombinant human and mouse CD81 LEL proteins were incubated with Huh-7.5 cells for 1 hour at 37°C, any unbound protein removed by washing, and the cells challenged with HCVpp and MLVpp. Alternatively, LEL proteins were incubated with HCVpp and MLVpp for 1 hour prior to infection. Human CD81 LEL inhibited HCVpp infectivity when incubated with the virus and not with the target cells, suggesting a mechanism of action dependent upon an interaction with the virus particle (Fig. 3A). Next we incubated a high-titer preparation of HCVpp with either CD81 LEL or a neutralizing anti-E2 MAb (C1) at 5 μg/ml for 1 hour at 37°C. As expected, a strong inhibition was observed with both treatments (Fig. 3B). To test the hypothesis that CD81 LEL and C1 inactivate virus particles through binding to them, we performed the same preincubation (no inhibitor, CD81 LEL, or C1 anti-E2) and then diluted the mixture 60-fold to reduce the CD81 LEL and C1 to an ineffective concentration of 0.08 μg/ml (Fig. 2C and data not shown) and tested HCVpp infectivity. After dilution the virus-CD81 mixture was as infectious as untreated virus, whereas minimal infectivity was detected in the virus-C1 mixture (Fig. 3C). Inhibition was observed when the CD81 LEL was added after the dilution step (to a final concentration of 5 μg/ml). These data suggest that CD81 LEL does not irreversibly neutralize HCVpp or that it must be present during the virus-cell incubation to inhibit infection. Finally, if the inhibitory effect(s) of recombinant CD81 LEL is dependent upon an interaction(s) with cell surface-expressed CD81, one may expect the antiviral activity of diverse CD81 LELs to differ according to the CD81 species present on the target cell. The human hepatoma cell line HepG2 does not express CD81 and can be rendered susceptible to HCVpp infection by expression of human CD81 (4, 49). As shown in the following section, several nonhuman CD81 sequences, including the AGM sequence, confer susceptibility to HCVpp upon HepG2 cells. Thus, we were able to test the inhibitory activity of human and AGM CD81 LELs for HCVpp infection of HepG2 cells expressing human CD81 (Fig. 3D) or AGM CD81 (Fig. 3E). In both cases, human CD81 LEL was a more potent inhibitor of viral infectivity than AGM CD81 LEL, suggesting that inhibition depends on the ability of the soluble CD81 to interact with the virus and not cell surface-expressed tetraspanins.
FIG. 3.
GST-CD81 inhibition of HCVpp infectivity. (A) Human and mouse GST-CD81 LEL (hLEL and mLEL, respectively) proteins were either added to Huh-7.5 cells (“cells”) and incubated for 1 h at 37°C before cells were washed and pseudoparticles added, or CD81 LEL was mixed with pseudoparticles (“virus”) for 1 h at 37°C before adding the mixture to cells. In both cases the final concentration of CD81 LEL was 10 μg/ml. White bars represent HCVpp-H77; shaded bars represent MLVpp. Three days after infection, cells were lysed and luciferase activity determined. Cells infected with no-Env control virus gave a mean luciferase reading of 2,134 ± 308 (data not shown). (B and C) A high-titer infectious preparation of HCVpp-H77 was incubated with no inhibitor or with 5 μg/ml of human CD81 LEL and C1 anti-E2 MAb for 1 h at 37°C before infection. (B) Aliquots of the untreated and treated virus mixtures were tested for infectivity on Huh-7.5 cells. (C) The remaining virus was diluted 60-fold (to reduce the CD81 LEL and C1 to a biologically inactive concentration) and tested for infectivity in the presence and absence of additional human CD81 LEL and C1 MAb at a final concentration of 5 μg/ml. Three days after infection, cells were lysed and luciferase activity determined. Cells infected with no-Env control virus gave mean luciferase readings of 5,006 ± 715 and 856 ± 92 at the two respective dilutions (data not shown). (D and E) Human and AGM CD81 LEL inhibition of HCVpp-H77 infection of HepG2 cells expressing either human (D) or AGM (E) CD81. All infections were performed in triplicate, and the data are representative of two independent experiments (mean ± standard deviation). RLU, relative light units.
Diverse CD81 proteins support HCVpp infection.
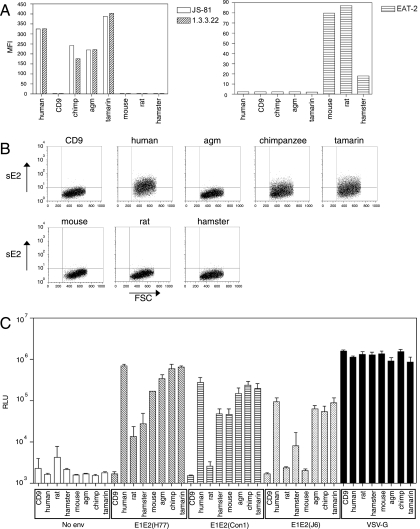
Each of the cloned CD81 sequences (human, AGM, chimpanzee, tamarin, mouse, rat, or hamster) was expressed in HepG2 cells by using the pTRIP expression system and assessed for its ability to support HCVpp infection. Although the human and chimpanzee CD81 protein sequences are predicted to be identical (Fig. 1), cells were transduced to express the chimpanzee cDNA to rule out any effects that might arise from differing nucleotide sequences. As a negative control, cells were transduced to express CD9, a tetraspanin that does not support HCVpp infection (49). Flow cytometric analysis confirmed expression of each species of CD81 at the cell surface (Fig. 4A). sE2 binding to the panel of full-length CD81 proteins was assessed by flow cytometry (Fig. 4B). sE2 bound to HepG2 cells expressing human, tamarin, and chimpanzee CD81 proteins, whereas negligible binding was observed with HepG2 cells expressing CD9, AGM, mouse, rat, or hamster CD81.
FIG. 4.
CD81s from different species support HCVpp-glycoprotein mediated infection. (A) HepG2 cells were transduced with packaged lentiviruses expressing different CD81 sequences or CD9 as a negative control, and stained with anti-CD81 MAbs (1.3.3.22, JS-81, or EAT-2), and analyzed for CD81 expression by flow cytometry. (B) CD81-transduced cell populations were incubated with HCV strain H sE2 protein, and binding was detected with anti-E2 MAbs. FSC, forward scatter. (C) HepG2 cells transduced to express the different species of CD81 were infected with HCVpp generated without a viral envelope (no env) or bearing H77, Con1, or J6 HCV E1E2 or VSV G glycoproteins. Cells were lysed and luciferase activity determined 3 days after infection. All infections were performed in triplicate, and the mean luciferase activity is shown with standard deviation bars. Open bars, infection with no Env-pp; diagonally hatched bars, infection with HCVpp-H77; horizontally hatched bars, infection with HCVpp-Con1; stippled bars, infection with HCVpp-J6; black bars, infection with VSV-Gpp pseudotype virus. RLU, relative light units.
The susceptibility of each CD81-transduced cell line to HCVpp infection was determined (Fig. 4C). As expected, expression of human CD81 conferred susceptibility to infection by HCVpp bearing E1E2 from H77 (genotype 1a), Con1 (genotype 1b), and J6 (genotype 2a); incorporation of the H77 glycoproteins gave rise to the most infectious HCVpp, whereas HCVpp-Con1 was of intermediate infectivity and HCVpp-J6 the least infectious. HCVpp bearing the strain H glycoproteins gave results similar to those for the HCVpp-H77 (data not shown). Despite the lack of interaction of sE2 with the AGM CD81-transduced cells, these cells supported levels of infection similar to those for cells transduced with the human, chimpanzee, and tamarin CD81 sequences. The pattern of infection was more complex for the rodent CD81 sequences: mouse, hamster, and rat CD81 proteins supported HCVpp-H77 infection; mouse and hamster, but not rat, proteins supported HCVpp-Con1 infection; and mouse, hamster, and rat proteins failed to support HCVpp-J6 infection (Fig. 4C). For each HCVpp, the rodent sequences supported lower levels of infection than their primate homologues, likely reflecting a reduced affinity between the HCV glycoproteins and the rodent CD81.
Diverse CD81 proteins support HCVcc infection.
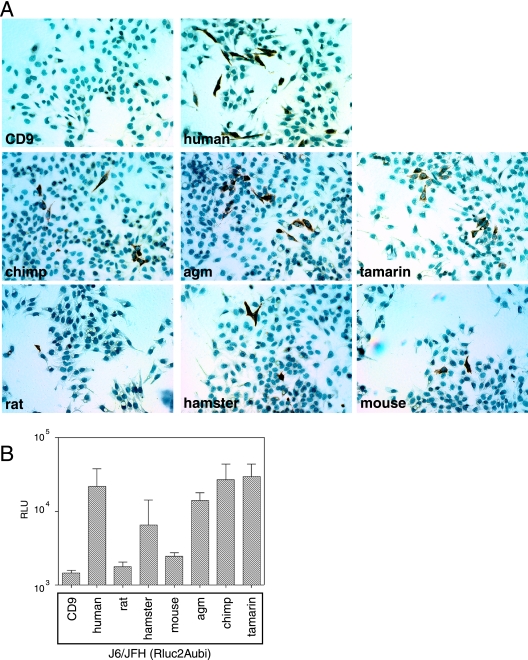
It has recently become possible to propagate infectious HCV particles based on the JFH genotype 2a sequence in cell culture (HCVcc) (25, 46, 50). We reported that chimeric J6/JFH virus was able to infect HepG2 cells expressing human CD81, confirming that CD81 is critical for HCVcc infection (25). We used this chimeric J6/JFH virus to determine whether our panel of CD81 proteins could support HCVcc infection. At 72 h postinfection, the cells were immunostained for the HCV nonstructural protein, NS5A (Fig. 5A). Multiple foci of NS5A-positive cells were present in the HepG2 cells expressing human, chimpanzee, AGM, and tamarin CD81 proteins; fewer foci were present in HepG2 cells expressing mouse or hamster CD81; and a single positive cell was seen in the rat CD81-expressing population (Fig. 5A). This suggests that rodent CD81 sequences are less able to support HCV entry than the primate sequences, consistent with the HCVpp infection data (Fig. 4C). Thus, HCVpp infection reflects HCVcc entry, at least with regard to CD81 sequence requirements.
FIG. 5.
HCVcc infection is supported by multiple CD81 sequences. (A) HepG2 cells transduced to express CD81s from different species were infected with J6/JFH HCVcc. At 72 h postinfection, cells were fixed and immunostained for NS5A. (B) HepG2 cells transduced to express CD81s from different species were infected with HCVcc derived from the J6/JFH genotype 2a chimeric virus expressing the Renilla luciferase, J6/JFH(Rluc2Aubi). Cells were lysed and luciferase activity determined 3 days after infection. All infections were performed in triplicate, and the mean luciferase activity, after deduction of the mean background value from triplicate uninfected wells, is shown with standard deviation bars. RLU, relative light units.
To obtain a quantitative measurement of the efficiency with which different CD81 sequences support HCVcc infection, we used a reporter virus, J6/JFH(Rluc2Aubi), based on the J6/JFH chimeric virus but expressing the Renilla luciferase gene. The CD81-transduced HepG2 cells were infected with J6/JFH(Rluc2Aubi) virus, and Renilla luciferase was measured after 72 h (Fig. 5B). While luciferase signals were low compared to those in HCVpp infections, HCVcc infection was supported by human, AGM, chimpanzee, and tamarin CD81 proteins. Levels of infection were not clearly different from background in HepG2 cells expressing rodent CD81 sequences, with mouse and hamster CD81 proteins marginally above rat CD81 and the CD9 negative control.
Variant CD81 proteins support HCVpp infection.
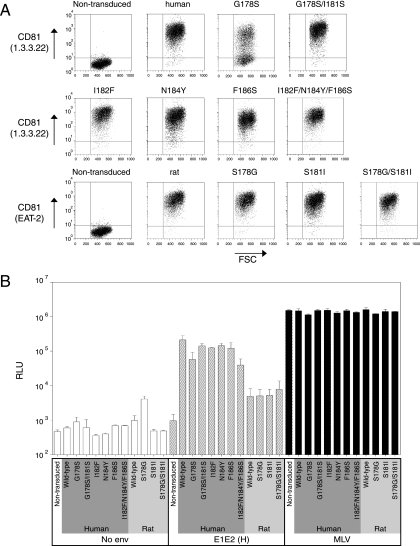
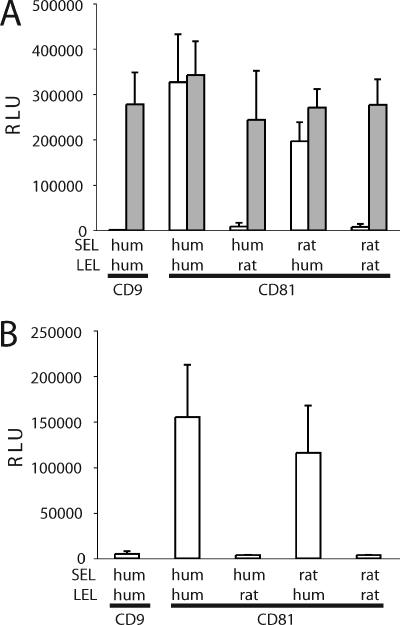
The majority of CD81 sequences tested conferred some degree of susceptibility to infection by HCVpp or HCVcc (Fig. 4 and 5). The least efficient CD81 sequence was rat, which conferred modest levels of infection by HCVpp-H77/H, but not HCVpp-Con1 or -J6 or HCVcc. Within the CD81 LEL, amino acids 178, 181, and 184 are unique to the rat sequence (Fig. 1). Position 184 was previously reported to be important for sE2 binding (11). To evaluate whether amino acids at positions 178 and 181 are critical for HCV infection, we expressed CD81 molecules altered at these residues. The rat residues, either singly or in combination, were introduced into the context of the human CD81 (human CD81 G178S and G178S/I181S), and the human residues were introduced into the context of the rat sequence (rat CD81 S178G, S181I, and S178G/S181I). To further our understanding of the importance of particular CD81 residues in HCV infection, amino acid changes previously reported to abrogate the sE2-CD81 LEL interaction were introduced into the human CD81 (human CD81 I182F, N184Y, and F186S and the three in combination, I182F/N184Y/F186S) (11). The variant CD81 proteins were expressed in HepG2 cells, cell surface expression was confirmed by flow cytometry (Fig. 6A), and cells were tested for infection by pseudotype viruses. Surprisingly, when the individual changes, or their combinations, were introduced into the human CD81 sequence, only moderate reductions in HCVpp-H infection were observed, and the luciferase signal for each was clearly above that observed for rat CD81 (Fig. 6B). Conversely, replacement of the residues unique to the rat sequence with their human counterparts (S178G, S181I, and S178G/S181I) did not significantly modulate the levels of infection above that conferred by wild-type rat CD81. One interpretation is that amino acid changes outside the LEL contribute to the minimal receptor activity of rat CD81. To assess the role of regions outside the LEL, chimeric human and rat CD81 proteins expressing the heterologous species LEL (human-rat and rat-human) were generated, transduced into HepG2 cells, and tested for their ability to support HCVpp-H77 and HCVcc infection. Both chimeric proteins were expressed at the cell surface as determined by flow cytometry (data not shown). HCVpp infected cells expressing the chimeric CD81 containing the human but not rat LEL (Fig. 7A). This pattern was confirmed using HCVcc encoding a luciferase reporter, indicating that the LEL is the region of CD81 defining HCV entry (Fig. 7B).
FIG. 6.
Mutated CD81 sequences confer susceptibility to HCVpp infection. (A) HepG2 cells were transduced with packaged lentiviruses expressing wild-type or mutant CD81 sequences. Transduced cells were stained with anti-CD81 MAbs (1.3.3.22 or EAT-2) and analyzed for CD81 expression by flow cytometry. FSC, forward scatter. (B) HepG2 cells transduced to express mutant CD81 proteins were infected with HCVpp. Cells were lysed and luciferase activity determined 3 days after infection. All infections were performed in triplicate, and mean luciferase activity is shown with standard deviation bars. Depicted are results from a representative of two separate experiments. Open bars, infection with no-Env pseudotype virus; hatched bars, infection with HCVpp-H; black bars, infection with MLVpp. RLU, relative light units.
FIG. 7.
Chimeric rat-human SEL-LEL CD81 proteins confer susceptibility to HCVpp and HCVcc infection. HepG2 cells were transduced with packaged lentiviruses expressing parental or human-rat and rat-human SEL-LEL swap proteins. HepG2 cells transduced to express human CD9 or the chimeric human CD81 proteins were infected with (A) HCVpp-H77 (white bars) or VSV-Gpp (shaded bars) or (B) HCVcc encoding a luciferase reporter, J6/JFH(Rluc2Aubi). All infections were performed in triplicate, and mean luciferase activity is shown with standard deviation bars.
DISCUSSION
Initially identified as a binding partner for sE2, CD81 has recently been shown to be essential for HCVcc infection (25, 46, 50). The interaction between CD81 and the HCV glycoproteins is a potential target for therapeutic intervention. A greater understanding of the structural requirements for CD81 to support infection may aid in the design of inhibitors of this process. In this study, we used a series of CD81 sequences derived from different species to investigate these requirements.
We studied CD81 sequences for their interaction with the HCV glycoproteins in five independent assays: (i) sE2 binding to GST-CD81 LEL proteins (Fig. 2B); (ii) GST-CD81 LEL inhibition of HCVpp infection (Fig. 2C and 3); (iii) sE2 binding to cell surface-expressed, full-length CD81 proteins (Fig. 4B); and (iv) HCVpp (Fig. 4C, 6B, and 7A) and (v) HCVcc (Fig. 5 and 7B) infection of cells expressing full-length CD81. Consistent results were obtained in the first two assays using recombinant CD81 LEL proteins; however, this was not predictive of the ability of full-length CD81 to support HCVpp and HCVcc infection. sE2 interaction with cell surface-expressed CD81 also failed to predict virus-receptor interactions leading to HCV entry. One explanation for these observations is that recombinant sE2 and GST-CD81 LEL fail to mimic their full-length counterparts. Truncated soluble forms of strain H or HCV-1 genotype 1 E2 proteins have been reported to bind CD81 with an affinity of 10−9 M (34), whereas E2 proteins cloned from other genotypes show minimal interaction with CD81 (38, 40, 43). In contrast, it has been demonstrated that HCVpp bearing diverse glycoproteins infect cells in a CD81-dependent manner, despite the differing affinities of their sE2s for CD81 (22, 29). Compared to E1E2 complexes, sE2 has an increased rate of dissociation from CD81 LEL and may be an inadequate model for multivalent viral particle interactions with CD81 (6, 32). These data suggest that care should be taken in interpreting the biological relevance of studies using sE2 as a model for virus-CD81 interactions.
We previously reported that a GST-AGM CD81 LEL fusion protein failed to interact with sE2 or to inhibit HCVpp infection (13, 15, 18). However, in this report we demonstrate a low-affinity interaction between sE2 and GST-AGM CD81 LEL (Fig. 2B), as reported by others (1, 32). This difference may reflect the extent of native folding of the recombinant CD81 proteins used in the various studies. Characterization of the recombinant CD81 proteins used in this study, with antibodies whose recognition was dependent on disulfide bond formation, indicated that disulfide bonds were formed in at least a proportion of the human, mouse, and rat GST-CD81 LEL proteins (Fig. 2A). However, the GST-mouse CD81 LEL failed to inhibit HCVpp infection (Fig. 2B), whereas the full-length mouse CD81 was able to support infection by HCVpp (Fig. 4C), suggesting that recombinant LEL proteins may not accurately mimic native CD81 conformation. Several reports have used recombinant tetraspanin LELs to study cell adhesion and fusion, suggesting an interaction between soluble LEL and target cell-expressed tetraspanins (16, 17, 44). Our experiments demonstrate that incubation of human CD81 LEL with Huh-7.5 cells does not affect HCV infectivity, that recombinant LEL has to be present during the virus-cell incubation period to block HCV entry, and that recombinant LEL inhibition of HCVpp infectivity is independent of the CD81 sequence expressed on the target cell (Fig. 3). These data are consistent with the observation that anti-CD81 antibodies neutralize cell surface-bound HCVpp, suggesting that CD81 is not the primary attachment factor but is a critical cofactor (7).
Another factor to consider is that HCVpp and HCVcc infection assays are likely to be more sensitive than the other assays employed, where multivalent virus particles may interact with cellular molecules in a cooperative manner, allowing CD81 molecules with a low affinity for the viral glycoproteins to function as a receptor. CD81 density is also likely to play a role in HCV entry, such that overexpression of CD81 sequences at the surface of HepG2 cells may allow suboptimal sequences to function as receptors. Cell surface receptor, and coreceptor, densities have been reported to be important determinants of HIV infection (8, 19, 21, 36). For example, CCR3 and STRL33/Bonzo function as HIV type 1 coreceptors only when expression exceeds a threshold level (39, 42). Consistent with CD81 cell surface density being important for HCVpp entry, silencing of expression in Huh-7.5 cells resulted in a threefold reduction in surface CD81 but abrogated HCVpp infection (49), suggesting that a threshold density is required for infection. Importantly, while we cannot infer that physiological levels of mouse, hamster, or AGM CD81 are capable of supporting infection by HCV, it is clear that these sequences can support HCV infection in the context of a human liver cell background.
The rodent CD81 sequences showed differences in their ability to support HCVpp infection of particles bearing H77, Con1, or J6 glycoproteins, suggesting differences in the affinity of these CD81 proteins for diverse HCV glycoproteins (Fig. 4C). This conclusion is supported by the observation that human CD81 LEL neutralizes HCVpp-H77, -Con1, and -J6 infection with various efficiencies (50% inhibitory concentrations of 1.2 μg/ml for H77, 5.6 μg/ml for Con1, and 12.4 μg/ml for J6) (data not shown), consistent with a model where glycoproteins of different genotypes have different affinities for CD81 (29). Interestingly, the infectivity of HCVpp correlated with their sensitivity to neutralization by human CD81 LEL.
All of the tested CD81 sequences were able to support some level of HCV infection, though the rodent sequences were less efficient than primate sequences. For both HCVpp-H77 and J6/JFH HCVcc, the level of infection supported by the mouse CD81 was approximately 10-fold less than that supported by the human sequence (Fig. 4C). In contrast, Bertaux and Dragic reported that transient expression of murine CD81 in HepG2 cells failed to allow HCVpp entry (5). This discrepancy may be explained by differences in CD81 expression levels and the infectivity of the inoculum used for challenge experiments. CD81 is not the sole determinant of HCV host range, as transgenic mice expressing human CD81 are not susceptible to HCV infection (28). It is unclear whether the inefficiency of murine CD81 would be sufficient to preclude propagation of HCV in mice. Nonetheless, the markedly reduced efficiency of rodent CD81 sequences may be an important consideration in efforts to develop a small animal model of HCV infection.
Given that rat CD81 was the least efficient of the tested sequences, we aimed to define whether amino acid residues 178 and 181 within the LEL, which are unique to the rat sequence, are critical for receptor function. Introduction of the rat residues into human CD81 reduced, but did not abolish, HCVpp infection (Fig. 6). Similarly, substitution of the human residues into the rat CD81 had a minimal effect on HCVpp infectivity. Chimeric human CD81 with rat LEL was unable to support HCVpp infection, whereas the rat CD81 with human LEL allowed HCVpp and HCVcc infection, demonstrating the critical role of the LEL in HCV entry (Fig. 7). Hence, the low efficiency of rat CD81 may be attributable not to one or two single residues unique to the rat sequence but to several differences from the human sequence that, in combination, result in a low-affinity or suboptimal interaction(s) with the virus particle.
Previous reports on the HCV-CD81 interaction studied binding of sE2 to recombinant LEL or to full-length CD81 expressed at the cell surface. Those studies identified several amino acid changes that inhibited sE2 binding: I182F, N184Y, F186L, F186S, and D196E (11, 15). We previously demonstrated that human CD81 bearing either F186L or D196E could support HCVpp infection (49). Here, we analyzed a number of additional residues important for sE2 binding, by expressing human CD81 with I182F, N184Y, F186S, or all three changes in combination. These changes, particularly in combination, reduced but did not abrogate HCVpp infection (Fig. 6), underscoring the robustness of the virus-CD81 interaction and the limited predictive power of sE2 binding with regard to HCV glycoprotein-mediated cell entry.
Our findings highlight the limitation of biochemical assays for studying HCV glycoprotein interactions with cellular receptors. Neither the binding of sE2 to recombinant or full-length CD81 nor the ability of recombinant CD81 LEL to inhibit HCVpp infection predicts the ability of CD81 sequences to support HCV entry. Thus, small molecules designed to inhibit the interaction between recombinant proteins may be inadequate to inhibit viral entry. Furthermore, our data indicate a remarkable plasticity in the sequence of CD81 required to support infection. We were unable to identify single amino acid positions in CD81 that are critical for HCVpp or HCVcc entry. Finally, the observation that murine CD81 can support HCV entry, albeit with reduced efficiency, may be important information for the development of a small animal model of HCV infection.
Acknowledgments
We thank Robert Lanford and Yoshi Matsuura for kindly providing RNA and cells, respectively. We are indebted to Carine Logvinoff for helpful advice and discussions and to Merna Torres, Hernan Jaramillo, and Jack Hietpas for expert technical assistance.
This work was supported by Public Health Service grants CA57973 and AI50798, the Greenberg Medical Research Institute, and the Wellcome Trust. T.V.H. is supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 30 August 2006.
REFERENCES
- 1.Allander, T., X. Forns, S. U. Emerson, R. H. Purcell, and J. Bukh. 2000. Hepatitis C virus envelope protein E2 binds to CD81 of tamarins. Virology 277:358-367. [DOI] [PubMed] [Google Scholar]
- 2.Bartosch, B., J. Bukh, J. C. Meunier, C. Granier, R. E. Engle, W. C. Blackwelder, S. U. Emerson, F. L. Cosset, and R. H. Purcell. 2003. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc. Natl. Acad. Sci. USA 100:14199-14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. [DOI] [PubMed] [Google Scholar]
- 5.Bertaux, C., and T. Dragic. 2006. Different domains of CD81 mediate distinct stages of hepatitis C virus pseudoparticle entry. J. Virol. 80:4940-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocquerel, L., C. C. Kuo, J. Dubuisson, and S. Levy. 2003. CD81-dependent binding of hepatitis C virus E1E2 heterodimers. J. Virol. 77:10677-10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cormier, E. G., F. Tsamis, F. Kajumo, R. J. Durso, J. P. Gardner, and T. Dragic. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 101:7270-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doms, R. W. 2000. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology 276:229-237. [DOI] [PubMed] [Google Scholar]
- 9.Drummer, H. E., A. Maerz, and P. Poumbourios. 2003. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett. 546:385-390. [DOI] [PubMed] [Google Scholar]
- 10.Drummer, H. E., and P. Poumbourios. 2004. Hepatitis C virus glycoprotein E2 contains a membrane-proximal heptad repeat sequence that is essential for E1E2 glycoprotein heterodimerization and viral entry. J. Biol. Chem. 279:30066-30072. [DOI] [PubMed] [Google Scholar]
- 11.Drummer, H. E., K. A. Wilson, and P. Poumbourios. 2002. Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J. Virol. 76:11143-11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flint, M., C. Logvinoff, C. M. Rice, and J. A. McKeating. 2004. Characterization of infectious retroviral pseudotype particles bearing hepatitis C virus glycoproteins. J. Virol. 78:6875-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner, J. P., R. J. Durso, R. R. Arrigale, G. P. Donovan, P. J. Maddon, T. Dragic, and W. C. Olson. 2003. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 100:4498-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higginbottom, A., E. R. Quinn, C. C. Kuo, M. Flint, L. H. Wilson, E. Bianchi, A. Nicosia, P. N. Monk, J. A. McKeating, and S. Levy. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74:3642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higginbottom, A., Y. Takahashi, L. Bolling, S. A. Coonrod, J. M. White, L. J. Partridge, and P. N. Monk. 2003. Structural requirements for the inhibitory action of the CD9 large extracellular domain in sperm/oocyte binding and fusion. Biochem. Biophys. Res. Commun. 311:208-214. [DOI] [PubMed] [Google Scholar]
- 17.Ho, S. H., F. Martin, A. Higginbottom, L. J. Partridge, V. Parthasarathy, G. W. Moseley, P. Lopez, C. Cheng-Mayer, and P. N. Monk. 2006. Recombinant extracellular domains of tetraspanin proteins are potent inhibitors of the infection of macrophages by human immunodeficiency virus type 1. J. Virol. 80:6487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabat, D., S. L. Kozak, K. Wehrly, and B. Chesebro. 1994. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J. Virol. 68:2570-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitadokoro, K., D. Bordo, G. Galli, R. Petracca, F. Falugi, S. Abrignani, G. Grandi, and M. Bolognesi. 2001. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 20:12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak, S. L., E. J. Platt, N. Madani, F. E. Ferro, Jr., K. Peden, and D. Kabat. 1997. CD4, CXCR-4, and CCR-5 dependencies for infections by primary patient and laboratory-adapted isolates of human immunodeficiency virus type 1. J. Virol. 71:873-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavillette, D., A. W. Tarr, C. Voisset, P. Donot, B. Bartosch, C. Bain, A. H. Patel, J. Dubuisson, J. K. Ball, and F. L. Cosset. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41:265-274. [DOI] [PubMed] [Google Scholar]
- 23.Levy, S., and T. Shoham. 2005. The tetraspanin web modulates immune-signalling complexes. Nat Rev. Immunol. 5:136-148. [DOI] [PubMed] [Google Scholar]
- 24.Levy, S., S. C. Todd, and H. T. Maecker. 1998. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16:89-109. [DOI] [PubMed] [Google Scholar]
- 25.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 26.Logvinoff, C., M. E. Major, D. Oldach, S. Heyward, A. Talal, P. Balfe, S. M. Feinstone, H. Alter, C. M. Rice, and J. A. McKeating. 2004. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 101:10149-10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lozach, P. Y., H. Lortat-Jacob, A. de Lacroix de Lavalette, I. Staropoli, S. Foung, A. Amara, C. Houles, F. Fieschi, O. Schwartz, J. L. Virelizier, F. Arenzana-Seisdedos, and R. Altmeyer. 2003. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 278:20358-20366. [DOI] [PubMed] [Google Scholar]
- 28.Masciopinto, F., G. Freer, V. L. Burgio, S. Levy, L. Galli-Stampino, M. Bendinelli, M. Houghton, S. Abrignani, and Y. Uematsu. 2002. Expression of human CD81 in transgenic mice does not confer susceptibility to hepatitis C virus infection. Virology 304:187-196. [DOI] [PubMed] [Google Scholar]
- 29.McKeating, J. A., L. Q. Zhang, C. Logvinoff, M. Flint, J. Zhang, J. Yu, D. Butera, D. D. Ho, L. B. Dustin, C. M. Rice, and P. Balfe. 2004. Diverse hepatitis C virus glycoproteins mediate viral infection in a CD81-dependent manner. J. Virol. 78:8496-8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meola, A., A. Sbardellati, B. Bruni Ercole, M. Cerretani, M. Pezzanera, A. Ceccacci, A. Vitelli, S. Levy, A. Nicosia, C. Traboni, J. McKeating, and E. Scarselli. 2000. Binding of hepatitis C virus E2 glycoprotein to CD81 does not correlate with species permissiveness to infection. J. Virol. 74:5933-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meunier, J. C., R. E. Engle, K. Faulk, M. Zhao, B. Bartosch, H. Alter, S. U. Emerson, F. L. Cosset, R. H. Purcell, and J. Bukh. 2005. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc. Natl. Acad. Sci. USA 102:4560-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakajima, H., L. Cocquerel, N. Kiyokawa, J. Fujimoto, and S. Levy. 2005. Kinetics of HCV envelope proteins' interaction with CD81 large extracellular loop. Biochem. Biophys. Res. Commun. 328:1091-1100. [DOI] [PubMed] [Google Scholar]
- 33.Op De Beeck, A., C. Voisset, B. Bartosch, Y. Ciczora, L. Cocquerel, Z. Keck, S. Foung, F. L. Cosset, and J. Dubuisson. 2004. Characterization of functional hepatitis C virus envelope glycoproteins. J. Virol. 78:2994-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petracca, R., F. Falugi, G. Galli, N. Norais, D. Rosa, S. Campagnoli, V. Burgio, E. Di Stasio, B. Giardina, M. Houghton, S. Abrignani, and G. Grandi. 2000. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J. Virol. 74:4824-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 36.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pohlmann, S., J. Zhang, F. Baribaud, Z. Chen, G. J. Leslie, G. Lin, A. Granelli-Piperno, R. W. Doms, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 77:4070-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roccasecca, R., H. Ansuini, A. Vitelli, A. Meola, E. Scarselli, S. Acali, M. Pezzanera, B. B. Ercole, J. McKeating, A. Yagnik, A. Lahm, A. Tramontano, R. Cortese, and A. Nicosia. 2003. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J. Virol. 77:1856-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rucker, J., A. L. Edinger, M. Sharron, M. Samson, B. Lee, J. F. Berson, Y. Yi, B. Margulies, R. G. Collman, B. J. Doranz, M. Parmentier, and R. W. Doms. 1997. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J. Virol. 71:8999-9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seigneuret, M. 2006. Complete predicted three-dimensional structure of the facilitator transmembrane protein and hepatitis C virus receptor CD81: conserved and variable structural domains in the tetraspanin superfamily. Biophys. J. 90:212-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharron, M., S. Pohlmann, K. Price, E. Lolis, M. Tsang, F. Kirchhoff, R. W. Doms, and B. Lee. 2000. Expression and coreceptor activity of STRL33/Bonzo on primary peripheral blood lymphocytes. Blood 96:41-49. [PubMed] [Google Scholar]
- 43.Shaw, M. L., J. McLauchlan, P. R. Mills, A. H. Patel, and E. A. McCruden. 2003. Characterisation of the differences between hepatitis C virus genotype 3 and 1 glycoproteins. J. Med. Virol. 70:361-372. [DOI] [PubMed] [Google Scholar]
- 44.Takeda, Y., I. Tachibana, K. Miyado, M. Kobayashi, T. Miyazaki, T. Funakoshi, H. Kimura, H. Yamane, Y. Saito, H. Goto, T. Yoneda, M. Yoshida, T. Kumagai, T. Osaki, S. Hayashi, I. Kawase, and E. Mekada. 2003. Tetraspanins CD9 and CD81 function to prevent the fusion of mononuclear phagocytes. J. Cell Biol. 161:945-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tscherne, D. M., C. T. Jones, M. J. Evans, B. D. Lindenbach, J. A. McKeating, and C. M. Rice. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, M. Y., B. Bartosch, P. Zhang, Z. P. Guo, P. M. Renzi, L. M. Shen, C. Granier, S. M. Feinstone, F. L. Cosset, and R. H. Purcell. 2004. Neutralizing antibodies to hepatitis C virus (HCV) in immune globulins derived from anti-HCV-positive plasma. Proc. Natl. Acad. Sci. USA 101:7705-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]