Abstract
Aminoglycoside antibiotics have had a major impact on our ability to treat bacterial infections for the past half century. Whereas the interest in these versatile antibiotics continues to be high, their clinical utility has been compromised by widespread instances of resistance. The multitude of mechanisms of resistance is disconcerting but also illuminates how nature can manifest resistance when bacteria are confronted by antibiotics. This article reviews the most recent knowledge about the mechanisms of aminoglycoside action and the mechanisms of resistance to these antibiotics.
INTRODUCTION
The first of the aminoglycoside antibiotics, streptomycin, was reported in 1944 (255), a seminal contribution that paved the way to discovery of a series of other aminoglycosides and other antibiotics from natural sources in the next two decades. Streptomycin proved to be effective for the first time in the treatment of tuberculosis, a property that has endured to the present. Subsequent discoveries of novel aminoglycoside antibiotics provided a host of therapeutic agents that showed broad spectra of antibiotic activity. The activity was often improved by medicinal chemists who were able to exploit the structures of the parental molecules in developing semisynthetic variants that resisted the activities of the aminoglycoside resistance enzymes or expanded the breadth of activity. These and related developments have been the subjects of a number of recent reviews on aminoglycoside antibiotics (10, 111, 112, 151, 317). In this report, we address recent discoveries on the mechanisms of activity of aminoglycosides, the mechanisms of resistance, and the epidemiology of the resistance of bacteria to these anti-infective agents.
ANTIMICROBIAL ACTIVITY AND CLINICAL INDICATIONS
Aminoglycoside antibiotics exhibit in vitro activity against a wide variety of clinically important gram-negative bacilli such as Escherichia coli, Salmonella spp., Shigella spp., Enterobacter spp., Citrobacter spp., Acinetobacter spp., Proteus spp., Klebsiella spp., Serratia spp., Morganella spp., and Pseudomonas spp. as well as Staphylococcus aureus and some streptococci. However, they lack predictable in vitro activity against Bacteroides spp. and other anaerobic microorganisms, Streptococcus pneumoniae, Neisseria gonorrhoeae, Burkholderia cepacia, and Stenotrophomonas maltophilia, and their activity against enterococci is adequate only when they are used synergistically with a cell wall-active antibiotic, such as β-lactams and vancomycin (101, 211).
As with other classes of antibiotics, significant differences in the spectrum of antimicrobial activity exist among the various aminoglycosides. Numerous aminoglycoside-modifying enzymes that inactivate various antibiotics of this class contribute significantly to differences in their in vitro activities. Generally, newer aminoglycosides such as gentamicin, tobramycin, amikacin, netilmicin, isepamicin, dibekacin, and arbekacin have broader spectra of activity than the older compounds like streptomycin and kanamycin. Among the newer aminoglycosides, gentamicin is more active than tobramycin against Serratia spp., while tobramycin has superior activity against Pseudomonas aeruginosa. In a recent study, isepamicin exhibited two- to fourfold-higher in vitro activity than amikacin against members of the family Enterobacteriaceae, and both antibiotics produced similar activities against Pseudomonas aeruginosa (18). Arbekacin, an aminoglycoside that is widely used in Japan, demonstrates the widest spectrum of antibacterial activity. Most impressive is its remarkable activity against methicillin-resistant Staphylococcus aureus (7), including isolates that exhibit resistance to other aminoglycosides (57, 113, 118).
Despite their potential nephrotoxicity and ototoxicity and problems associated with aminoglycoside-resistant organisms (see below), aminoglycoside antibiotics remain valuable and sometimes indispensable for the treatment of various infections and prophylaxis in special situations. Aminoglycosides exhibit several characteristics that make them useful as antimicrobial agents. Among them are concentration-dependent bactericidal activity, postantibiotic effect, relatively predictable pharmacokinetics, and synergism with other antibiotics. In contrast to β-lactams, the bactericidal activity of aminoglycosides depends more on their concentration than on the duration of bacterial exposure to inhibitory concentrations of antibiotic and is also significantly less dependent on the bacterial inoculum size (17, 61, 77, 79, 193, 198, 309). The killing potential of aminoglycosides is concentration dependent and increases with increasing concentrations of the antibiotic (124, 326). Significantly, aminoglycosides exhibit the postantibiotic effect (62, 63, 183). They continue to kill bacteria even after the aminoglycoside has been removed following a short incubation with the microorganism (35, 85, 144, 326). Concentration-dependent bactericidal activity and postantibiotic effect in combination with the attenuated risk of nephrotoxicity and ototoxicity are the major reasons for once-daily dosing with aminoglycosides in patients with normal renal function (95, 101, 102, 233).
Another important characteristic of aminoglycosides is their ability to produce synergistic bactericidal activity in combination with antimicrobial agents inhibiting cell wall biosynthesis, including β-lactams and vancomycin. Synergism presumably arises as the result of enhanced intracellular uptake of aminoglycosides caused by the increased permeability of bacteria after incubation with cell wall synthesis inhibitors (82, 194). The synergistic effect between aminoglycosides and β-lactam antibiotics is well established for enterococci (39, 134, 150). The issue of synergism against other microorganisms is less clear. Despite the fact that such a synergistic effect was also demonstrated for some strains of Enterobacteriaceae (80, 148, 219), Pseudomonas aeruginosa (29, 56, 147, 209), staphylococci (33, 34, 252, 310), including methicillin-resistant Staphylococcus aureus (MRSA) (125), and other microorganisms (80), many of these microorganisms were not inhibited by a combination of aminoglycoside and cell wall-active compounds (131, 153, 245). In a disconcerting development, antagonism between aminoglycosides and β-lactams was recently described (135). This antagonism resulted from induction of the aminoglycoside-modifying enzyme AAC(6′)-APH(2") by β-lactam antibiotics and is particularly troublesome because it was detected in MRSA.
Aminoglycoside antibiotics are useful for empirical treatment of febrile neutropenic patients and patients with serious infections caused by aerobic gram-negative microorganisms, including Enterobacteriaceae and Pseudomonas aeruginosa (101, 211). Because of the potential risk of failure in patients infected with resistant microorganisms, aminoglycosides are often administered in combination with other antibacterial agents (132, 133). Combination with antipseudomonal penicillins or cephalosporins is recommended for treatment of systemic pseudomonal infections. Such combinations are beneficial because of the probability of not only achieving the synergistic effect but also preventing rapid development of resistance to β-lactam antibiotics. For urinary tract infections, monotherapy with an aminoglycoside is possible because intravenously administered antibiotic is excreted exclusively by the kidneys.
Aminoglycosides are often used in combination with β-lactams and glycopeptides for the treatment of patients with bacterial endocarditis caused by enterococci and less often for streptococcal endocarditis (104, 264). Such combinations are synergistic despite the fact that enterococci are intrinsically resistant to aminoglycosides and β-lactams. Of great concern is the discovery of new aminoglycoside-modifying enzymes in enterococci that are able to inactivate most of the clinically available aminoglycoside antibiotics, including gentamicin (51, 300). Because of the lack of cross-resistance, streptomycin or arbekacin can be used instead of gentamicin against enterococci that produce such gentamicin-modifying enzymes (51, 140, 322). When considering therapeutic options, it is important to remember that microorganisms growing in an anaerobic environment such as an abscess have diminished susceptibility to aminoglycosides.
Streptomycin, the first aminoglycoside, was also the first antibiotic that was active against Mycobacterium tuberculosis (255). It still shows excellent activity against Mycobacterium tuberculosis and remains a first-line drug in combination chemotherapy for drug-resistant tuberculosis (103, 206). Aminoglycosides are also active against Yersinia pestis (94), Francisella tularensis (11) and Brucella spp. (4, 200), the etiological agents of plague, tularemia, and brucellosis, respectively. Streptomycin alone or in combination with other antimicrobial agents is a first-line antibiotic for therapy of these life-threatening infections, whereas other aminoglycosides can be used as alternatives (83, 181, 226, 272, 273, 323).
Aminoglycosides are mainly administered parenterally. Under certain circumstances, alternative drug delivery approaches are used to increase the concentration of the antibiotic at the site of infection or to reduce the risk of nephrotoxicity and ototoxicity. One example is the use of aerosolized tobramycin or gentamicin solutions for inhalation as a promising addition to the traditional therapy and prophylaxis of respiratory tract infections, including those occurring in cystic fibrosis patients (59, 122, 201, 202, 238, 243). Another interesting approach is the development and use of liposome-encapsulated aminoglycosides (256), used for both local applications (14, 227, 234, 235, 325) and intravenous administration (283, 285). The activity of liposome-encapsulated aminoglycosides against intracellular microorganisms is of special interest (8, 88, 97, 174, 176).
MECHANISM OF ACTION
Although it is well known that aminoglycosides bind to the bacterial ribosome and inhibit protein synthesis, the precise mechanisms of their antimicrobial activity are still a subject of study. The ribosome is a complex structure comprising three RNA molecules and more than 50 proteins. The complex catalyzes protein synthesis with the assistance of several GTP-hydrolyzing protein factors. The bacterial ribosome consists of two subunits with relative sedimentation rates of 50S and 30S (28, 45, 314). The large subunit comprises two RNA molecules, referred to as the 5S and 23S RNAs, and 33 proteins, while the small subunit is made up of a single 16S RNA and 20 to 21 proteins. Aminoglycoside antibiotics bind to the 30S ribosomal subunit, which plays a crucial role in providing high-fidelity translation of genetic material.
Recently, atomic structures for both the large and the small ribosomal subunits and high-resolution crystal structures of the 30S subunit with streptomycin, spectinomycin, paromomycin, and hygromycin B have been solved (12, 27, 45, 117, 257, 306, 315). Together with several available nuclear magnetic resonance structures for the ribosomal constituents (89-91, 320, 321), these structures provide valuable information not only on processes during translation but also on molecular mechanisms of interaction of aminoglycosides with bacterial ribosome. The ribosome has three functionally important tRNA binding sites, designated A (for aminoacyl), P (for peptidyl), and E (for exit) (reviewed previously [107]). During protein synthesis, the ribosome decodes information stored in the mRNA and catalyzes sequential incorporation of amino acids into a growing polypeptide chain. High fidelity of translation (the ribosome misincorporates only one amino acid per approximately 3,000 peptide bonds synthesized [25]) is achieved by the ability to discriminate between conformational changes in the ribosome induced by binding of cognate (correct) and noncognate (incorrect) tRNAs at the A site (168). Although the precise mechanism of discrimination is presently unknown, correct codon-anticodon recognition by the ribosome is believed to be achieved in a two-step process—an initial recognition of base pairing between the codon on the mRNA and the anticodon of the tRNA and subsequent proofreading (129, 212).
Although all aminoglycosides bind to the 30S ribosomal subunit and interfere with protein synthesis, the details of interactions for different classes of aminoglycosides are different. Paromomycin and other aminoglycosides that contain the 2-deoxystreptamine ring increase the error rate of the ribosome by allowing incorporation of the noncognate (or near-cognate) tRNAs. The structure of the 30S subunit indicates that two universally conserved adenine residues (A1492 and A1493) are directly involved in the decoding process during normal translation (213). In the native structure of the ribosome, these adenine residues are stacked in the interior of helix 44. Binding of the tRNA to the A site flips A1493 and A1492 out from their stacked position; it also flips G530 out from the syn to the anti conformation. The N1 of adenines interacts with the 2′-OH groups of the tRNA residues that are in the first and second positions of the codon-anticodon triplet. Because the distance between the 2′-OH groups will depend on the geometry of base pairing, such hydrogen bonding might allow discrimination between cognate and near-cognate or noncognate tRNAs (45).
The following mechanism was proposed to explain how the 2-deoxystreptamine-containing aminoglycosides decrease accuracy of translation. Analysis of the structures indicates that paromomycin and other aminoglycosides that contain the 2-deoxystreptamine ring bind in the major groove of helix H44 of 16S RNA. Binding of the antibiotic results in flipping out of the same conserved A1492 and A1493 residues that are normally displaced upon binding of the cognate tRNA. Conformational changes induced in the 30S subunit by binding of cognate tRNA are energetically favorable because they allow the ribosome to participate in a greater number of compensating interactions between the codon and anticodon double helixes (213). Because the flipping-out of the adenine residues that takes place during normal translation might require energy expenditure, paromomycin-induced flipping-out can reduce energetic cost, allowing binding of near-cognate tRNAs and subsequent mistranslation of mRNA. Recently, it had been demonstrated that paromomycin inhibits not only protein synthesis but also assembly of the 30S ribosomal subunit (185).
Streptomycin differs structurally from other aminoglycosides because its aminocyclitol ring is streptidine instead of 2-deoxystreptamine. Despite this difference, streptomycin binds at the functional center of the ribosome in close proximity to the binding site of paromomycin. Tight binding of antibiotic is facilitated by interaction with nucleotides 13, 526, 915, and 1490 of 16S rRNA and protein S12 (45, 105, 186, 191, 196). Like 2-deoxystreptamine-containing aminoglycosides, streptomycin induces misreading of the genetic code (142, 143, 250), but the underlying mechanism seems to be different. It has been shown that during translation, the 30S subunit switches between two distinct conformations. These conformational switches involve two alternating base pairing schemes between the nucleotides of helix 27, facilitated by ribosomal proteins S5 and S12 (168). In one of the two conformations, nucleotides C912, U911, and C910 are paired with residues G885, G886, and G887 (the 912-885 conformation). The alternative conformation results from base pairing between nucleotides 912 to 910 and G888, A889, and G890 (the 912-888 conformation).
It has been demonstrated that mutational stabilization of one of the conformations over the other results in two distinct fidelity phenotypes. Mutations that enhance stability of the 912-888 conformation increase fidelity of translation and were designated restrictive or hyperaccurate, while mutations favoring the alternative 912-885 conformation decreased fidelity and were designated ram (ribosomal ambiguity) or error-prone. It was proposed that the interaction of streptomycin with the ribosome preferentially stabilizes the ram state (45). Such stabilization would not only render the A site more promiscuous by lowering its affinity for tRNAs and allowing binding of near-cognate tRNAs, but could also affect the proposed proofreading by making transition to the restrictive site more difficult. It was hypothesized that various ribosomal mutations producing streptomycin resistance or streptomycin dependence shift the delicate balance that exists between the two conformational states (45).
Spectinomycin, a nonaminoglycoside aminocyclitol, binds in the minor groove at the end of helix 34 in close proximity to helix H28 and a loop formed by the protein S5, and interacts with residues G1064, C1066, G1068, and C1192 (45). It inhibits translocation of the peptidyl-tRNA from the A site to the P site. It was proposed that translocation of tRNA from the A site to the P site requires the movement of various elements of the ribosome, including helix 34. The X-ray structure of the 30S subunit with spectinomycin provided evidence that binding of the antibiotic at the end of helix 34 interferes sterically with movement of the ribosome and restricts conformational changes of helix 34. The authors suggested that ribosomal mutations producing spectinomycin resistance may destabilize the network of interactions between the head and the body of the 30S subunit, allowing the head to move even in the presence of the antibiotic (45). Thus, in contrast to other aminoglycosides, spectinomycin does not cause misreading, and as a consequence, it has only a bacteriostatic effect on microbial cells.
UPTAKE OF AMINOGLYCOSIDES
Despite the impressive progress that has been achieved in the last several years in elucidating the details of the interaction of aminoglycoside antibiotics with the ribosome and their consequent influence on the translational machinery, many aspects of the interaction of aminoglycosides with the bacterial cell still remain unclear. It has been proposed that aminoglycoside antibiotics penetrate aerobically growing bacteria in three consecutive steps (71, 114, 284). The first step is energy-independent binding of the positively charged aminoglycosides to the negatively charged moieties of phospholipids, lipopolysaccharides, and outer membrane proteins in gram-negative bacteria and to phospholipids and teichoic acids in gram-positive microorganisms (284). It is believed that binding of aminoglycosides results in displacement of Mg2+ and Ca2+ ions that link adjacent lipopolysaccharide molecules, a process that damages the outer membrane and enhances its permeability (114, 178). The initial electrostatic surface binding is followed by the energy-dependent phase I of uptake. Uptake of aminoglycosides during energy-dependent phase I requires a threshold transmembrane potential generated by a membrane-bound respiratory chain. That is why microorganisms with deficient electron transport systems, such as anaerobes, are intrinsically resistant to aminoglycosides. For the same reason, enterococci and other facultative anaerobes are resistant to low concentrations of aminoglycosides.
It is thought that during energy-dependent phase I, only a small quantity of antibiotic molecules cross the cytoplasmic membrane (32, 69). The binding of incoming antibiotic to the ribosome results in misreading of mRNA and production of misfolded proteins. Some of these proteins are incorporated in the cytoplasmic membrane, leading to the loss of membrane integrity. This in turn triggers a cascade of events called energy-dependent phase II of uptake. During this phase, additional quantities of aminoglycosides are transported across the damaged cytoplasmic membrane. As a consequence, antibiotics accumulate rapidly in the cytoplasm and virtually irreversibly saturate all ribosomes, resulting in the death of the cell. The higher the concentration of the aminoglycoside, the more rapid is the onset of energy-dependent phase II and subsequent bacterial death.
MECHANISMS OF RESISTANCE
Several mechanisms have been proposed for bacterial resistance to aminoglycoside antibiotics. They include decreased antibiotic uptake and accumulation, modification of the ribosomal target, efflux of antibiotic, and enzymatic modification of aminoglycosides.
As discussed above, the bacterial uptake of aminoglycoside antibiotics requires respiration, which generates an electrical potential across the cytoplasmic membrane. A low level of transmembrane potential or even its absence is responsible for the intrinsic resistance of anaerobic bacteria (31) and decreased susceptibility of facultative anaerobes such as enterococci to aminoglycosides (192). Chromosomal mutations in Staphylococcus aureus influencing transmembrane electrical potential have also been shown to produce aminoglycoside resistance (190). Such mutations lower growth rates and allow bacteria to survive during aminoglycoside therapy (30). Recently, various multidrug active efflux systems have been identified as mechanisms of natural resistance to aminoglycoside antibiotics in gram-negative bacteria such as Burkholderia pseudomallei (197), Pseudomonas aeruginosa (3, 312), Acinetobacter baumannii (175), and E. coli (248). Efflux pumps extrude the incoming antibiotic and produce low-level broad-spectrum resistance that often includes antibiotics of different classes.
Modifications of the target that produce aminoglycoside resistance include mutational changes in the ribosomal proteins or 16S rRNA and enzymatic methylation of the rRNA. The mutational changes are known to produce resistance mostly to streptomycin; also, mutation in the 16S rRNA could result in decreased susceptibility to spectinomycin (271). Streptomycin is still used for treatment of tuberculosis, and mutational resistance to streptomycin in clinical isolates of Mycobacterium tuberculosis is therefore of clinical importance (55). Streptomycin resistance due to mutational changes in 16S rRNA or ribosomal proteins has occasionally been demonstrated for other microorganisms (81, 155, 177, 231). It has been proposed that numerous mutations in the 16S rRNA and the 30S ribosomal subunit proteins disrupt a delicate balance that exists between the ram and the restrictive states and thus interfere with optimal translation (see above) (45). Methylation at N7 of guanine residues of the 16S rRNA produces high-level resistance to aminoglycoside antibiotics in aminoglycoside-producing microorganisms (64, 295), but this type of resistance has not been reported in clinically important microorganisms.
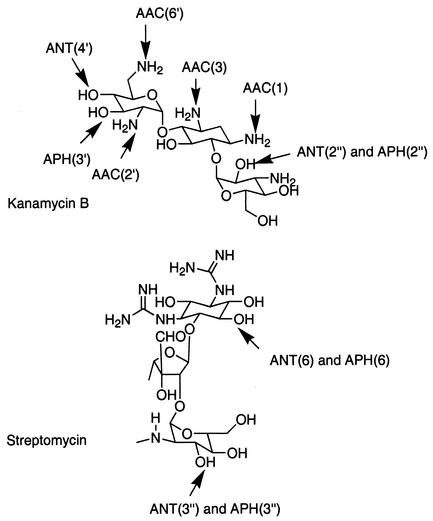
The major mechanism of aminoglycoside resistance in clinical isolates of gram-negative and gram-positive bacteria is enzymatic modification of the amino or hydroxyl groups of these antibiotics (Fig. 1). Modified aminoglycosides bind poorly to the ribosome (167) and fail to trigger energy-dependent phase II, allowing bacteria to survive in the presence of the drug.
FIG. 1.
Sites of modification by aminoglycoside-modifying enzymes.
Three families of enzymes that perform cofactor-dependent drug modification in the bacterial cytoplasm have been recognized; these are aminoglycoside phosphotransferases (APHs), aminoglycoside acetyltransferases (AACs), and aminoglycoside nucleotidyltransferases (ANTs). The sites that are modified by these enzymes for different classes of aminoglycosides were discussed in the article by Wright et al. (317). Figure 1 provides a sample of these enzymatic modifications for two aminoglycosides. The level of resistance produced differs significantly in various microorganisms and individual strains and depends on many factors, including the amount of enzyme produced, its catalytic efficiency, and the type of aminoglycoside. Whereas many of these enzymes give sufficient activities to result in effective resistance, generally only phosphotransferases produce high levels of resistance. Each of the three families of enzymes is further divided into classes, designated by the site of modification, which is indicated in parentheses. They are further subdivided into enzyme types (designated by Roman numerals) that specify unique resistance phenotypes. Finally, individual enzymes of the same class and type that produce the same phenotype but are encoded by different genes are designated by a lowercase letter (268). For example, the AAC(6′)-I enzymes AAC(6′)-Ia, AAC(6′)-Ib, AAC(6′)-Ic, etc., are aminoglycoside acetyltransferases that modify the antibiotic at position 6′ and produce the same phenotype (inactivation of tobramycin, amikacin, netilmicin, kanamycin, and dibekacin) but are encoded by different genes.
There are still inconsistencies in the nomenclature of genes for aminoglycoside-modifying enzymes. In some cases, these genes are named according to the site of modification, followed by a number to distinguish between genes for individual types of enzymes. According to another nomenclature, the genes for AAC(6′)-Ia and AAC(3)-Ia are referred to as aacA1and aacC1, respectively. This nomenclature is somewhat confusing, and we will follow the nomenclature proposed by Shaw and colleagues (268), which utilizes the same names for the enzymes and the corresponding genes, but the names of genes are in lowercase letters and italicized. According to this more convenient nomenclature, the genes for the AAC(6′)-Ia and AAC(3)-Ia enzymes are termed aac(6′)-Ia and aac(3)-Ia, respectively.
Aminoglycoside Phosphotransferases
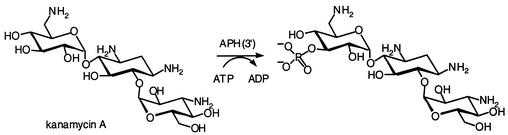
Aminoglycoside phosphotransferases (kinases) utilize ATP as the second substrate and are able to phosphorylate specific hydroxyl groups in all classes of aminoglycoside antibiotics. Seven classes of enzymes, APH(3′), APH(2"), APH(3"), APH(4), APH(7"), APH(6), and APH(9) have been identified in clinical isolates and aminoglycoside-producing organisms (268). The largest class of aminoglycoside phosphotransferases includes enzymes that modify the hydroxyl groups of antibiotics at the 3′ position. Seven different types of APH(3′), APH(3′)-I to APH(3′)-VII, have been identified among gram-negative and gram-positive bacteria and also aminoglycoside-producing microorganisms (268). Figure 2 depicts this reaction for the phosphorylation of kanamycin A by APH(3′)s. The product of phosphorylation lacks antibacterial activity.
FIG. 2.
Phosphorylation of kanamycin A by APH(3′)s.
APH(3′)-I produces resistance to kanamycin, neomycin, lividomycin, paromomycin, and ribostamycin. The gene for the first APH(3′)-I enzyme (aph(3′)-Ia) was discovered on transposon Tn903 in E. coli (215). Subsequently, it was identified on plasmids and transposons in many gram-negative bacteria, including Klebsiella pneumoniae (163), Salmonella enterica serovar Typhimurium (319), Salmonella enterica (92), Proteus vulgaris (204), Vibrio cholerae (24, 126), Campylobacter jejuni (216), and the fish pathogen Pasteurella piscicida (146). Recently the aph(3′)-I gene was identified in the gram-positive opportunistic human pathogen Corynebacterium striatum (287). The gene was flanked by divergently oriented IS26 insertion sequences and was located on the composite transposon Tn5715, exemplifying a case of horizontal transfer of antibiotic resistance genes between gram-negative and gram-positive bacteria. The high frequency of occurrence of APH(3′)-I and other enzymes producing kanamycin resistance in bacteria resulted in the clinical obsolescence of the kanamycins. Subsequent lack of use of kanamycins in the clinic has resulted in a corresponding decline in the importance of these enzymes in clinical isolates.
In contrast to APH(3′)-I, APH(3′)-II is rarely found in clinical isolates (268) despite the fact that it confers a similar spectrum of resistance and the gene for this enzyme is also located on a transposable element, Tn5 (16). The gene for APH(3′)-IIb has been identified in the chromosome of Pseudomonas aeruginosa (109, 275). APH(3′)-IIb shows 52% amino acid identity and 67% similarity with APH(3′)-IIa. Recently, another phosphotransferase that produces a spectrum of resistance characteristic of APH(3′)-II has been identified in Pseudomonas aeruginosa (244). The enzyme shows 36% amino acid identity with APH(3′)-IIa and 38% identity with APH(3′)-IIb and is the first phosphotransferase whose gene has been identified within an integron.
The genes for the APH(3′)-III phosphotransferases were originally isolated from Staphylococcus aureus (106) and Streptococcus faecalis (299). Later the gene was identified in Campylobacter coli, which became a precedent for antibiotic resistance gene transfer between gram-positive and gram-negative bacteria (220, 288). APH(3′)-III produces resistance to kanamycin, neomycin, lividomycin, paromomycin, butirosin, and ribostamycin. The enzyme is also able to modify amikacin and isepamicin (301). Furthermore, although the level of amikacin resistance that this enzyme produces is low, clinical enterococcal isolates that express APH(3′)-III are resistant to amikacin-ampicillin synergism (40, 162). A study of the epidemiology of aminoglycoside-modifying enzymes in various countries indicated the presence of the aph(3′)-IIIa gene in 9% of MRSA in Japan (136) and in 13% of MRSA in Europe; the percentage was even higher in methicillin-susceptible staphylococci (259). Almost 50% of the aminoglycoside-resistant isolates of Enterococcus faecalis and Enterococcus faecium studied in Japan were positive for aph(3′)-IIIa. Often, aph(3′)-IIIa in gram-positive bacteria is found in combination with genes for other aminoglycoside-modifying enzymes (302) or is genetically linked with resistance genes for other classes of antibiotics (23, 311).
The genes for APH(3′)-IV and -V were detected only in antibiotic-producing microorganisms (123, 130, 251, 294) and will not be addressed here. The genes for two other enzymes, APH(3′)-VI and APH(3′)-VII, were cloned correspondingly from Acinetobacter baumannii (179) and Campylobacter jejuni (291). APH(3′)-VI produces resistance to amikacin, isepamicin, kanamycin, neomycin, paromomycin, butirosin, and ribostamycin (158) and is associated primarily with Acinetobacter spp. (268). Earlier studies of the distribution of aminoglycoside resistance genes demonstrated the presence of aph(3′)-VIa in 83 to 95% of amikacin-resistant Acinetobacter strains (156, 268). In a subsequent survey, the gene was identified in 46% of aminoglycoside-resistant Acinetobacter strains (188). APH(3′)-VII produces resistance to kanamycin and neomycin (290), but the distribution of the gene has not been studied.
Four genes encoding APH(2") enzymes have been detected in gram-positive bacteria. The aph(2")-Ia gene, which is found downstream from aac(6′)-Ie, encodes the C-terminal part of the bifunctional enzyme AAC(6′)-Ie-APH(2")-Ia. The presence of acetyltransferase and phosphotransferase activities in the bifunctional enzyme found in enterococcal, streptococcal, and staphylococcal isolates renders them resistant to virtually all clinically available aminoglycoside antibiotics except streptomycin and, to some extent, arbekacin (51, 98-100, 162). The biochemical properties of this intriguing enzyme were investigated recently (9, 98-100). Three other enzymes, APH(2")-Ib, -Ic, and -Id, have recently been identified in enterococcal clinical isolates (53, 141, 300). APH(2")-Ic produce midlevel resistance to gentamicin, tobramycin, and kanamycin, while APH(2")-Ib and APH(2")-Id are responsible for higher levels of resistance to these antibiotics and also produce resistance to netilmicin and dibekacin (51). Although APH(2")-Ib, -Ic, and -Id are not as widely distributed among clinical isolates as the bifunctional enzyme, they all abolish synergism between gentamicin and cell wall-active agents such as ampicillin and vancomycin.
APH(3") and APH(6) modify the 3"- and 6-hydroxyl groups of streptomycin, respectively. Of the two genes for APH(3")-I, aph(3")-Ia was identified in a streptomycin-producing strain of Streptomyces griseus (121). The second gene, aph(3")-Ib, was cloned from a broad-host-range plasmid, RSF1010 (261), that is widely distributed among gram-negative bacteria. Despite the fact that the two enzymes are distributed among unrelated microorganisms, they show 50% amino acid identity and 68% similarity (268). Four enzymes that phosphorylate streptomycin at the 6 position have been identified. The genes for two of them, APH(6)-Ia and -Ib, were cloned from streptomycin producers, Streptomyces griseus and Streptomyces glaucescens (72, 308). The third enzyme, APH(6)-Ic, is encoded by a gene located within transposon Tn5, which is found infrequently in gram-negative bacteria (268).
The gene for APH(6)-Id was cloned from plasmid RSF1010, which was also the source for APH(3")-Ib (261). The genes encoding these two different phosphotransferases (referred to in the literature as strA and strB) are linked and have been reported in both pathogenic and environmental microorganisms. For the sake of consistency, we will call these linked genes aph(3")-Ib-aph(6)-Id, according to the nomenclature proposed by Shaw and colleagues (268). The aph(3")-Ib-aph(6)-Id genes from plant bacteria are usually associated with the Tn5393 or Tn5393-like transposons that reside on large conjugative plasmids, although the small nonconjugative plasmid RSF1010 that carries this tandem set of genes has also been detected in plant pathogens (278). Plant-associated bacteria in which the linked genes were identified include Erwinia amylovora and Erwinia herbicola (50), Pseudomonas syringae (50, 280), and Xanthomonas campestris (278, 281). In contrast to the majority of plant bacteria, the aph(3")-Ib-aph(6)-Id genes from human and animal pathogens were found to be associated with small nonconjugative plasmids of gram-negative microorganisms (73, 145, 279).
Recently, the aph(3")-Ib-aph(6)-Id genes were found on a Tn5393-like transposon from a fish pathogen (154), on an integron from Pseudomonas aeruginosa (208), and on a conjugative transposon-like mobile genetic element from Vibrio cholerae (15, 126). Moreover, the aph(3")-Ib-aph(6)-Id tandem was detected within the Tn5393 transposon on the large plasmid pTP10 from a multiresistant clinical isolate of the gram-positive opportunistic human pathogen Corynebacterium striatum (287). Analysis of the structure of pTP10 suggests that it evolved by acquisition of genetic information from different unrelated species, including gram-negative bacteria, to result in a modular plasmid producing aminoglycoside resistance in a new gram-positive host.
Aminoglycoside phosphotransferases APH(4) and APH(7") produce resistance to hygromycin (108, 324), while APH(9) produces resistance to spectinomycin (171, 282). These enzymes do not present any clinical significance and will not be discussed here in detail. Table 1 summarizes the substrate profiles of the aminoglycoside phosphotransferases.
TABLE 1.
Substrate profiles of aminoglycoside phosphotransferases
| Phosphotransferase | Substrate(s) |
|---|---|
| APH (3′) | |
| I | Kanamycin, neomycin, lividomycin, paromomycin, ribostamycin |
| II | Kanamycin, neomycin, butirosin, paromomycin, ribostamycin |
| III | Kanamycin, neomycin, lividomycin, paromomycin, ribostamycin, butirosin, amikacin, isepamicin |
| IV | Kanamycin, neomycin, butirosin, paromomycin, ribostamycin |
| V | Neomycin, paromomycin, ribostamycin |
| VI | Kanamycin, neomycin, paromomycin, ribostamycin, butirosin amikacin, isepamicin |
| VII | Kanamycin, neomycin |
| APH(2′′) | |
| Ia (bifunctional enzyme) | Kanamycin, gentamicin, tobramycin, sisomicin, dibekacin |
| Ib, Id | Kanamycin, gentamicin, tobramycin, netilmicin, dibekacin |
| Ic | Kanamycin, gentamicin, tobramycin |
| APH(3′′)-Ia, -Ib | Streptomycin |
| APH(7′′)-Ia | Hygromycin |
| APH(4)-Ia, -Ib | Hygromycin |
| APH(6)-Ia, -Ib, -Ic, -Id | Streptomycin |
| APH(9)-Ia, -Ib | Spectinomycin |
Aminoglycoside Acetyltransferases
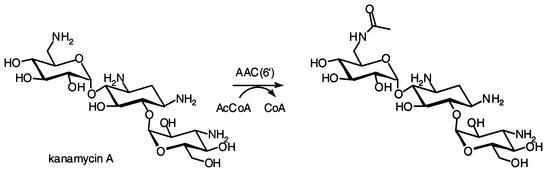
Aminoglycoside acetyltransferases comprise four classes of enzymes: AAC(1), AAC(3), AAC(2′), and AAC(6′). They utilize acetyl coenzyme A as the donor of the acetyl group in modifying aminoglycosides at positions 1 and 3 of the 2-deoxystreptamine ring and positions 2′ and 6′ of the 6-aminohexose ring. This enzymatic reaction is depicted in Fig. 3 for kanamycin A.
FIG. 3.
Acetylation of kanamycin A by AAC(6′)s.
Aminoglycoside 6′-acetyltransferases are broad-spectrum enzymes capable of modifying most of the clinically important aminoglycosides. AAC(6′)-I produces resistance to amikacin, tobramycin, netilmicin, kanamycin, isepamicin, dibekacin, and sisomicin. Genes for at least 24 AAC(6′)-I enzymes have been identified in both gram-negative and gram-positive microorganisms. The only two known AAC(6′)-II enzymes confer resistance to gentamicin, tobramycin, netilmicin, and sisomicin but not to amikacin. Comparison of the amino acid sequences of AAC(6′)s allowed their classification into three subfamilies (116). The first subfamily contains AAC(6′)-Ib, AAC(6′)-Ie, AAC(6′)-IIa, and AAC(6′)-IIb. The second subfamily includes AAC(6′)-Ia, AAC(6′)-Ii, and AAC(6′)-Ip (46, 116, 305). The subsequently discovered AAC(6′)-Iq (46) and AAC(6′)-Id (52) could also be included in this subfamily. The third subfamily originally consisted of AAC(6′)-Ic, AAC(6′)-Id, AAC(6′)-If, AAC(6′)-Ig, AAC(6′)-Ih, AAC(6′)-Ij, and AAC(6′)-Ik, but later AAC(6′)-Il, -Ir, -Is, -It, -Iu, -Iv, -Iw, -Ix, and -Iz were included as well (161).
Of all the known AAC(6′)s, AAC(6′)-Ib is the most prevalent among various gram-negative microorganisms (268, 305). It was identified in more then 70% of all gram-negative bacterial isolates showing the characteristic AAC(6′) resistance profile (268). The gene for AAC(6′)-Ib has been detected in either transposons or integrons (22, 165, 296, 297). It is reasonable to assume that the location of the aac(6′)-Ib gene on mobile genetic elements has facilitated its rapid dissemination in the presence of selective antibiotic pressure among a wide range of microorganisms. The genes for several AAC(6′)-I enzymes have been identified in the chromosomes of gram-negative bacteria. DNA-DNA hybridization experiments demonstrated that the genes for AAC(6′)-Ik from Acinetobacter sp. strain 6, AAC(6′)-If from Acinetobacter sp. strain 13, AAC(6′)-Ig from Acinetobacter haemolyticus, AAC(6′)-Ic from Serratia marcescens, and AAC(6′)-Iz from Stenotrophomonas maltophilia are species specific and are always present in the chromosome, whether or not the resistance phenotype is expressed (157, 159, 161, 249, 269).
The gene for another aminoglycoside 6′-I acetyltransferase, AAC(6′)-Ie, is part of the fused gene for the major determinant of aminoglycoside resistance in staphylococci and enterococci (51, 268). AAC(6′)-Ie is the N-terminal half of the aforementioned bifunctional enzyme AAC(6′)-Ie-APH(2")-Ia, which produces resistance to virtually all aminoglycosides, including gentamicin, amikacin, tobramycin, netilmicin, and kanamycin but not streptomycin (86, 162). The gene for this bifunctional enzyme is a part of the composite transposon Tn4001, which is widely distributedamong gram-positive microorganisms (222). Recently, the aac(6′)-Ie-aph(2")-Ia gene was detected in Lactobacillus and Pediococcus isolates of animal origin (289). All Enterococcus faecium strains produce a chromosomally encoded enzyme AAC(6′)-Ii that confers resistance to tobramycin, sisomicin, netilmicin, and kanamycin (58). Another aminoglycoside 6′-I acetyltransferase, AAC(6′)-Im, has been discovered in both Enterococcus faecium and E. coli (52). Its gene, aac(6′)-Im, exhibits 65% identity in DNA sequence with the aac(6′)-Ie portion of the fused gene encoding the bifunctional enzyme. Moreover, the gene for AAC(6′)-Im was found to be adjacent to that for APH(2")-Ib.
In contrast to the fused genes encoding the bifunctional enzyme, aac(6′)-Im and aph(2")-Ib are separate open reading frames that are probably transcribed from a single promoter. Hybridization experiments revealed the presence of the aac(6′)-Im gene not only in enterococci and E. coli, but also in several other genera of gram-negative bacteria, including Pseudomonas, Klebsiella, Citrobacter, Enterobacter, Serratia, and Aeromonas. These data provide strong evidence for the horizontal transfer of resistance genes between gram-positive and gram-negative bacteria (52).
AAC(6′)-IIa and -IIb were originally detected in Pseudomonas aeruginosa (265) and Pseudomonas fluorescens (268). They show 66% amino acid identity and 78% similarity and confer resistance to gentamicin, tobramycin, netilmicin, sisomicin, and dibekacin. Although in early studies the genes for AAC(6′)-II were detected only rarely, recent surveys indicate that production of these enzymes and the presence of low permeability and efflux pumps have become the major mechanisms of aminoglycoside resistance in clinical isolates of Pseudomonas aeruginosa (188, 218). Recently, the gene for a new bifunctional enzyme, ANT(3")-Ii-AAC(6′)-IId, was identified in an integron from a clinical isolate of Serratia marcescens (47). The aac(6′)-IId portion of the gene is identical to the previously identified aac(6′)-Ib′ acetyltransferase from Pseudomonas aeruginosa (160) and differs by only one base pair from the gene for aac(6′)-Ib. This difference results in the single Leu83Ser substitution in the enzyme, which in turn abolishes resistance to amikacin and changes the specificity of the AAC(6′)-I enzyme to that of AAC(6′)-II (160, 241). Therefore, despite the classification of the acetyltransferase activity from Serratia marcescens as AAC(6′)-II by its conferred resistance phenotype, at the amino acid level this enzyme is closer to AAC(6′)-Ib.
Two other aminoglycoside resistance genes, aacA29a and aacA29b, have recently been located in an integron from Pseudomonas aeruginosa, although the G+C content value of 56% suggests that the genes may have originated from a different microorganism (230). The two enzymes differed by only four amino acids, and they showed only 35% and 34% identity with the most closely related AAC(6′)-Il. Because of an apparent truncation of their carboxyl termini, these enzymes are 13 to 22 amino acids shorter than the more typical AAC(6′)s. They produce resistance to amikacin, tobramycin, kanamycin, dibekacin, and isepamicin and were named AAC(6′)-29a and -29b.
AAC(3)s are widely distributed among different bacterial genera, including aminoglycoside producers. They constitute the second largest group of aminoglycoside acetyltransferases and are the second most common mechanism of aminoglycoside acetyltransferase-mediated aminoglycoside resistance in clinical isolates after the AAC(6′)-I enzymes.
AAC(3)-I enzymes produce a narrow spectrum of resistance that includes gentamicin, sisomicin, and fortimicin. The genes for two enzymes, AAC(3)-Ia and AAC(3)-Ib, were identified alone or together in up to 30% of clinical isolates of gram-negative bacteria (188, 269, 292, 316). Comparison of the amino acid sequences of AAC(3)-Ia (316) and -Ib (262) demonstrated that they are 76% identical and 84% similar. The gene for AAC(3)-Ia was detected on conjugative plasmids and transposons and within gene cassettes in integrons from enterobacteria and Pseudomonas aeruginosa (47, 230, 232, 298). The aac(3)-Ib gene has recently been found fused to another aminoglycoside resistance gene, aac(6′)-Ib, located in an integron from Pseudomonas aeruginosa (78).
Three aac(3)-II genes encoding AAC(3)-IIa, -IIb, and -IIc have been identified. The aac(3)-IIc gene shows 97% identity and the aac(3)-IIb shows 72% identity with the aac(3)-IIa gene (240; S. Vakulenko, H. Entina, and S. Navashin, presentation at 17th Int. Congr. Chemother., 1991). AAC(3)-II enzymes produce resistance to gentamicin, tobramycin, sisomicin, netilmicin, and dibekacin. This resistance pattern had earlier been designated AAC(3)-III (5) and AAC(3)-V (13, 240), but later was renamed AAC(3)-II because all the genes turned out to be identical (268). The AAC(3)-II acetyltransferase is commonly seen in various clinical isolates of gram-negative bacteria (188, 218, 268). The frequency of the AAC(3)-II phenotype varies among different genera, from 18% in Pseudomonas spp. to 60% in some other gram-negative bacteria. It was demonstrated that 85% of the bacteria that show the AAC(3)-II phenotype produce AAC(3)-IIa and only 6% produce AAC(3)-IIb (268).
Three other AAC(3)s from clinical isolates, AAC(3)-III, AAC(3)-IV, and AAC(3)-VI, are uncommon. AAC(3)-IIIa, -IIIb, and -IIIc produce resistance to gentamicin, tobramycin, sisomicin, kanamycin, neomycin, paromomycin, lividomycin, and dibekacin and were identified in Pseudomonas aeruginosa (21, 268, 307). AAC(3)-IV acetylates gentamicin, tobramycin, netilmicin, sisomicin, apramycin, and dibekacin and has been observed, although rarely, in clinically important microorganisms (5, 48). Curiously, a gene encoding an enzyme that is more then 70% identical to AAC(3)-IV has been identified on the chromosome of the nitrogen-fixing bacterium Sinorhizobium meliloti (42). The AAC(3)-VI enzyme produces resistance to gentamicin and occurs rarely among clinical isolates. The gene for AAC(3)-VI has been cloned from a conjugative plasmid of Enterobacter cloacae and shown to have 50% amino acid identity with the gene for AAC(3)-IIa (239, 268). The genes for AAC(3)-VII, -VIII, -IX, and -X have been discovered in aminoglycoside-producing actinomycetes.
AAC(1) produces resistance to apramycin, paromomycin, lividomycin, and ribostamycin and has been identified in animal isolates of E. coli (119, 169). Probably because AAC(1) enzymes produce no clinically important resistance, the genes for these acetyltransferases have not been cloned, so the distribution of AAC(1) among clinical isolates has not been studied.
Representatives of the AAC(2′)-I subclass of aminoglycoside acetyltransferases are unique because they have been shown to be chromosomally encoded and universally present in the species in which they were first identified. The gene for AAC(2′)-Ia has been isolated from Providencia stuartii (242). Production of the AAC(2′)-Ia enzyme alone or, more often, in combination with other enzymes is the major mechanism of resistance to gentamicin, tobramycin, netilmicin, dibekacin, and neomycin in Providencia species (188). The gene for AAC(2′)-Ia is present in all clinical isolates of Providencia stuartii and is normally expressed at low levels (187, 242). Mutants producing clinically significant levels of aminoglycoside resistance were shown to have substantially increased levels of aac(2")-Ia mRNA. Subsequent studies demonstrated that regulation of AAC(2′)-Ia is an extremely complex process, involving at least seven regulatory genes acting in at least two pathways (172, 173).
Providencia stuartii, like many other microorganisms, has acetylated peptidoglycan. Mutants of Providencia stuartii that express aac(2")-Ia at various levels show not only different sensitivities to aminoglycosides but also altered cell morphologies due to changes in the degree of O-acetylation of peptidoglycan (223-225). These data and the observation that AAC(2′)-Ia can use components of O-acetylated peptidoglycan to acetylate gentamicin led to a proposal that this enzyme plays some role in peptidoglycan biosynthesis (223). In essence, it was proposed that the enzyme carries out peptidoglycan O-acetylation, and it would appear that acetylation of aminoglycosides is an adventitious activity (242). It has been argued that the enzyme and “acetate” are transported to the periplasm, where the peptidoglycan is modified. The absence of correlation between the kinetic parameters for AAC(2′)-Ia with the resistance phenotype for different aminoglycosides has been interpreted as evidence for lack of importance of these enzymes as resistance determinants (93). We hasten to add that AAC(2′)-Ia is not a major peptidoglycan O-acetyltransferase in Providencia stuartii, because in the absence of the enzyme, 42% of peptidoglycan is still O-acetylated, compared to 54% in the parental wild-type strain (224). It is plausible that AAC(2′)-Ia is actually cytoplasmic, like all its kin, where acetyl coenzyme A would be available to it. Since peptidoglycan is largely assembled inside the cytoplasm, it is likely that the enzyme may acetylate the components of the cell wall in this milieu, before their export to the periplasmic space for final cell wall assembly.
The genes for AAC(2′)-Ib, -Ic, -Id, and -Ie have been detected in mycobacterial species (1, 2). These enzymes exhibit 63 to 79% homology and are universally present in mycobacteria. Evaluation of the kinetic parameters for purified AAC(2′)-Ic from Mycobacterium tuberculosis with various aminoglycosides as the substrate demonstrated that this enzyme is able to perform both N- and O-acetylation of the antibiotics (120). In contrast to AAC(2′)-Ia in Providencia stuartii, AAC(2′)-I enzymes in mycobacteria do not produce detectable resistance to aminoglycoside antibiotics, and their function remains unknown. Table 2 summarizes the substrate profiles of the aminoglycoside acetyltransferases.
TABLE 2.
Substrate profiles of aminoglycoside acetyltransferases
| Acetyltransferase | Substrate(s) |
|---|---|
| AAC(6′) | |
| I (at least 24 different enzymes) | Tobramycin, amikacin, netilmicin, dibekacin, sisomicin, kanamycin, isepamicin |
| II | Tobramycin, gentamicin, netilmicin, dibekacin, sisomicin, kanamycin |
| AAC(3) | |
| Ia, Ib | Gentamicin, sisomicin, fortimicin |
| IIa, IIb, IIc | Tobramycin, gentamicin, netilmicin, dibekacin, sisomicin |
| IIIa, IIIb, IIIc | Tobramycin, gentamicin, dibekacin, sisomicin, kanamycin, neomycin, paromomycin, lividomycin |
| IV | Tobramycin, gentamicin, netilmicin, dibekacin, sisomicin, apramycin |
| VII | Gentamicin |
| AAC(1) | Paromomycin, lividomycin, ribostamycin, apramycin |
| AAC(2′)-Ia | Tobramycin, gentamicin, netilmicin, dibekacin, neomycin |
Aminoglycoside Nucleotidyltransferases
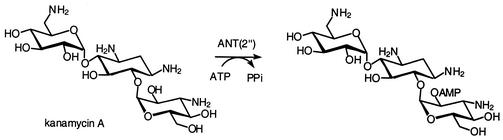
Aminoglycoside nucleotidyltransferases (ANTs) comprise five classes, ANT(2"), ANT(3"), ANT(4′), ANT(6), and ANT(9). They utilize ATP as the second substrate and modify aminoglycoside antibiotics by transferring AMP to their hydroxyl group at positions 2", 3", 4′, 6, and 9, respectively. Figure 4 depicts the reaction of ANT(2") with kanamycin A.
FIG. 4.
Adenylation of kanamycin A by ANT(2").
ANT(2")-Ia produces resistance to gentamicin, tobramycin, sisomicin, dibekacin, and kanamycin (41). The gene for ANT(2")-Ia is widespread among all gram-negative bacteria, but the frequency of its detection varies from one country to another. The ant(2")-Ia gene was found within various genetic backgrounds, including small nonconjugative plasmids (43, 263), conjugative plasmids (43), and various transposons and integrons (36, 47, 199, 229). Studies of the epidemiology of aminoglycoside resistance in different parts of the world clearly demonstrate a relationship between the type of aminoglycosides used and the prevalence in bacteria of a particular aminoglycoside-modifying enzyme. Therefore, it is not surprising that in the United States, where gentamicin accounts for about 80% of all aminoglycoside use whereas amikacin accounted for only 10%, the gene for ANT(2")-Ia was detected in 81% of all aminoglycoside-resistant enterobacteria. In contrast, in Japan, where amikacin has been the most commonly used aminoglycoside, the gene for ANT(2")-Ia was detected in less then 50% of all clinical isolates studied (188).
ANT(3")-I confers resistance to streptomycin by modifying its 3"-hydroxyl group and to spectinomycin by modifying it at position 9 (70, 127). At least eight individual ant(3")-I genes have been identified (221). The ANT(3")-I enzymes show between 59% and 95% amino acid sequence identity and are widely distributed among gram-negative microorganisms. For example, ANT(3")-Ia was detected in more than 90% of streptomycin-resistant clinical isolates (266). The ant(3")-Ia gene was detected within transposons and various plasmids in gram-negative bacteria (49, 87, 127, 258) and the gram-positive bacteria Staphylococcus aureus (60) and Corynebacterium glutamicum (210). Also, class I integrons harboring ant(3")-I genes have been identified frequently among various clinical isolates of Enterobacteriaceae (26, 165, 182, 253, 313), Pseudomonas aeruginosa (207), and Vibrio cholerae (66-68, 84). Recent detection of the ant(3")-Ia gene within integrons on the large conjugative plasmid of an Enterococcus clinical isolate (54) is of obvious concern, because streptomycin is used for synergistic therapy of serious enterococcal infections when high-level resistance to gentamicin is encountered.
ANT(4′)-Ia produces resistance to amikacin, tobramycin, dibekacin, isepamicin, and kanamycin, while ANT(4′)-IIa produces resistance to amikacin, tobramycin, isepamicin, and kanamycin (139, 254). The ant(4′)-Ia genes (often referred to in the literature as aadD) were cloned from the plasmids of both Staphylococcus aureus and thermophilic bacilli and turned out to be almost identical except for one base pair difference (180, 203). Recently, this gene was detected on the large conjugative plasmid from Staphylococcus aureus (20) and as a part of the mec region from methicillin-resistant staphylococci (138, 277). It is the predominant mechanism of aminoglycoside resistance in Japan among strains of Staphylococcus aureus that produce aminoglycoside-modifying enzymes (136) and has been identified alone or in combination with other enzymes in more then 50% of MRSA in Europe (259). This enzyme is also responsible for aminoglycoside resistance in enterococci (44, 259). In contrast to ANT(4′)-I, ANT(4′)-IIa is found only among gram-negative bacteria, including many representatives of the Enterobacteriaceae and Pseudomonas spp. (267). The gene for ANT(4′)-IIa adenyltransferase was originally cloned from Pseudomonas aeruginosa (139), and its nucleotide sequence has been determined (267). No homology was found between ANT(4′)-IIa and ANT(4′)-Ia.
ANT(6)-I produces resistance to streptomycin, and the ant(6)-Ia gene has been found by DNA-DNA hybridization in more then 80% of enterococcal and staphylococcal clinical isolates in Europe (217). In a more recent study in Japan, enterococcal strains were investigated by PCR for the presence of genes for aminoglycoside-modifying enzymes, and the gene for ANT(6)-Ia was identified in almost 50% of Enterococcus faecalis and Enterococcus faecium isolates (149). Most of these isolates produced other aminoglycoside-modifying enzymes, rendering the strains resistant to almost all clinically available aminoglycoside antibiotics. Another two genes encoding chromosomal aminoglycoside 6-adenylyltransferase were identified in Bacillus subtilis (214, 274) and Bacillus halodurans (286). The amino acid sequence of the enzyme from Bacillus subtilis is 58% identical and 74% similar to that of ANT(6)-Ia, while the enzyme from Bacillus halodurans is 41% identical and 56% similar to ANT(6)-Ia. The similarity between two Bacillus enzymes is even lower.
The ANT(9)-I adenyltransferase is limited to Staphylococcus aureus (205), produces resistance only to spectinomycin, and therefore has limited clinical importance. Table 3 summarizes the substrate profiles of the aminoglycoside nucleotidyltransferases.
TABLE 3.
Substrate profiles of aminoglycoside nucleotidyltransferases
| Nucleotidyltransferase | Substrate(s) |
|---|---|
| ANT(2′′)-I | Tobramycin, gentamicin, dibekacin, sisomicin, kanamycin |
| ANT(3′)-I | Streptomycin, spectinomycin |
| ANT(4′)-Ia | Tobramycin, amikacin, dibekacin, kanamycin, isepamicin |
| ANT(4′)-IIa | Tobramycin, amikacin, kanamycin, isepamicin |
| ANT(6′)-I | Streptomycin |
| ANT(9)-I | Spectinomycin |
EPIDEMIOLOGY OF AMINOGLYCOSIDE RESISTANCE
The resistance of clinical isolates to aminoglycoside antibiotics varies with the specific drug, the microorganism, its mechanism of resistance, the geographic area, and many other factors. Resistance to aminoglycosides is generally associated with enzymatic modification of the antibiotic. One exception is the high proportion of clinical isolates of Pseudomonas aeruginosa that exhibit low-level aminoglycoside resistance as a result of diminished penetration of the drug into the cell. Furthermore, in Mycobacterium tuberculosis, the principal mechanism of aminoglycoside resistance is mutational alteration of the ribosomal target. Selection of microorganisms producing aminoglycoside-modifying enzymes depends on the amount of antibiotic usage in particular hospitals and the standards of clinical treatment within various countries.
In 1997, the SENTRY surveillance program was established to monitor predominant pathogens and antimicrobial resistance patterns of nosocomial and community-acquired infections over a broad network of hospitals in the United States, Canada, South America, and Europe (228). Analysis of more then 4,000 clinical isolates from patients with bloodstream infections in the United States revealed that the four most common gram-negative bacteria (E. coli, Klebsiella spp., Pseudomonas aeruginosa, and Enterobacter spp.) were quite susceptible to the aminoglycosides tested: 96 to 100% to amikacin, 90 to 96% to gentamicin, and 94 to 97% to tobramycin (228). Among gram-positive bacteria, 14% of Staphylococcus aureus strains and 40% of coagulase-negative staphylococci were resistant to gentamicin (228).
In a similar study performed under the European SENTRY program, the in vitro activities of several aminoglycoside antibiotics against more then 7,000 clinical isolates from 20 university hospitals were evaluated (260). The susceptibility to amikacin, gentamicin, and tobramycin among E. coli was 99.6%, 95.4%, and 95.7%, respectively; among Klebsiella spp., it was 95.6%, 88.1%, and 85.8%, respectively; among Pseudomonas aeruginosa isolates, 91.8%, 78.8%, and 80.8%, respectively; and among Enterobacter spp., it was 95.8%, 90.1%, and 82.1%, respectively. Among clinical isolates belonging to Acinetobacter spp., only 58%, 43%, and 60% were susceptible to amikacin, gentamicin, and tobramycin, respectively, whereas the susceptibility levels among isolates of Stenotrophomonas maltophilia for the same antibiotics were 76%, 54%, and 58%, respectively. Furthermore, only 7% of methicillin-susceptible Staphylococcus aureus (MSSA) and 17% of methicillin-susceptible coagulase-negative staphylococci were resistant to gentamicin. Resistance levels for gentamicin among MRSA, methicillin-resistant coagulase-negative staphylococci, and Enterococcus spp. reached 80%, 62%, and 39%, respectively. Forty-nine percent of Enterococcus isolates also exhibited high-level resistance to streptomycin. Comparison with previous surveillance studies (19, 75, 76) revealed that during a 10- to 15-year period, resistance to gentamicin in Europe increased up to 5% among some gram-negative bacterial species and more then 10% in Staphylococcus aureus (260).
When trends in the antimicrobial susceptibility of more than 6,000 Pseudomonas aeruginosa isolates were monitored between 1997 and 1999 in various regions of the world, decreases in susceptibility to aminoglycosides were seen over the 3-year period in Europe (from 89% to 79% for amikacin and from 76% to 68% for tobramycin) and Latin America (from 78% to 70% for amikacin and from 68% to 64% for tobramycin). At the same time, the susceptibility of Pseudomonas aeruginosa to amikacin and tobramycin in the United Stated remained the same and increased somewhat in Canada (96). A similar study with 5,000 enterococcal isolates from the United States, Canada, Latin America, Europe, and the Asia-Pacific region demonstrated that from 14% to 40% of them were highly resistant to gentamicin and from 28% to 45% were highly resistant to streptomycin (170). Isolates from Latin America were more susceptible to aminoglycosides than isolates from other regions.
The epidemiology of aminoglycoside resistance is becoming more complex, in part because of the multitude of aminoglycoside-modifying enzymes that exist for these antibiotics and also from the presence of disparate additional mechanisms for antibiotic resistance other than enzymatic resistance determinants. Based on the variations in intensity and patterns of aminoglycoside usage, different combinations of aminoglycoside-modifying enzymes have been selected in clinical strains in different countries and sometimes even in different hospitals within a given country. Because the genes for the aminoglycoside-modifying enzymes are often located on plasmids or transposons, together with the genes encoding resistance to other classes of antibacterials, the total consumption of nonaminoglycoside antibiotics can also significantly influence the epidemiological features of aminoglycoside resistance.
Despite the increasing complexity of mechanisms for resistance to antibiotics, the epidemiology for resistance to aminoglycosides, particularly that of gram-negative bacteria, have been studied extensively. Seminal contributions have been made by researchers at the Schering-Plough Corp., who developed the methods for determination of aminoglycoside resistance mechanisms in gram-negative microorganisms. These methods involve disk susceptibility testing of the microorganisms for 12 selected aminoglycosides followed by subsequent DNA hybridization with up to 19 different aminoglycoside resistance gene probes (189, 293). Comparison of the data on mechanisms of aminoglycoside resistance in bacteria isolated from various regions of the world over a period of almost 30 years revealed that they became more complex over time (188). This complexity is the result of combinations in individual bacterial isolates of various plasmid-mediated aminoglycoside-modifying enzymes. Additionally, chromosomal enzymes in Providencia stuartii and Serratia marcescens and permeability resistance in Pseudomonas aeruginosa contribute further to the complexity of resistance in these microorganisms.
Analysis of data on the use of various aminoglycoside antibiotics in different countries and regions of the world indicates that a correlation exists between the selective pressure of antibiotics and the patterns of combinations of aminoglycoside resistance mechanisms (188). For example, in the 1970s and 1980s, gentamicin was the most frequently used aminoglycoside (around 80%) in the United States, while amikacin was used more heavily in Japan. During the same time period, the principal mechanisms of aminoglycoside resistance in the United States were production of ANT(2")-I, an enzyme that confers resistance to gentamicin, tobramycin, kanamycin, and dibekacin but not to amikacin, and AAC(3)-I (resistance to gentamicin), whereas in Japan, in addition to ANT(2")-I, AAC(6′)-I (produces resistance to amikacin, netilmicin, tobramycin, dibekacin, and kanamycin but not to gentamicin) was identified in 50% of isolates (236, 270).
More recent surveys have demonstrated that broadening of aminoglycoside resistance spectra to include most of the clinically available aminoglycosides, such as gentamicin, tobramycin, amikacin, and netilmicin, occurred in countries and hospitals where these antibiotics were used more extensively (218). In Belgium, France, and Greece, where amikacin has been used more extensively than in other European countries, the incidence of AAC(6′)-I, an enzyme that produces resistance to amikacin, was also much higher. In contrast, gentamicin accounted for about 80% of all the aminoglycosides consumed in Germany, and the major resistance mechanism was ANT(2")-I, followed by AAC(3)-IV, enzymes that produce resistance to gentamicin (188). Despite the existence of more than 50 aminoglycoside-modifying enzymes, only several of them, ANT(2")-I, AAC(6′)-I, and, to a lesser extent, AAC(3)-I, AAC(3)-II, AAC(3)-III, AAC(3)-IV, and AAC(3)-VI, have been selected in gram-negative bacteria to produce the majority of aminoglycoside resistance.
The epidemiology of aminoglycoside resistance has also been studied in gram-positive bacteria such as staphylococci and streptococci, although much less extensively. Three types of enzymes, bifunctional AAC(6′)-I-APH(2"), ANT(4′)-I, and APH(3′)-III, are of particular significance because they produce resistance to clinically important aminoglycosides. AAC(6′)-I-APH(2") produces resistance to gentamicin, tobramycin, and amikacin among other aminoglycosides, ANT(4′)-I to tobramycin and amikacin, and APH(3′)-III to amikacin. Earlier surveys of aminoglycoside resistance in clinical isolates from Europe demonstrated that the bifunctional AAC(6′)-I-APH(2") enzyme was the most common in staphylococci, occurring in 87 to 91% of the resistant isolates tested (74, 293). This is not surprising if we consider that gentamicin was the most commonly used aminoglycoside, and neither ANT(4′)-I nor APH(3′)-III produces gentamicin resistance.
In a recent European survey, 363 staphylococcal isolates resistant to at least one aminoglycoside antibiotic were screened by multiplex PCR for the presence of the genes for the same three aminoglycoside-modifying enzymes. Although the occurrence of AAC(6′)-I-APH(2") among isolates was still very high (68% of all isolates), the proportions of two other enzymes in staphylococci, ANT(4′)-I and APH(3′)-III, increased significantly, to 48% and 14%, respectively, in comparison with earlier studies (259). The bifunctional AAC(6′)-I-APH(2") enzyme was most prevalent among MRSA isolates and was found in 76% of MRSA, 67% of methicillin-resistant, coagulase-negative staphylococci, and only 50% of MSSA and 32% of methicillin-sensitive, coagulase-negative staphylococci. The same was true for the ANT(4′)-I resistance mechanism. The gene for ANT(4′)-I was detected in 53% of MRSA and 42% of MSSA. At the same time, the least common aminoglycoside-modifying enzyme, APH(3′)-III, was found more frequently in MSSA, and only 7% of MRSA harbored the gene for this enzyme (259).
In a similar study of the distribution of these three genes among MRSA, MSSA, and coagulase-negative staphylococci from Kuwait, the gene for AAC(6′)-I-APH(2") was detected in all but two gentamicin-resistant isolates, always in combination with the genes for ANT(4′)-I and APH(3′)-III (302). In contrast to the European isolates, the most prevalent aminoglycoside-modifying enzyme in 381 MRSA isolates from Japanese hospitals was ANT(4′)-I (136). The gene for this enzyme was identified in almost 85% of all MRSA strains. The frequencies of occurrence of the genes for the other two enzymes, 62% for AAC(6′)-I-APH(2") and 9% for APH(3′)-III, were similar to those detected in MRSA strains from Europe. Production of AAC(6′)-I-APH(2") alone or in combination with ANT(4′)-I and/or APH(3′)-III renders MRSA resistant to all aminoglycosides in clinical use. A recent study in Japan (136) confirmed excellent activity of arbekacin, a derivative of dibekacin, against aminoglycoside-resistant staphylococci (113, 137). Among aminoglycoside-resistant MRSA isolates, only 7% that produced AAC(6′)-I-APH(2"), 6% that produced AAC(6′)-I-APH(2") in combination with APH(3′)-III, and 12% that produced the bifunctional enzyme in combination with ANT(4′)-I were resistant to arbekacin.
All enterococci are intrinsically resistant to aminoglycosides due to the facultative anaerobic mechanism that results in impaired uptake of aminoglycosides. Combination with inhibitors of cell wall synthesis, such as β-lactams or vancomycin, results in synergistic effects, in part because of increased aminoglycoside uptake. Gentamicin is the most commonly used aminoglycoside in the United States in combination with cell wall inhibitors for synergistic treatment of enterococcal infections. Production by enterococci of enzymes that modify aminoglycoside antibiotics has important consequences for antibiotic therapy of enterococcal infections. Among at least 10 aminoglycoside-modifying enzymes that were identified in enterococci (51, 52), the bifunctional AAC(6′)-I-APH(2") has the greatest clinical importance because it produces resistance to virtually all aminoglycosides except streptomycin (86, 162). Therefore, only streptomycin can be used in combination therapy against AAC(6′)-I-APH(2")-producing strains, provided that they are not also highly resistant to streptomycin. Based on the phenotype produced by the bifunctional enzyme, the current methods for predicting synergism of cell wall inhibitors with aminoglycosides requires only susceptibility testing for high-level resistance to gentamicin and streptomycin.
Three other enzymes, APH(2")-Ib, APH(2")-Ic, and APH(2")-Id, which produce gentamicin resistance, have recently been identified in enterococci. These enzymes also produce resistance to various aminoglycosides but not to streptomycin and amikacin (51). Therefore, amikacin can be used successfully in synergistic combinations against streptomycin-resistant enterococci producing APH(2")-Ib, APH(2")-Ic, and APH(2")-Id but not AAC(6′)-I-APH(2"). It should also be mentioned that arbekacin (an aminoglycoside that is not used clinically in the United States) is active against strains producing APH(2")-Ib, -Ic, and -Id and against 40% of isolates producing the bifunctional enzyme (51). Identification by PCR of aminoglycoside-modifying enzymes in 121 enterococcal isolates highly resistant to gentamicin revealed the gene for the AAC(6′)-I-APH(2") enzyme in 79% and the genes for APH(2")-Ib, APH(2")-Ic, and APH(2")-Id enzymes in 5%, 1.6%, and 14%, respectively, of the isolates that were studied (51, 141, 300). The appearance of these new gentamicin-modifying enzymes in enterococci requires reevaluation of the current strategy for predicting synergistic combinations with aminoglycosides.
The presence of the genes for seven aminoglycoside-modifying enzymes in 279 clinical isolates of enterococci from a university hospital in Japan was studied recently (149). Almost half of the isolates produced ANT(6)-Ia, the streptomycin-modifying enzyme. The gene for the bifunctional enzyme AAC(6′)-I-APH(2") was detected in 42.5% of Enterococcus faecalis and only 4.3% of Enterococcus faecium isolates. Almost half of Enterococcus faecalis and Enterococcus faecium isolates produced APH(3′)-IIIa, an enzyme that produces resistance to amikacin and kanamycin but not to other aminoglycosides. The high incidence of APH(3′)-IIIa in Japan can be explained by the extensive use of amikacin in that country.
STRATEGIES TO COUNTER RESISTANCE
As discussed in this report, the primary mechanism of resistance to aminoglycosides is enzymatic modification of these drugs in the resistant organisms. In principle, if the resistance mechanisms were to be inhibited, the antibacterial activity of various aminoglycosides could be restored. This concept is inspired by the successful introduction into clinical use of the first inhibitor of β-lactamases, enzymes that produce resistance to β-lactam antibiotics (164, 247). The combination of the inhibitor, clavulanic acid, and amoxicillin, a penicillin, has been marketed for two decades. The clinical benefit of this strategy led to other β-lactamase inhibitor-penicillin combinations (38). The success of the approach depends on the high prevalence of class A β-lactamases, enzymes that are inhibited by clavulanic acid, among bacterial pathogens.
There are no clinical success stories for inhibition of aminoglycoside resistance enzymes. For that matter, not many inhibitors of these enzymes are known. The difficulty arises from the presence of so many different enzymes to inhibit and also from the fact that the biochemical mechanism of many of these enzymes has not been studied in any meaningful detail. The mechanisms for many of these enzymes are still unknown (10). Nonetheless, a number of recent studies have attempted to circumvent the activities of aminoglycoside resistance enzymes. The majority of these studies have been carried out with aminoglycoside phosphotransferases, which have been scrutinized in more enzymological detail.
Most of the studies on inhibitors of aminoglycoside-modifying enzymes have centered on aminoglycoside-based molecules. Indeed, if the site of the enzymatic modification in the aminoglycoside is altered, one would in principle have a nonsubstrate molecule that retains affinity for binding to the active sites of these enzymes. For example, tobramycin and dibekacin, both of which lack the 3′-hydroxyl moiety, are competitive inhibitors for APH(3′)s (184). Similarly, paromomycin and lividomycin A, both of which have 6′-hydroxyl groups instead of amines, are competitive inhibitors of AAC(6′)s (318). 6′-Methylated derivatives of kanamycins are also effective against organisms that harbor AAC(6′)s, presumably because the site of transfer of the acetyl group is now more hindered and cannot serve its function effectively (303).
Roestamadji et al. reported the first mechanism-based inhibitors for the APH(3′)s (inhibitors 1 and 2) (246). These compounds (1 and 2) were based on the structures of neamine and kanamycin B, respectively (Fig. 5). Each possesses a nitro (NO2) group in place of an amine at the 2′ position. These compounds serve as substrates for the APH(3′)s, but upon transfer of the phosphate (species 3), the phosphate is eliminated rapidly to generate a reactive entity (species 4) that covalently and irreversibly modifies the active sites of the enzymes.
FIG. 5.
Mechanism-based inhibition of APH(3′)s.
Inhibitors of protein kinases have been shown to inhibit APH(3′)s (37, 65). The realization that these inhibitors might also affect aminoglycoside phosphotransferases was made when the Wright and Berghuis laboratories noted that the X-ray structure of APH(3′)-IIIa was very similar to that of eukaryotic protein kinases, especially in the ATP-binding domain (128). The inhibitors bind to the ATP-binding domains of these proteins. Furthermore, bisubstrate analogues have been explored as inhibitors of these enzymes (166). The concept behind the inhibitors was to occupy both subsites, the aminoglycoside- and ATP-binding subsites, within the active site of these two-substrate enzymes. A set of molecules that covalently linked an aminoglycoside to adenosine were shown to inhibit APH(3′) in a competitive manner (166). A set of dimeric aminoglycosides have been reported recently (276). These molecules bind to the A site of the ribosome, where they manifest their antibacterial activity, but since their structures have been altered dramatically, they are not recognized by several of the aminoglycoside-modifying enzymes (276). This is a promising strategy to counter the resistant phenotype.
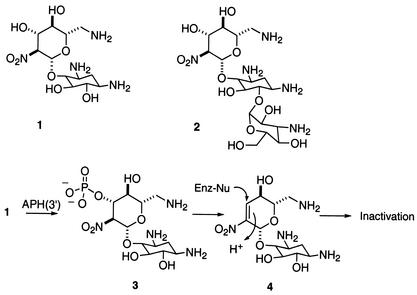
Two alternative strategies to overcoming aminoglycoside inactivation have been explored recently (110, 246). One is based on the principle that interactions between substrates and enzymes at sites remote from the seat of the reaction would significantly influence the catalytic facility of the reaction (246). Accordingly, removal of a series of amines throughout the framework of the structures for neamine and kanamycin B showed considerable attenuation in the catalytic prowess of APH(3′)s against these antibiotics, despite the fact that the hydroxyl that serves as the recipient of the phosphate group in these enzymatic reactions had not been altered (246).
Another strategy exploited the catalytic reaction of APH(3′)s in turnover of a novel molecule (5) that was engineered to accept the phosphate group and immediately release it into the milieu (Fig. 6) (110). In essence, the resistance enzyme would be converted into an ATP hydrolase, as ATP is consumed to give rise to the formation of ADP and inorganic phosphate. This strategy is depicted below. In lieu of a hydroxyl group in position 3′, a ketone was introduced into the structure of kanamycin A, which prefers to be hydrated (species 6). The hydrated species serves as a substrate to APH(3′) to give rise to species 7, which is inherently unstable and releases the inorganic phosphate from the antibiotic. Hence, the antibiotic is not inactivated as a phosphorylated derivative.
FIG. 6.
Cyclic process of phosphorylation and release of inorganic phosphate for derivative 5 of kanamycin A.
CONCLUDING REMARKS
Aminoglycoside antibiotics have been available for over 50 years. Although their clinical use has been extensive, their toxicity and the prevalence of resistance in clinical strains have prompted the pharmaceutical industry to look for alternatives. Whereas the search for novel targets for antibiotics from the genomic information is ongoing (6, 115, 152, 195, 237), no antibacterial agent based on these efforts has so far entered clinical trials. Meanwhile, structural knowledge of the ribosome, the target for aminoglycosides, has invigorated the field of antibiotic development. It is expected that knowledge of the binding interactions of aminoglycosides and the ribosome would lead to concepts in drug design that would take us away from the parental structures of aminoglycosides in the direction of different structural classes that bind to the same ribosomal target sites as aminoglycosides. Indeed, one such example was reported recently (110). Only time will tell if these investigations will prove fruitful.
REFERENCES
- 1.Ainsa, J. A., C. Martin, B. Gicquel, and R. Gomez-Lus. 1996. Characterization of the chromosomal aminoglycoside 2′-N-acetyltransferase gene from Mycobacterium fortuitum. Antimicrob. Agents Chemother. 40:2350-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ainsa, J. A., E. Perez, V. Pelicic, F. X. Berthet, B. Gicquel, and C. Martin. 1997. Aminoglycoside 2′-N-acetyltransferase genes are universally present in mycobacteria: characterization of the aac(2′)-Ic gene from Mycobacterium tuberculosis and the aac(2′)-Id gene from Mycobacterium smegmatis. Mol. Microbiol. 24:431-441. [DOI] [PubMed] [Google Scholar]
- 3.Aires, J. R., T. Kohler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akova, M., D. Gur, D. M. Livermore, T. Kocagoz, and H. E. Akalin. 1999. In vitro activities of antibiotics alone and in combination against Brucella melitensis at neutral and acidic pHs. Antimicrob. Agents Chemother. 43:1298-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allmansberger, R., B. Brau, and W. Piepersberg. 1985. Genes for gentamicin-(3)-N-acetyl-transferases III and IV. II. Nucleotide sequences of three AAC(3)-III genes and evolutionary aspects. Mol. Gen. Genet. 198:514-520. [DOI] [PubMed] [Google Scholar]
- 6.Allsop, A. E. 1998. New antibiotic discovery, novel screens, novel targets and impact of microbial genomics. Curr. Opin. Microbiol. 1:530-534. [DOI] [PubMed] [Google Scholar]
- 7.Aoki, Y. 1994. Bactericidal activity of arbekacin against methicillin-resistant Staphylococcus aureus. Comparison with that of vancomycin. Jpn. J. Antibiot. 47:640-646. [PubMed] [Google Scholar]
- 8.Ashtekar, D., N. Duzgunes, and P. R. Gangadharam. 1991. Activity of free and liposome encapsulated streptomycin against Mycobacterium avium complex (MAC) inside peritoneal macrophages. J. Antimicrob. Chemother. 28:615-617. [DOI] [PubMed] [Google Scholar]
- 9.Azucena, E., I. Grapsas, and S. Mobashery. 1997. Properties of a bifunctional bacterial antibiotic resistance enzyme that catalyzes ATP-dependent 2"-phosphorylation and acetyl coenzyme A-dependent 6′-acetylation of aminoglycosides. J. Am. Chem. Soc. 119:2317-2318. [Google Scholar]
- 10.Azucena, E., and S. Mobashery. 2001. Aminoglycoside-modifying enzymes: mechanisms of catalytic processes and inhibition. Drug Resist. Update 4:106-117. [DOI] [PubMed] [Google Scholar]
- 11.Baker, C. N., D. G. Hollis, and C. Thornsberry. 1985. Antimicrobial susceptibility testing of Francisella tularensis with a modified Mueller-Hinton broth. J. Clin. Microbiol. 22:212-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 13.Barg, N. L. 1988. Construction of a probe for the aminoglycoside 3-V-acetyltransferase gene and detection of the gene among endemic clinical isolates. Antimicrob. Agents Chemother. 32:1834-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barza, M., M. Stuart, and F. Szoka, Jr. 1987. Effect of size and lipid composition on the pharmacokinetics of intravitreal liposomes. Investig. Ophthalmol. Vis. Sci. 28:893-900. [PubMed] [Google Scholar]
- 15.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck, E., G. Ludwig, E. A. Auerswald, B. Reiss, and H. Schaller. 1982. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene 19:327-336. [DOI] [PubMed] [Google Scholar]
- 17.Begg, E. J., B. A. Peddie, S. T. Chambers, and D. R. Boswell. 1992. Comparison of gentamicin dosing regimens using an in-vitro model. J. Antimicrob. Chemother. 29:427-433. [DOI] [PubMed] [Google Scholar]
- 18.Belgian Isepamicin Multicentre Study Group. 2001. Comparative in vitro activity of isepamicin and other antibiotics against Gram-negative bacilli from intensive care units in Belgium. Acta Clin. Belg. 56:307-315. [DOI] [PubMed] [Google Scholar]
- 19.Bengtsson, S., S. Bernander, J. E. Brorson, K. Dornbusch, A. Forsgren, H. Hallander, C. Henning, S. Holm, A. S. Malmborg, and L. Nilsson. 1986. In vitro aminoglycoside resistance of gram-negative bacilli and staphylococci isolated from blood in Sweden, 1980-1984. Scand. J. Infect. Dis. 18:257-263. [DOI] [PubMed] [Google Scholar]
- 20.Berg, T., N. Firth, S. Apisiridej, A. Hettiaratchi, A. Leelaporn, and R. A. Skurray. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J. Bacteriol. 180:4350-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biddlecome, S., M. Haas, J. Davies, G. H. Miller, D. F. Rane, and P. J. Daniels. 1976. Enzymatic modification of aminoglycoside antibiotics: a new 3-N-acetylating enzyme from a Pseudomonas aeruginosa isolate. Antimicrob. Agents Chemother. 9:951-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bissonnette, L., and P. H. Roy. 1992. Characterization of In0 of Pseudomonas aeruginosa plasmid pVS1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria. J. Bacteriol. 174:1248-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boerlin, P., A. P. Burnens, J. Frey, P. Kuhnert, and J. Nicolet. 2001. Molecular epidemiology and genetic linkage of macrolide and aminoglycoside resistance in Staphylococcus intermedius of canine origin. Vet. Microbiol. 79:155-169. [DOI] [PubMed] [Google Scholar]
- 24.Boltner, D., C. MacMahon, J. T. Pembroke, P. Strike, and A. M. Osborn. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184:5158-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouadloun, F., D. Donner, and C. G. Kurland. 1983. Codon-specific missense errors in vivo. EMBO J. 2:1351-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyd, D., A. Cloeckaert, E. Chaslus-Dancla, and M. R. Mulvey. 2002. Characterization of variant Salmonella genomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob. Agents Chemother. 46:1714-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brodersen, D. E., W. M. Clemons, Jr., A. P. Carter, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143-1154. [DOI] [PubMed] [Google Scholar]
- 28.Brodersen, D. E., W. M. Clemons, Jr., A. P. Carter, B. T. Wimberly, and V. Ramakrishnan. 2002. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 316:725-768. [DOI] [PubMed] [Google Scholar]
- 29.Brumfitt, W., A. Percival, and D. A. Leigh. 1967. Clinical and laboratory studies with carbenicillin. A new penicillin active against Pseudomonas pyocyanea. Lancet 1:1289-1293. [DOI] [PubMed] [Google Scholar]
- 30.Bryan, L. E. 1984. Aminoglycoside resistance, p. 241-277. In L. E. Bryan (ed.), Antimicrobial drug resistance. Academic Press, Orlando, Fla.
- 31.Bryan, L. E., S. K. Kowand, and H. M. van den Elzen. 1979. Mechanism of aminoglycoside antibiotic resistance in anaerobic bacteria: Clostridium perfringens and Bacteroides fragilis. Antimicrob. Agents Chemother. 15:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bryan, L. E., and H. M. van den Elzen. 1977. Effects of membrane-energy mutations and cations on streptomycin and gentamicin accumulation by bacteria: a model for entry of streptomycin and gentamicin in susceptible and resistant bacteria. Antimicrob. Agents Chemother. 12:163-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bulger, R. J. 1967. Antibiotic combination in staphylococcal infections. Br. Med. J. 4:805.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulger, R. J. 1967. In-vitro activity of cephalothin/kanamycin and methicillin/kanamycin combinations against methicillin-resistant Staphylococcus aureus. Lancet 1:17-19. [DOI] [PubMed] [Google Scholar]
- 35.Bundtzen, R. W., A. U. Gerber, D. L. Cohn, and W. A. Craig. 1981. Post-antibiotic suppression of bacterial growth. Rev. Infect. Dis. 3:28-37. [DOI] [PubMed] [Google Scholar]
- 36.Bunny, K. L., R. M. Hall, and H. W. Stokes. 1995. New mobile gene cassettes containing an aminoglycoside resistance gene, aacA7, and a chloramphenicol resistance gene, catB3, in an integron in pBWH301. Antimicrob. Agents Chemother. 39:686-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burk, D., and A. Berghuis. 2002. Protein kinase inhibitors and antibiotic resistance. Pharmacol. Ther. 93:283-292. [DOI] [PubMed] [Google Scholar]
- 38.Bush, K., and S. Mobashery. 1998. How β-lactamases have driven pharmaceutical drug discovery: from mechanistic knowledge to clinical circumvention, p. 71-98. In B. P. Rosen and S. Mobashery (ed.), Resolving the antibiotic paradox. Kluwer Akademic/Plenum Publishers, New York, N.Y. [PubMed]
- 39.Calderwood, S. A., C. Wennersten, R. C. Moellering, Jr., L. J. Kunz, and D. J. Krogstad. 1977. Resistance to six aminoglycosidic aminocyclitol antibiotics among enterococci: prevalence, evolution, and relationship to synergism with penicillin. Antimicrob. Agents Chemother. 12:401-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calderwood, S. B., C. Wennersten, and R. C. Moellering, Jr. 1981. Resistance to antibiotic synergism in Streptococcus faecalis: further studies with amikacin and with a new amikacin derivative, 4′-deoxy-6′-N-methylamikacin. Antimicrob. Agents Chemother. 19:549-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cameron, F. H., D. J. Groot Obbink, V. P. Ackerman, and R. M. Hall. 1986. Nucleotide sequence of the AAD(2") aminoglycoside adenylyltransferase determinant aadB. Evolutionary relationship of this region with those surrounding aadA in R538-1 and dhfrII in R388. Nucleic Acids Res. 14:8625-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Puhler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carattoli, A., F. Tosini, W. P. Giles, M. E. Rupp, S. H. Hinrichs, F. J. Angulo, T. J. Barrett, and P. D. Fey. 2002. Characterization of plasmids carrying CMY-2 from expanded-spectrum cephalosporin-resistant Salmonella strains isolated in the United States between 1996 and 1998. Antimicrob. Agents Chemother. 46:1269-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carlier, C., and P. Courvalin. 1990. Emergence of 4′,4"-aminoglycoside nucleotidyltransferase in enterococci. Antimicrob. Agents Chemother. 34:1565-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carter, A. P., W. M. Clemons, D. E. Brodersen, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340-348. [DOI] [PubMed] [Google Scholar]
- 46.Centron, D., and P. H. Roy. 1998. Characterization of the 6′-N-aminoglycoside acetyltransferase gene aac(6′)-Iq from the integron of a natural multiresistance plasmid. Antimicrob. Agents Chemother. 42:1506-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Centron, D., and P. H. Roy. 2002. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob. Agents Chemother. 46:1402-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaslus-Dancla, E., Y. Glupcznski, G. Gerbaud, M. Lagorce, J. P. Lafont, and P. Courvalin. 1989. Detection of apramycin resistant Enterobacteriaceae in hospital isolates. FEMS Microbiol. Lett. 52:261-265. [DOI] [PubMed] [Google Scholar]
- 49.Chinault, A. C., V. A. Blakesley, E. Roessler, D. G. Willis, C. A. Smith, R. G. Cook, and R. G. Fenwick, Jr. 1986. Characterization of transferable plasmids from Shigella flexneri 2a that confer resistance to trimethoprim, streptomycin, and sulfonamides. Plasmid 15:119-131. [DOI] [PubMed] [Google Scholar]
- 50.Chiou, C. S., and A. L. Jones. 1993. Nucleotide sequence analysis of a transposon (Tn5393) carrying streptomycin resistance genes in Erwinia amylovora and other gram-negative bacteria. J. Bacteriol. 175:732-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chow, J. W. 2000. Aminoglycoside resistance in enterococci. Clin. Infect. Dis. 31:586-589. [DOI] [PubMed] [Google Scholar]
- 52.Chow, J. W., V. Kak, I. You, S. J. Kao, J. Petrin, D. B. Clewell, S. A. Lerner, G. H. Miller, and K. J. Shaw. 2001. Aminoglycoside resistance genes aph(2")-Ib and aac(6′)-Im detected together in strains of both Escherichia coli and Enterococcus faecium. Antimicrob. Agents Chemother. 45:2691-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chow, J. W., M. J. Zervos, S. A. Lerner, L. A. Thal, S. M. Donabedian, D. D. Jaworski, S. Tsai, K. J. Shaw, and D. B. Clewell. 1997. A novel gentamicin resistance gene in Enterococcus. Antimicrob. Agents Chemother. 41:511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark, N. C., O. Olsvik, J. M. Swenson, C. A. Spiegel, and F. C. Tenover. 1999. Detection of a streptomycin/spectinomycin adenylyltransferase gene (aadA) in Enterococcus faecalis. Antimicrob. Agents Chemother. 43:157-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cohn, D. L., F. Bustreo, and M. C. Raviglione. 1997. Drug-resistant tuberculosis: review of the worldwide situation and the W.H.O./IUATLD Global Surveillance Project. International Union Against Tuberculosis and Lung Disease. Clin. Infect. Dis. 24(Suppl. 1):S121-S130. [DOI] [PubMed] [Google Scholar]
- 56.Comber, K. R., M. J. Basker, C. D. Osborne, and R. Sutherland. 1977. Synergy between ticarcillin and tobramycin against Pseudomonas aeruginosa and Enterobacteriaceae in vitro and in vivo. Antimicrob. Agents Chemother. 11:956-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cordeiro, J. C., A. O. Reis, E. A. Miranda, and H. S. Sader. 2001. In vitro antimicrobial activity of the aminoglycoside arbekacin tested against oxacillin-resistant Staphylococcus aureus isolated in Brazilian hospitals. Braz. J. Infect. Dis. 5:130-135. [DOI] [PubMed] [Google Scholar]
- 58.Costa, Y., M. Galimand, R. Leclercq, J. Duval, and P. Courvalin. 1993. Characterization of the chromosomal aac(6′)-Ii gene specific for Enterococcus faecium. Antimicrob. Agents Chemother. 37:1896-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Couch, L. A. 2001. Treatment with tobramycin solution for inhalation in bronchiectasis patients with Pseudomonas aeruginosa. Chest 120(Suppl.):114S-117S. [DOI] [PubMed]
- 60.Courvalin, P., and M. Fiandt. 1980. Aminoglycoside-modifying enzymes of Staphylococcus aureus: expression in Escherichia coli. Gene 9:247-269. [DOI] [PubMed] [Google Scholar]
- 61.Craig, W. A., and S. C. Ebert. 1990. Killing and regrowth of bacteria in vitro: a review. Scand. J. Infect. Dis. 74:63-70. [PubMed] [Google Scholar]
- 62.Craig, W. A., and S. Gudmundsson. 1996. Postantibiotic effect, p. 296-329. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. Williams and Wilkins, Baltimore, Md.
- 63.Craig, W. A., and B. Vogelman. 1987. The postantibiotic effect. Ann. Intern. Med. 106:900-902. [DOI] [PubMed] [Google Scholar]
- 64.Cundliffe, E. 1989. How antibiotic-producing organisms avoid suicide. Annu. Rev. Microbiol. 43:207-233. [DOI] [PubMed] [Google Scholar]
- 65.Daigle, D. M., G. A. McKay, and G. D. Wright. 1997. Inhibition of aminoglycoside antibiotic resistance enzymes by protein kinase inhibitors. J. Biol. Chem. 272:24755-24758. [DOI] [PubMed] [Google Scholar]
- 66.Dalsgaard, A., A. Forslund, D. Sandvang, L. Arntzen, and K. Keddy. 2001. Vibrio cholerae O1 outbreak isolates in Mozambique and South Africa in 1998 are multiple-drug resistant, contain the SXT element and the aadA2 gene located on class 1 integrons. J. Antimicrob. Chemother. 48:827-838. [DOI] [PubMed] [Google Scholar]
- 67.Dalsgaard, A., A. Forslund, O. Serichantalergs, and D. Sandvang. 2000. Distribution and content of class 1 integrons in different Vibrio cholerae O-serotype strains isolated in Thailand. Antimicrob. Agents Chemother. 44:1315-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dalsgaard, A., A. Forslund, N. V. Tam, D. X. Vinh, and P. D. Cam. 1999. Cholera in Vietnam: changes in genotypes and emergence of class I integrons containing aminoglycoside resistance gene cassettes in Vibrio cholerae O1 strains isolated from 1979 to 1996. J. Clin. Microbiol. 37:734-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Damper, P. D., and W. Epstein. 1981. Role of the membrane potential in bacterial resistance to aminoglycoside antibiotics. Antimicrob. Agents Chemother. 20:803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davies, J., and D. I. Smith. 1978. Plasmid-determined resistance to antimicrobial agents. Annu. Rev. Microbiol. 32:469-518. [DOI] [PubMed] [Google Scholar]
- 71.Davis, B. D. 1987. Mechanism of bactericidal action of aminoglycosides. Microbiol. Rev. 51:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Distler, J., A. Ebert, K. Mansouri, K. Pissowotzki, M. Stockmann, and W. Piepersberg. 1987. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 15:8041-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dixon, L. G., W. L. Albritton, and P. J. Willson. 1994. An analysis of the complete nucleotide sequence of the Haemophilus ducreyi broad-host-range plasmid pLS88. Plasmid 32:228-232. [DOI] [PubMed] [Google Scholar]
- 74.Dornbusch, K. 1987. In vitro susceptibility to aminoglycoside antibiotics in blood and urine isolates consecutively collected in twenty-nine European laboratories. European Study Group on Antibiotic Resistance. Eur. J. Clin. Microbiol. 6:378-385. [DOI] [PubMed] [Google Scholar]
- 75.Dornbusch, K., S. Bengtsson, J. E. Brorson, H. Fritz, C. Henning, G. Kronvall, P. Larsson, A. S. Malmborg, M. Thore, and A. Tarnvik. 1988. Susceptibility to β-lactam antibiotics and gentamicin of gram-negative bacilli isolated from hospitalized patients: a Swedish multicenter study. Scand. J. Infect. Dis. 20:641-647. [DOI] [PubMed] [Google Scholar]
- 76.Dornbusch, K., G. H. Miller, R. S. Hare, and K. J. Shaw. 1990. Resistance to aminoglycoside antibiotics in gram-negative bacilli and staphylococci isolated from blood. Report from a European collaborative study. The European Study Group on Antibiotic Resistance. J. Antimicrob. Chemother. 26:131-144. [DOI] [PubMed] [Google Scholar]
- 77.Drusano, G. L. 1988. Role of pharmacokinetics in the outcome of infections. Antimicrob. Agents Chemother. 32:289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dubois, V., L. Poirel, C. Marie, C. Arpin, P. Nordmann, and C. Quentin. 2002. Molecular characterization of a novel class 1 integron containing bla (GES-1) and a fused product of aac3-Ib/aac6′-Ib′ gene cassettes in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:638-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ebert, S. C., and W. A. Craig. 1990. Pharmacodynamic properties of antibiotics: application to drug monitoring and dosage regimen design. Infect. Control Hosp. Epidemiol. 11:319-326. [DOI] [PubMed] [Google Scholar]
- 80.Eliopoulos, G. M., and C. T. Eliopoulos. 1988. Antibiotic combinations: should they be tested? Clin. Microbiol. Rev. 1:139-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Eliopoulos, G. M., B. F. Farber, B. E. Murray, C. Wennersten, and R. C. Moellering, Jr. 1984. Ribosomal resistance of clinical enterococcal isolates to streptomycin. Antimicrob. Agents Chemother. 25:398-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eliopoulos, G. M., and R. C. Moellering, Jr. 1996. Antimicrobial combinations, p. 330-396. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. Williams and Wilkins, Baltimore, Md.
- 83.Enderlin, G., L. Morales, R. F. Jacobs, and J. T. Cross. 1994. Streptomycin and alternative agents for the treatment of tularemia: review of the literature. Clin. Infect. Dis. 19:42-47. [DOI] [PubMed] [Google Scholar]
- 84.Falbo, V., A. Carattoli, F. Tosini, C. Pezzella, A. M. Dionisi, and I. Luzzi. 1999. Antibiotic resistance conferred by a conjugative plasmid and a class I integron in Vibrio cholerae O1 El Tor strains isolated in Albania and Italy. Antimicrob. Agents Chemother. 43:693-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fantin, B., S. Ebert, J. Leggett, B. Vogelman, and W. A. Craig. 1991. Factors affecting duration of in-vivo postantibiotic effect for aminoglycosides against gram-negative bacilli. J. Antimicrob. Chemother. 27:829-836. [DOI] [PubMed] [Google Scholar]
- 86.Ferretti, J. J., K. S. Gilmore, and P. Courvalin. 1986. Nucleotide sequence analysis of the gene specifying the bifunctional 6′-aminoglycoside acetyltransferase 2"-aminoglycoside phosphotransferase enzyme in Streptococcus faecalis and identification and cloning of gene regions specifying the two activities. J. Bacteriol. 167:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fling, M. E., J. Kopf, and C. Richards. 1985. Nucleotide sequence of the transposon Tn7 gene encoding an aminoglycoside-modifying enzyme, 3"-(9)-O-nucleotidyltransferase. Nucleic Acids Res. 13:7095-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fountain, M. W., S. J. Weiss, A. G. Fountain, A. Shen, and R. P. Lenk. 1985. Treatment of Brucella canis and Brucella abortus in vitro and in vivo by stable plurilamellar vesicle-encapsulated aminoglycosides. J. Infect. Dis. 152:529-535. [DOI] [PubMed] [Google Scholar]
- 89.Fourmy, D., M. I. Recht, S. C. Blanchard, and J. D. Puglisi. 1996. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science 274:1367-1371. [DOI] [PubMed] [Google Scholar]
- 90.Fourmy, D., M. I. Recht, and J. D. Puglisi. 1998. Binding of neomycin-class aminoglycoside antibiotics to the A-site of 16 S rRNA. J. Mol. Biol. 277:347-362. [DOI] [PubMed] [Google Scholar]
- 91.Fourmy, D., S. Yoshizawa, and J. D. Puglisi. 1998. Paromomycin binding induces a local conformational change in the A-site of 16 S rRNA. J. Mol. Biol. 277:333-345. [DOI] [PubMed] [Google Scholar]
- 92.Frana, T. S., S. A. Carlson, and R. W. Griffith. 2001. Relative distribution and conservation of genes encoding aminoglycoside-modifying enzymes in Salmonella enterica serotype Typhimurium phage type DT104. Appl. Environ. Microbiol. 67:445-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Franklin, K., and A. J. Clarke. 2001. Overexpression and characterization of the chromosomal aminoglycoside 2′-N-acetyltransferase of Providencia stuartii. Antimicrob. Agents Chemother. 45:2238-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Frean, J. A., L. Arntzen, T. Capper, A. Bryskier, and K. P. Klugman. 1996. In vitro activities of 14 antibiotics against 100 human isolates of Yersinia pestis from a southern African plague focus. Antimicrob. Agents Chemother. 40:2646-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Freeman, C. D., D. P. Nicolau, P. P. Belliveau, and C. H. Nightingale. 1997. Once-daily dosing of aminoglycosides: review and recommendations for clinical practice. J. Antimicrob. Chemother. 39:677-686. [DOI] [PubMed] [Google Scholar]
- 96.Gales, A. C., R. N. Jones, J. Turnidge, R. Rennie, and R. Ramphal. 2001. Characterization of Pseudomonas aeruginosa isolates: occurrence rates, antimicrobial susceptibility patterns, and molecular typing in the global SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32(Suppl. 2):S146-S155. [DOI] [PubMed] [Google Scholar]
- 97.Gangadharam, P. R., D. R. Ashtekar, D. L. Flasher, and N. Duzgunes. 1995. Therapy of Mycobacterium avium complex infections in beige mice with streptomycin encapsulated in sterically stabilized liposomes. Antimicrob. Agents Chemother. 39:725-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gates, C. A., and D. B. Northrop. 1988. Alternative substrate and inhibition kinetics of aminoglycoside nucleotidyltransferase 2"-I in support of a Theorell-Chance kinetic mechanism. Biochemistry 27:3826-3833. [DOI] [PubMed] [Google Scholar]
- 99.Gates, C. A., and D. B. Northrop. 1988. Determination of the rate-limiting segment of aminoglycoside nucleotidyltransferase 2"-I by pH and viscosity-dependent kinetics. Biochemistry 27:3834-3842. [DOI] [PubMed] [Google Scholar]
- 100.Gates, C. A., and D. B. Northrop. 1988. Substrate specificities and structure-activity relationships for the nucleotidylation of antibiotics catalyzed by aminoglycoside nucleotidyltransferase 2"-I. Biochemistry 27:3820-3825. [DOI] [PubMed] [Google Scholar]
- 101.Gilbert, D. N. 2000. Aminoglycosides, p. 307-336. In G. E. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases, vol. 1. Churchill Livingstone, New York, N.Y.
- 102.Gilbert, D. N. 1991. Once-daily aminoglycoside therapy. Antimicrob. Agents Chemother. 35:399-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gillespie, S. H. 2002. Evolution of drug resistance in Mycobacterium tuberculosis: clinical and molecular perspective. Antimicrob. Agents Chemother. 46:267-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Graham, J. C., and F. K. Gould. 2002. Role of aminoglycosides in the treatment of bacterial endocarditis. J. Antimicrob. Chemother. 49:437-444. [DOI] [PubMed] [Google Scholar]
- 105.Gravel, M., P. Melancon, and L. Brakier-Gingras. 1987. Cross-linking of streptomycin to the 16S ribosomal RNA of Escherichia coli. Biochemistry 26:6227-6232. [DOI] [PubMed] [Google Scholar]
- 106.Gray, G. S., and W. M. Fitch. 1983. Evolution of antibiotic resistance genes: the DNA sequence of a kanamycin resistance gene from Staphylococcus aureus. Mol. Biol. Evol. 1:57-66. [DOI] [PubMed] [Google Scholar]
- 107.Green, R., and H. F. Noller. 1997. Ribosomes and translation. Annu. Rev. Biochem. 66:679-716. [DOI] [PubMed] [Google Scholar]
- 108.Gritz, L., and J. Davies. 1983. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene 25:179-188. [DOI] [PubMed] [Google Scholar]
- 109.Hachler, H., P. Santanam, and F. H. Kayser. 1996. Sequence and characterization of a novel chromosomal aminoglycoside phosphotransferase gene, aph(3′)-IIb, in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:1254-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Haddad, J., L. P. Kotra, B. Llano-Sotelo, C. Kim, E. F. Azucena, Jr., M. Liu, S. B. Vakulenko, C. S. Chow, and S. Mobashery. 2002. Design of novel antibiotics that bind to the ribosomal acyltransfer site. J. Am. Chem. Soc. 124:3229-3237. [DOI] [PubMed] [Google Scholar]
- 111.Haddad, J., L. P. Kotra, and S. Mobashery. 2001. Aminoglycoside antibiotics: structures and mechanism of action, p. 307-351. In G. Wang and C. Bertozzi (ed.), Glycochemistry: principles, synthesis, and applications. Marcel Dekker, Inc., New York, N.Y.
- 112.Haddad, J., M. Z. Liu, and S. Mobashery. 2001. Methodologies in syntheses of aminoglycoside antibiotics, p. 353-424. In G. Wang and C. Bertozzi (ed.), Glycochemistry: principles, synthesis, and applications. Marcel Dekker, Inc., New York, N.Y.
- 113.Hamilton-Miller, J. M., and S. Shah. 1995. Activity of the semisynthetic kanamycin B derivative arbekacin against methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 35:865-868. [DOI] [PubMed] [Google Scholar]
- 114.Hancock, R. E. 1984. Alterations in outer membrane permeability. Annu. Rev. Microbiol. 38:237-264. [DOI] [PubMed] [Google Scholar]
- 115.Haney, S. A., L. E. Alksne, P. M. Dunman, E. Murphy, and S. J. Projan. 2002. Genomics in anti-infective drug discovery — getting to endgame. Curr. Pharm. Des. 8:1099-1118. [DOI] [PubMed] [Google Scholar]
- 116.Hannecart-Pokorni, E., F. Depuydt, L. de wit, E. van Bossuyt, J. Content, and R. Vanhoof. 1997. Characterization of the 6′-N-aminoglycoside acetyltransferase gene aac(6′)-Il associated with a sulI-type integron. Antimicrob. Agents Chemother. 41:314-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Harms, J., F. Schluenzen, R. Zarivach, A. Bashan, S. Gat, I. Agmon, H. Bartels, F. Franceschi, and A. Yonath. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107:679-688. [DOI] [PubMed] [Google Scholar]
- 118.Hayashi, I., M. Inoue, and H. Hashimoto. 1994. Nationwide investigation in Japan on the efficacy of arbekacin in methicillin-resistant Staphylococcus aureus infections. Drug. Exp. Clin. Res. 20:225-232. [PubMed] [Google Scholar]
- 119.Hedges, R. W., and K. P. Shannon. 1984. Resistance to apramycin in Escherichia coli isolated from animals: detection of a novel aminoglycoside-modifying enzyme. J. Gen. Microbiol. 130:473-482. [DOI] [PubMed] [Google Scholar]
- 120.Hegde, S. S., F. Javid-Majd, and J. S. Blanchard. 2001. Overexpression and mechanistic analysis of chromosomally encoded aminoglycoside 2′-N-acetyltransferase (AAC(2′)-Ic) from Mycobacterium tuberculosis. J. Biol. Chem. 276:45876-45881. [DOI] [PubMed] [Google Scholar]
- 121.Heinzel, P., O. Werbitzky, J. Distler, and W. Piepersberg. 1988. A second streptomycin resistance gene from Streptomyces griseus codes for streptomycin-3"-phosphotransferase. Relationships between antibiotic and protein kinases. Arch. Microbiol. 150:184-192. [DOI] [PubMed] [Google Scholar]
- 122.Heinzl, B., E. Eber, B. Oberwaldner, G. Haas, and M. S. Zach. 2002. Effects of inhaled gentamicin prophylaxis on acquisition of Pseudomonas aeruginosa in children with cystic fibrosis: a pilot study. Pediatr. Pulmonol. 33:32-37. [DOI] [PubMed] [Google Scholar]
- 123.Herbert, C. J., M. Sarwar, S. S. Ner, I. G. Giles, and M. Akhtar. 1986. Sequence and interspecies transfer of an aminoglycoside phosphotransferase gene (APH) of Bacillus circulans. Self-defence mechanism in antibiotic-producing organisms. Biochem. J. 233:383-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hessen, M. T., P. G. Pitsakis, and M. E. Levison. 1989. Postantibiotic effect of penicillin plus gentamicin versus Enterococcus faecalis in vitro and in vivo. Antimicrob. Agents Chemother. 33:608-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hiramatsu, K. 1998. Vancomycin resistance in staphylococci. Drug Resist. Update 1:135-150. [DOI] [PubMed] [Google Scholar]
- 126.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hollingshead, S., and D. Vapnek. 1985. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid 13:17-30. [DOI] [PubMed] [Google Scholar]
- 128.Hon, W. C., G. A. McKay, P. R. Thompson, R. M. Sweet, D. S. Yang, G. D. Wright, and A. M. Berghuis. 1997. Structure of an enzyme required for aminoglycoside antibiotic resistance reveals homology to eukaryotic protein kinases. Cell 89:887-895. [DOI] [PubMed] [Google Scholar]
- 129.Hopfield, J. J. 1974. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. USA 71:4135-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hoshiko, S., C. Nojiri, K. Matsunaga, K. Katsumata, E. Satoh, and K. Nagaoka. 1988. Nucleotide sequence of the ribostamycin phosphotransferase gene and of its control region in Streptomyces ribosidificus. Gene 68:285-296. [DOI] [PubMed] [Google Scholar]
- 131.Hubner, J., D. Hartung, A. Kropec, and F. D. Daschner. 1991. Combination effect of SCE-2787 and cefepime with aminoglycosides on nosocomial gram-negative bacteria. Infection 19:186-189. [DOI] [PubMed] [Google Scholar]
- 132.Hughes, W. T., D. Armstrong, G. P. Bodey, E. J. Bow, A. E. Brown, T. Calandra, R. Feld, P. A. Pizzo, K. V. Rolston, J. L. Shenep, and L. S. Young. 2002. 2002 guidelines for the use of antimicrobial agents in neutropenic patients with cancer. Clin. Infect. Dis. 34:730-751. [DOI] [PubMed] [Google Scholar]
- 133.Hughes, W. T., D. Armstrong, G. P. Bodey, A. E. Brown, J. E. Edwards, R. Feld, P. Pizzo, K. V. Rolston, J., L. Shenep, and L. S. Young. 1997. 1997 guidelines for the use of antimicrobial agents in neutropenic patients with unexplained fever. I. Clin. Infect. Dis. 25:551-573. [DOI] [PubMed] [Google Scholar]
- 134.Iannini, P. B., J. Ehret, and T. C. Eickhoff. 1976. Effects of ampicillin-amikacin and ampicillin-rifampin on enterococci. Antimicrob. Agents Chemother. 9:448-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ida, T., R. Okamoto, M. Nonoyama, K. Irinoda, M. Kurazono, and M. Inoue. 2002. Antagonism between aminoglycosides and β−lactams in a methicillin-resistant Staphylococcus aureus isolate involves induction of an aminoglycoside-modifying enzyme. Antimicrob. Agents Chemother. 46:1516-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ida, T., R. Okamoto, C. Shimauchi, T. Okubo, A. Kuga, and M. Inoue. 2001. Identification of aminoglycoside-modifying enzymes by susceptibility testing: epidemiology of methicillin-resistant Staphylococcus aureus in Japan. J. Clin. Microbiol. 39:3115-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Inoue, M., M. Nonoyama, R. Okamoto, and T. Ida. 1994. Antimicrobial activity of arbekacin, a new aminoglycoside antibiotic, against methicillin-resistant Staphylococcus aureus. Drug. Exp. Clin. Res. 20:233-239. [PubMed] [Google Scholar]
- 138.Ito, T., Y. Katayama, and K. Hiramatsu. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jacoby, G. A., M. J. Blaser, P. Santanam, H. Hachler, F. H. Kayser, R. S. Hare, and G. H. Miller. 1990. Appearance of amikacin and tobramycin resistance due to 4′-aminoglycoside nucleotidyltransferase [ANT(4′)-II] in gram-negative pathogens. Antimicrob. Agents Chemother. 34:2381-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kak, V., S. M. Donabedian, M. J. Zervos, R. Kariyama, H. Kumon, and J. W. Chow. 2000. Efficacy of ampicillin plus arbekacin in experimental rabbit endocarditis caused by an Enterococcus faecalis strain with high-level gentamicin resistance. Antimicrob. Agents Chemother. 44:2545-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kao, S. J., I. You, D. B. Clewell, S. M. Donabedian, M. J. Zervos, J. Petrin, K. J. Shaw, and J. W. Chow. 2000. Detection of the high-level aminoglycoside resistance gene aph(2")-Ib in Enterococcus faecium. Antimicrob. Agents Chemother. 44:2876-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Karimi, R., and M. Ehrenberg. 1994. Dissociation rate of cognate peptidyl-tRNA from the A-site of hyper-accurate and error-prone ribosomes. Eur. J. Biochem. 226:355-360. [DOI] [PubMed] [Google Scholar]
- 143.Karimi, R., and M. Ehrenberg. 1996. Dissociation rates of peptidyl-tRNA from the P-site of E. coli ribosomes. EMBO J. 15:1149-1154. [PMC free article] [PubMed] [Google Scholar]
- 144.Karlowsky, J. A., G. G. Zhanel, R. J. Davidson, and D. J. Hoban. 1994. Postantibiotic effect in Pseudomonas aeruginosa following single and multiple aminoglycoside exposures in vitro. J. Antimicrob. Chemother. 33:937-947. [DOI] [PubMed] [Google Scholar]
- 145.Kehrenberg, C., and S. Schwarz. 2002. Nucleotide sequence and organization of plasmid pMVSCS1 from Mannheimia varigena: identification of a multiresistance gene cluster. J. Antimicrob. Chemother. 49:383-386. [DOI] [PubMed] [Google Scholar]
- 146.Kim, E. H., and T. Aoki. 1994. The transposon-like structure of IS26 — tetracycline and kanamycin resistance determinant derived from transferable R plasmid of fish pathogen Pasteurella piscicida. Microbiol. Immunol. 38:31-38. [DOI] [PubMed] [Google Scholar]
- 147.Klastersky, J., J. Beumer, and D. Daneau. 1972. Susceptibility of Pseudomonas aeruginosa to combinations of antibiotics. Pathol. Biol. 20:643-647. [PubMed] [Google Scholar]
- 148.Klastersky, J., R. Cappel, G. Swings, and L. Vandenborre. 1971. Bacteriological and clinical activity of the ampicillin-gentamicin and cephalothin-gentamicin combinations. Am. J. Med. Sci. 262:283-290. [DOI] [PubMed] [Google Scholar]
- 149.Kobayashi, N., M. Alam, Y. Nishimoto, S. Urasawa, N. Uehara, and N. Watanabe. 2001. Distribution of aminoglycoside resistance genes in recent clinical isolates of Enterococcus faecalis, Enterococcus faecium and Enterococcus avium. Epidemiol. Infect. 126:197-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Korzeniowski, O. M., C. Wennersten, R. C. Moellering, Jr., and M. A. Sande. 1978. Penicillin-netilmicin synergism against Streptococcus faecalis. Antimicrob. Agents Chemother. 13:430-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kotra, L. P., J. Haddad, and S. Mobashery. 2000. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob. Agents Chemother. 44:3249-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kotra, L. P., S. Vakulenko, and S. Mobashery. 2000. From genes to sequences to antibiotics: prospects for future developments from microbial genomics. Microbes Infect. 2:651-658. [DOI] [PubMed] [Google Scholar]
- 153.Kropec, A., S. Lemmen, M. Wursthorn, and F. D. Daschner. 1994. Combination effect of meropenem with aminoglycosides and teicoplanin on Pseudomonas and enterococci. Infection 22:306-308. [DOI] [PubMed] [Google Scholar]
- 154.L'Abee-Lund, T. M., and H. Sorum. 2000. Functional Tn5393-like transposon in the R plasmid pRAS2 from the fish pathogen Aeromonas salmonicida subspecies salmonicida isolated in Norway. Appl. Environ. Microbiol. 66:5533-5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lacey, R. W., and I. Chopra. 1972. Evidence for mutation to streptomycin resistance in clinical strains of Staphylococcus aureus. J. Gen. Microbiol. 73:175-180. [DOI] [PubMed] [Google Scholar]
- 156.Lambert, T., G. Gerbaud, P. Bouvet, J. F. Vieu, and P. Courvalin. 1990. Dissemination of amikacin resistance gene aphA6 in Acinetobacter spp. Antimicrob. Agents Chemother. 34:1244-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lambert, T., G. Gerbaud, and P. Courvalin. 1994. Characterization of the chromosomal aac(6′)-Ij gene of Acinetobacter sp. strain 13 and the aac(6′)-Ih plasmid gene of Acinetobacter baumannii. Antimicrob. Agents Chemother. 38:1883-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lambert, T., G. Gerbaud, and P. Courvalin. 1988. Transferable amikacin resistance in Acinetobacter spp. due to a new type of 3′-aminoglycoside phosphotransferase. Antimicrob. Agents Chemother. 32:15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lambert, T., G. Gerbaud, M. Galimand, and P. Courvalin. 1993. Characterization of Acinetobacter haemolyticus aac(6′)-Ig gene encoding an aminoglycoside, 6′-N-acetyltransferase, which modifies amikacin. Antimicrob. Agents Chemother. 37:2093-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lambert, T., M. C. Ploy, and P. Courvalin. 1994. A spontaneous point mutation in the aac(6′)-Ib′ gene results in altered substrate specificity of aminoglycoside 6′-N-acetyltransferase of a Pseudomonas fluorescens strain. FEMS Microbiol. Lett. 115:297-304. [DOI] [PubMed] [Google Scholar]
- 161.Lambert, T., M. C. Ploy, F. Denis, and P. Courvalin. 1999. Characterization of the chromosomal aac(6′)-Iz gene of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 43:2366-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Leclercq, R., S. Dutka-Malen, A. Brisson-Noel, C. Molinas, E. Derlot, M. Arthur, J. Duval, and P. Courvalin. 1992. Resistance of enterococci to aminoglycosides and glycopeptides. Clin. Infect. Dis. 15:495-501. [DOI] [PubMed] [Google Scholar]
- 163.Lee, K. Y., J. D. Hopkins, and M. Syvanen. 1991. Evolved neomycin phosphotransferase from an isolate of Klebsiella pneumoniae. Mol. Microbiol. 5:2039-2046. [DOI] [PubMed] [Google Scholar]
- 164.Leigh, D. A., K. Bradnock, and J. M. Marriner. 1981. Augmentin (amoxicillin and clavulanic acid) therapy in complicated infections due to β-lactamase-producing bacteria. J. Antimicrob. Chemother. 7:229-236. [DOI] [PubMed] [Google Scholar]
- 165.Levesque, C., L. Piche, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu, M., J. Haddad, E. Azucena, L. P. Kotra, M. Kirzhner, and S. Mobashery. 2000. Tethered bisubstrate derivatives as probes for mechanism and as inhibitors of aminoglycoside 3′-phosphotransferases. J. Org. Chem. 65:7422-7431. [DOI] [PubMed] [Google Scholar]
- 167.Llano-Sotelo, B., E. F. Azucena, L. P. Kotra, S. Mobashery, and C. S. Chow. 2002. Aminoglycosides modified by resistance enzymes display diminished binding to the bacterial ribosomal aminoacyl-tRNA site. Chem. Biol. 9:455-463. [DOI] [PubMed] [Google Scholar]
- 168.Lodmell, J. S., and A. E. Dahlberg. 1997. A conformational switch in Escherichia coli 16S ribosomal RNA during decoding of messenger RNA. Science 277:1262-1267. [DOI] [PubMed] [Google Scholar]
- 169.Lovering, A. M., L. O. White, and D. S. Reeves. 1987. AAC(1): a new aminoglycoside-acetylating enzyme modifying the Cl aminogroup of apramycin. J. Antimicrob. Chemother. 20:803-813. [DOI] [PubMed] [Google Scholar]
- 170.Low, D. E., N. Keller, A. Barth, and R. N. Jones. 2001. Clinical prevalence, antimicrobial susceptibility, and geographic resistance patterns of enterococci: results from the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32(Suppl. 2):S133-S145. [DOI] [PubMed] [Google Scholar]
- 171.Lyutzkanova, D., J. Distler, and J. Altenbuchner. 1997. A spectinomycin resistance determinant from the spectinomycin producer Streptomyces flavopersicus. Microbiology 143:2135-2143. [DOI] [PubMed] [Google Scholar]
- 172.Macinga, D. R., M. R. Paradise, M. M. Parojcic, and P. N. Rather. 1999. Activation of the 2′-N-acetyltransferase gene [aac(2′)-Ia] in Providencia stuartii by an interaction of AarP with the promoter region. Antimicrob. Agents Chemother. 43:1769-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Macinga, D. R., and P. N. Rather. 1999. The chromosomal 2′-N-acetyltransferase of Providencia stuartii: physiological functions and genetic regulation. Front. Biosci. 4:D132-D140. [DOI] [PubMed] [Google Scholar]
- 174.MacLeod, D. L., and J. F. Prescott. 1988. The use of liposomally entrapped gentamicin in the treatment of bovine Staphylococcus aureus mastitis. Can. J. Vet. Res. 52:445-450. [PMC free article] [PubMed] [Google Scholar]
- 175.Magnet, S., P. Courvalin, and T. Lambert. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45:3375-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Majumdar, S., D. Flasher, D. S. Friend, P. Nassos, D. Yajko, W. K. Hadley, and N. Duzgunes. 1992. Efficacies of liposome-encapsulated streptomycin and ciprofloxacin against Mycobacterium avium-M. intracellulare complex infections in human peripheral blood monocyte/macrophages. Antimicrob. Agents Chemother. 36:2808-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Maness, M. J., G. C. Foster, and P. F. Sparling. 1974. Ribosomal resistance to streptomycin and spectinomycin in Neisseria gonorrhoeae. J. Bacteriol. 120:1293-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Martin, N. L., and T. J. Beveridge. 1986. Gentamicin interaction with Pseudomonas aeruginosa cell envelope. Antimicrob. Agents Chemother. 29:1079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Martin, P., E. Jullien, and P. Courvalin. 1988. Nucleotide sequence of Acinetobacter baumannii aphA-6 gene: evolutionary and functional implications of sequence homologies with nucleotide-binding proteins, kinases and other aminoglycoside-modifying enzymes. Mol. Microbiol. 2:615-625. [DOI] [PubMed] [Google Scholar]
- 180.Matsumura, M., Y. Katakura, T. Imanaka, and S. Aiba. 1984. Enzymatic and nucleotide sequence studies of a kanamycin-inactivating enzyme encoded by a plasmid from thermophilic bacilli in comparison with that encoded by plasmid pUB110. J. Bacteriol. 160:413-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Maurin, M., and D. Raoult. 2001. Use of aminoglycosides in treatment of infections due to intracellular bacteria. Antimicrob. Agents Chemother. 45:2977-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mazel, D., B. Dychinco, V. A. Webb, and J. Davies. 2000. Antibiotic resistance in the ECOR collection: integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 44:1568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.McDonald, P. J., W. A. Craig, and C. M. Kunin. 1977. Persistent effect of antibiotics on Staphylococcus aureus after exposure for limited periods of time. J. Infect. Dis. 135:217-223. [DOI] [PubMed] [Google Scholar]
- 184.McKay, G. A., and G. D. Wright. 1995. Kinetic mechanism of aminoglycoside phosphotransferase type IIIa. Evidence for a Theorell-Chance mechanism. J. Biol. Chem. 270:24686-24692. [DOI] [PubMed] [Google Scholar]
- 185.Mehta, R., and W. S. Champney. 2002. 30S ribosomal subunit assembly is a target for inhibition by aminoglycosides in Escherichia coli. Antimicrob. Agents Chemother. 46:1546-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Melancon, P., C. Lemieux, and L. Brakier-Gingras. 1988. A mutation in the 530 loop of Escherichia coli 16S ribosomal RNA causes resistance to streptomycin. Nucleic Acids Res. 16:9631-9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Miller, G., F. Sabatelli, R. Hare, and J. Waitz. 1980. Survey of aminoglycoside resistance patterns. Dev. Ind. Microbiol. 8:91-104. [Google Scholar]
- 188.Miller, G. H., F. J. Sabatelli, R. S. Hare, Y. Glupczynski, P. Mackey, D. Shlaes, K. Shimizu, and K. J. Shaw. 1997. The most frequent aminoglycoside resistance mechanisms—changes with time and geographic area: a reflection of aminoglycoside usage patterns? Aminoglycoside Resistance Study Groups. Clin. Infect. Dis. 24(Suppl. 1):S46-S62. [DOI] [PubMed] [Google Scholar]
- 189.Miller, G. H., F. J. Sabatelli, L. Naples, R. S. Hare, and K. J. Shaw. 1995. The changing nature of aminoglycoside resistance mechanisms and the role of isepamicin — a new broad-spectrum aminoglycoside. The Aminoglycoside Resistance Study Groups. J. Chemother. 7(Suppl. 2):31-44. [PubMed] [Google Scholar]
- 190.Miller, M. H., S. C. Edberg, L. J. Mandel, C. F. Behar, and N. H. Steigbigel. 1980. Gentamicin uptake in wild-type and aminoglycoside-resistant small-colony mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 18:722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389-394. [DOI] [PubMed] [Google Scholar]
- 192.Moellering, R. C., Jr. 1991. The enterococcus: a classic example of the impact of antimicrobial resistance on therapeutic options. The Garrod Lecture. J. Antimicrob. Chemother. 28:1-12. [DOI] [PubMed] [Google Scholar]
- 193.Moellering, R. C., Jr. 1983. In vitro antibacterial activity of the aminoglycoside antibiotics. Rev. Infect. Dis. 5(Suppl.):S212-S232. [Google Scholar]
- 194.Moellering, R. C., Jr., and A. N. Weinberg. 1971. Studies on antibiotic synergism against enterococci. II. Effect of various antibiotics on the uptake of 14C-labeled streptomycin by enterococci. J. Clin. Investig. 50:2580-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Moir, D. T., K. J. Shaw, R. S. Hare, and G. F. Vovis. 1999. Genomics and antimicrobial drug discovery. Antimicrob. Agents Chemother. 43:439-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Montandon, P. E., R. Wagner, and E. Stutz. 1986. E. coli ribosomes with a C912 to U base change in the 16S rRNA are streptomycin resistant. EMBO J. 5:3705-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Moore, R. A., D. DeShazer, S. Reckseidler, A. Weissman, and D. E. Woods. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43:465-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Moore, R. D., P. S. Lietman, and C. R. Smith. 1987. Clinical response to aminoglycoside therapy: importance of the ratio of peak concentration to minimal inhibitory concentration. J. Infect. Dis. 155:93-99. [DOI] [PubMed] [Google Scholar]
- 199.Morabito, S., R. Tozzoli, A. Caprioli, H. Karch, and A. Carattoli. 2002. Detection and characterization of class 1 integrons in enterohemorrhagic Escherichia coli. Microb. Drug Resist. 8:85-91. [DOI] [PubMed] [Google Scholar]
- 200.Mortensen, J. E., D. G. Moore, J. E. Clarridge, and E. J. Young. 1986. Antimicrobial susceptibility of clinical isolates of Brucella. Diagn. Microbiol. Infect. Dis. 5:163-169. [DOI] [PubMed] [Google Scholar]
- 201.Moss, R. B. 2001. Administration of aerosolized antibiotics in cystic fibrosis patients. Chest 120:107S-113S. [DOI] [PubMed]
- 202.Moss, R. B. 2002. Long-term benefits of inhaled tobramycin in adolescent patients with cystic fibrosis. Chest 121:55-63. [DOI] [PubMed] [Google Scholar]
- 203.Muller, R. E., T. Ano, T. Imanaka, and S. Aiba. 1986. Complete nucleotide sequences of Bacillus plasmids pUB110dB, pRBH1 and its copy mutants. Mol. Gen. Genet. 202:169-171. [DOI] [PubMed] [Google Scholar]
- 204.Murata, T., M. Ohnishi, T. Ara, J. Kaneko, C. G. Han, Y. F. Li, K. Takashima, H. Nojima, K. Nakayama, A. Kaji, Y. Kamio, T. Miki, H. Mori, E. Ohtsubo, Y. Terawaki, and T. Hayashi. 2002. Complete nucleotide sequence of plasmid Rts1: implications for evolution of large plasmid genomes. J. Bacteriol. 184:3194-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Murphy, E. 1985. Nucleotide sequence of a spectinomycin adenyltransferase AAD(9) determinant from Staphylococcus aureus and its relationship to AAD(3")(9). Mol. Gen. Genet. 200:33-39. [DOI] [PubMed] [Google Scholar]
- 206.Musser, J. M. 1995. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 8:496-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Naas, T., L. Poirel, and P. Nordmann. 1999. Molecular characterisation of In51, a class 1 integron containing a novel aminoglycoside adenylyltransferase gene cassette, aadA6, in Pseudomonas aeruginosa. Biochim. Biophys. Acta 1489:445-451. [DOI] [PubMed] [Google Scholar]
- 208.Naas, T., W. Sougakoff, A. Casetta, and P. Nordmann. 1998. Molecular characterization of OXA-20, a novel class D β-lactamase, and its integron from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:2074-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Nakamura, A., M. Hosoda, T. Kato, Y. Yamada, M. Itoh, K. Kanazawa, and H. Nouda. 2000. Combined effects of meropenem and aminoglycosides on Pseudomonas aeruginosa in vitro. J. Antimicrob. Chemother. 46:901-904. [DOI] [PubMed] [Google Scholar]
- 210.Nesvera, J., J. Hochmannova, and M. Patek. 1998. An integron of class 1 is present on the plasmid pCG4 from gram-positive bacterium Corynebacterium glutamicum. FEMS Microbiol. Lett. 169:391-395. [DOI] [PubMed] [Google Scholar]
- 211.Nicolau, D. P., and R. Quintiliani, Jr. 1999. Aminoglycosides, p. 621-638. In V. L. Yu, T. C. Merigan, Jr., and S. L. Barriere (ed.), Antimicrobial therapy and vaccines. Williams and Wilkins, Baltimore, Md.
- 212.Ninio, J. 1975. Kinetic amplification of enzyme discrimination. Biochimie 57:587-595. [DOI] [PubMed] [Google Scholar]
- 213.Ogle, J. M., D. E. Brodersen, W. M. Clemons, Jr., M. J. Tarry, A. P. Carter, and V. Ramakrishnan. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897-902. [DOI] [PubMed] [Google Scholar]
- 214.Ohmiya, K., T. Tanaka, N. Noguchi, K. O'Hara, and M. Kono. 1989. Nucleotide sequence of the chromosomal gene coding for the aminoglycoside 6-adenylyltransferase from Bacillus subtilis Marburg 168. Gene 78:377-378. [DOI] [PubMed] [Google Scholar]
- 215.Oka, A., H. Sugisaki, and M. Takanami. 1981. Nucleotide sequence of the kanamycin resistance transposon Tn903. J. Mol. Biol. 147:217-226. [DOI] [PubMed] [Google Scholar]
- 216.Ouellette, M., G. Gerbaud, T. Lambert, and P. Courvalin. 1987. Acquisition by a Campylobacter-like strain of aphA-1, a kanamycin resistance determinant from members of the family Enterobacteriaceae. Antimicrob. Agents Chemother. 31:1021-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ounissi, H., E. Derlot, C. Carlier, and P. Courvalin. 1990. Gene homogeneity for aminoglycoside-modifying enzymes in gram-positive cocci. Antimicrob. Agents Chemother. 34:2164-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Over, U., D. Gur, S. Unal, and G. H. Miller. 2001. The changing nature of aminoglycoside resistance mechanisms and prevalence of newly recognized resistance mechanisms in Turkey. Clin. Microbiol. Infect. 7:470-478. [DOI] [PubMed] [Google Scholar]
- 219.Owens, R. C., Jr., M. A. Banevicius, D. P. Nicolau, C. H. Nightingale, and R. Quintiliani. 1997. In vitro synergistic activities of tobramycin and selected β-lactams against 75 gram-negative clinical isolates. Antimicrob. Agents Chemother. 41:2586-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Papadopoulou, B., and P. Courvalin. 1988. Dispersal in Campylobacter spp. of aphA-3, a kanamycin resistance determinant from gram-positive cocci. Antimicrob. Agents Chemother. 32:945-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Partridge, S. R., C. M. Collis, and R. M. Hall. 2002. Class 1 integron containing a new gene cassette, aadA10, associated with Tn1404 from R151. Antimicrob. Agents Chemother. 46:2400-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Paulsen, I. T., N. Firth, and R. A. Skurray. 1997. Resistance to antimicrobial agents other than β-lactams., p. 175-212. In K. B. Krossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 223.Payie, K. G., and A. J. Clarke. 1997. Characterization of gentamicin 2′-N-acetyltransferase from Providencia stuartii: its use of peptidoglycan metabolites for acetylation of both aminoglycosides and peptidoglycan. J. Bacteriol. 179:4106-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Payie, K. G., P. N. Rather, and A. J. Clarke. 1995. Contribution of gentamicin 2′-N-acetyltransferase to the O acetylation of peptidoglycan in Providencia stuartii. J. Bacteriol. 177:4303-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Payie, K. G., H. Strating, and A. J. Clarke. 1996. The role of O-acetylation in the metabolism of peptidoglycan in Providencia stuartii. Microb. Drug Resist. 2:135-140. [DOI] [PubMed] [Google Scholar]
- 226.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Peyman, G. A., H. C. Charles, K. R. Liu, B. Khoobehi, and M. Niesman. 1988. Intravitreal liposome-encapsulated drugs: a preliminary human report. Int. Ophthalmol. 12:175-182. [DOI] [PubMed] [Google Scholar]
- 228.Pfaller, M. A., R. N. Jones, G. V. Doern, and K. Kugler. 1998. Bacterial pathogens isolated from patients with bloodstream infection: frequencies of occurrence and antimicrobial susceptibility patterns from the SENTRY Antimicrobial Surveillance Program (United States and Canada, 1997). Antimicrob. Agents Chemother. 42:1762-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Ploy, M. C., F. Denis, P. Courvalin, and T. Lambert. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 44:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Poirel, L., T. Lambert, S. Turkoglu, E. Ronco, J. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the bla (VIM-2) carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Prammananan, T., P. Sander, B. A. Brown, K. Frischkorn, G. O. Onyi, Y. Zhang, E. C. Bottger, and R. J. Wallace, Jr. 1998. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J. Infect. Dis. 177:1573-1581. [DOI] [PubMed] [Google Scholar]
- 232.Preston, K. E., M. A. Kacica, R. J. Limberger, W. A. Archinal, and R. A. Venezia. 1997. The resistance and integrase genes of pACM1, a conjugative multiple-resistance plasmid, from Klebsiella oxytoca. Plasmid 37:105-118. [DOI] [PubMed] [Google Scholar]
- 233.Preston, S. L., and L. L. Briceland. 1995. Single daily dosing of aminoglycosides. Pharmacotherapy 15:297-316. [PubMed] [Google Scholar]
- 234.Price, C. I., J. W. Horton, and C. R. Baxter. 1992. Liposome delivery of aminoglycosides in burn wounds. Surg. Gynecol. Obstet. 174:414-418. [PubMed] [Google Scholar]
- 235.Price, C. I., J. W. Horton, and C. R. Baxter. 1994. Liposome encapsulation: a method for enhancing the effectiveness of local antibiotics. Surgery 115:480-487. [PubMed] [Google Scholar]
- 236.Price, K. E., P. A. Kresel, L. A. Farchione, S. B. Siskin, and S. A. Karpow. 1981. Epidemiological studies of aminoglycoside resistance in the USA J. Antimicrob. Chemother. 8(Suppl. A):89-105. [DOI] [PubMed] [Google Scholar]
- 237.Projan, S. 2002. New (and not so new) antibacterial targets — from where and when will the novel drugs come? Curr. Opin. Pharmacol. 2:513.. [DOI] [PubMed] [Google Scholar]
- 238.Quittner, A. L., and A. Buu. 2002. Effects of tobramycin solution for inhalation on global ratings of quality of life in patients with cystic fibrosis and Pseudomonas aeruginosa infection. Pediatr. Pulmonol. 33:269-276. [DOI] [PubMed] [Google Scholar]
- 239.Rather, P. N., P. A. Mann, R. Mierzwa, R. S. Hare, G. H. Miller, and K. J. Shaw. 1993. Analysis of the aac(3)-VIa gene encoding a novel 3-N-acetyltransferase. Antimicrob. Agents Chemother. 37:2074-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Rather, P. N., R. Mierzwa, R. S. Hare, G. H. Miller, and K. J. Shaw. 1992. Cloning and DNA sequence analysis of an aac(3)-Vb gene from Serratia marcescens. Antimicrob. Agents Chemother. 36:2222-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Rather, P. N., H. Munayyer, P. A. Mann, R. S. Hare, G. H. Miller, and K. J. Shaw. 1992. Genetic analysis of bacterial acetyltransferases: identification of amino acids determining the specificities of the aminoglycoside 6′-N-acetyltransferase Ib and IIa proteins. J. Bacteriol. 174:3196-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Rather, P. N., E. Orosz, K. J. Shaw, R. Hare, and G. Miller. 1993. Characterization and transcriptional regulation of the 2′-N-acetyltransferase gene from Providencia stuartii. J. Bacteriol. 175:6492-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ratjen, F., G. Doring, and W. H. Nikolaizik. 2001. Effect of inhaled tobramycin on early Pseudomonas aeruginosa colonisation in patients with cystic fibrosis. Lancet 358:983-984. [DOI] [PubMed] [Google Scholar]
- 244.Riccio, M. L., L. Pallecchi, R. Fontana, and G. M. Rossolini. 2001. In70 of plasmid pAX22, a bla (VIM-1)-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrob. Agents Chemother. 45:1249-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Rochon-Edouard, S., M. Pestel-Caron, J. F. Lemeland, and F. Caron. 2000. In vitro synergistic effects of double and triple combinations of β-lactams, vancomycin, and netilmicin against methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 44:3055-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Roestamadji, J., and S. Mobashery. 1998. The use of neamine as a molecular template: inactivation of bacterial antibiotic resistance enzyme aminoglycoside 3′-phosphotransferase type IIa. Bioorg. Med. Chem. Lett. 8:3483-3488. [DOI] [PubMed] [Google Scholar]
- 247.Roselle, G. A., R. Bode, B. Hamilton, M. Bibler, R. Sullivan, R. Douce, J. L. Staneck, and W. E. Bullock. 1985. Clinical trial of the efficacy and safety of ticarcillin and clavulanic acid. Antimicrob. Agents Chemother. 27:291-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Rosenberg, E. Y., D. Ma, and H. Nikaido. 2000. AcrD of Escherichia coli is an aminoglycoside efflux pump. J. Bacteriol. 182:1754-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Rudant, E., P. Bourlioux, P. Courvalin, and T. Lambert. 1994. Characterization of the aac(6′)-Ik gene of Acinetobacter sp. 6. FEMS Microbiol. Lett. 124:49-54. [DOI] [PubMed] [Google Scholar]
- 250.Ruusala, T., and C. G. Kurland. 1984. Streptomycin preferentially perturbs ribosomal proofreading. Mol. Gen. Genet. 198:100-104. [DOI] [PubMed] [Google Scholar]
- 251.Salauze, D., and J. Davies. 1991. Isolation and characterisation of an aminoglycoside phosphotransferase from neomycin-producing Micromonospora chalcea: comparison with that of Streptomyces fradiae and other producers of 4,6-disubstituted 2-deoxystreptamine antibiotics. J. Antibiot. (Tokyo) 44:1432-1443. [DOI] [PubMed] [Google Scholar]
- 252.Sande, M. A., and K. B. Courtney. 1976. Nafcillin-gentamicin synergism in experimental staphylococcal endocarditis. J. Lab. Clin. Med. 88:118-124. [PubMed] [Google Scholar]
- 253.Sandvang, D. 1999. Novel streptomycin and spectinomycin resistance gene as a gene cassette within a class 1 integron isolated from Escherichia coli. Antimicrob. Agents Chemother. 43:3036-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Santanam, P., and F. H. Kayser. 1978. Purification and characterization of an aminoglycoside inactivating enzyme from Staphylococcus epidermidis FK109 that nucleotidylates the 4′- and 4"-hydroxyl groups of the aminoglycoside antibiotics. J. Antibiot. (Tokyo) 31:343-351. [DOI] [PubMed] [Google Scholar]
- 255.Schatz, A., and S. A. Waksman. 1944. Effect of streptomycin and other antibiotic substances upon Mycobacterium tuberculosis and related organisms. Proc. Soc. Exp. Biol. Med. 57:244-248. [Google Scholar]
- 256.Schiffelers, R., G. Storm, and I. Bakker-Woudenberg. 2001. Liposome-encapsulated aminoglycosides in preclinical and clinical studies. J. Antimicrob. Chemother. 48:333-344. [DOI] [PubMed] [Google Scholar]
- 257.Schluenzen, F., A. Tocilj, R. Zarivach, J. Harms, M. Gluehmann, D. Janell, A. Bashan, H. Bartels, I. Agmon, F. Franceschi, and A. Yonath. 2000. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 102:615-623. [DOI] [PubMed] [Google Scholar]
- 258.Schmidt, F. R., E. J. Nucken, and R. B. Henschke. 1988. Nucleotide sequence analysis of 2"-aminoglycoside nucleotidyl-transferase ANT(2") from Tn4000: its relationship with AAD(3") and impact on Tn21 evolution. Mol. Microbiol. 2:709-717. [DOI] [PubMed] [Google Scholar]
- 259.Schmitz, F. J., A. C. Fluit, M. Gondolf, R. Beyrau, E. Lindenlauf, J. Verhoef, H. P. Heinz, and M. E. Jones. 1999. The prevalence of aminoglycoside resistance and corresponding resistance genes in clinical isolates of staphylococci from 19 European hospitals. J. Antimicrob. Chemother. 43:253-259. [PubMed] [Google Scholar]
- 260.Schmitz, F. J., J. Verhoef, and A. C. Fluit. 1999. Prevalence of aminoglycoside resistance in 20 European university hospitals participating in the European SENTRY Antimicrobial Surveillance Programme. Eur. J. Clin. Microbiol. 18:414-421. [DOI] [PubMed] [Google Scholar]
- 261.Scholz, P., V. Haring, B. Wittmann-Liebold, K. Ashman, M. Bagdasarian, and E. Scherzinger. 1989. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75:271-288. [DOI] [PubMed] [Google Scholar]
- 262.Schwocho, L. R., C. P. Schaffner, G. H. Miller, R. S. Hare, and K. J. Shaw. 1995. Cloning and characterization of a 3-N-aminoglycoside acetyltransferase gene, aac(3)-Ib, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1790-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Segal, H., and B. G. Elisha. 1997. Identification and characterization of an aadB gene cassette at a secondary site in a plasmid from Acinetobacter. FEMS Microbiol. Lett. 153:321-326. [DOI] [PubMed] [Google Scholar]
- 264.Shanson, D. C. 1998. New guidelines for the antibiotic treatment of streptococcal, enterococcal, and staphylococcal endocarditis. J. Antimicrob. Chemother. 42:292-296. [DOI] [PubMed] [Google Scholar]
- 265.Shaw, K. J., C. A. Cramer, M. Rizzo, R. Mierzwa, K. Gewain, G. H. Miller, and R. S. Hare. 1989. Isolation, characterization, and DNA sequence analysis of an AAC(6′)-II gene from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 33:2052-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Shaw, K. J., R. S. Hare, F. J. Sabatelli, M. Rizzo, C. A. Cramer, L. Naples, S. Kocsi, H. Munayyer, P. Mann, G. H. Miller, et al. 1991. Correlation between aminoglycoside resistance profiles and DNA hybridization of clinical isolates. Antimicrob. Agents Chemother. 35:2253-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Shaw, K. J., H. Munayyer, P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Nucleotide sequence analysis and DNA hybridization studies of the ant(4′)-IIa gene from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 37:708-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Shaw, K. J., P. N. Rather, F. J. Sabatelli, P. Mann, H. Munayyer, R. Mierzwa, G. L. Petrikkos, R. S. Hare, G. H. Miller, P. Bennett, et al. 1992. Characterization of the chromosomal aac(6′)-Ic gene from Serratia marcescens. Antimicrob. Agents Chemother. 36:1447-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Shimizu, K., T. Kumada, W. C. Hsieh, H. Y. Chung, Y. Chong, R. S. Hare, G. H. Miller, F. J. Sabatelli, and J. Howard. 1985. Comparison of aminoglycoside resistance patterns in Japan, Formosa, Korea, Chile, and the United States. Antimicrob. Agents Chemother. 28:282-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Sigmund, C. D., M. Ettayebi, and E. A. Morgan. 1984. Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 12:4653-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Solera, J. 2000. Treatment of human brucellosis. Leban. Med. J. 48:255-263. [PubMed] [Google Scholar]
- 273.Solera, J., J. L. Beato, E. Martinez-Alfaro, J. C. Segura, and E. de Tomas. 2001. Azithromycin and gentamicin therapy for the treatment of humans with brucellosis. Clin. Infect. Dis. 32:506-509. [DOI] [PubMed] [Google Scholar]
- 274.Sorokin, A., A. Bolotin, B. Purnelle, H. Hilbert, J. Lauber, A. Dusterhoft, and S. D. Ehrlich. 1997. Sequence of the Bacillus subtilis genome region in the vicinity of the lev operon reveals two new extracytoplasmic function RNA polymerase sigma factors, SigV and SigZ. Microbiology 143:2939-2943. [DOI] [PubMed] [Google Scholar]
- 275.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 276.Sucheck, S. J., A. L. Wong, K. M. Koeller, D. D. Boehr, K.-A. Draker, P. Sears, G. D. Wright, and C.-H. Wong. 2000. Design of bifunctional antibiotics that target bacterial rRNA and inhibit resistance-causing enzymes. J. Am. Chem. Soc. 122:5230-5231. [Google Scholar]
- 277.Sugiyama, M., K. Yuasa, M. Z. Bhuiyan, Y. Iwai, N. Masumi, and K. Ueda. 1996. IS43 mec-mediated integration of a bleomycin-resistance gene into the chromosome of a methicillin-resistant Staphylococcus aureus strain isolated in Japan. Appl. Microbiol. Biotechnol. 46:61-66. [DOI] [PubMed] [Google Scholar]
- 278.Sundin, G. W. 2000. Examination of base pair variants of the strA-strB streptomycin resistance genes from bacterial pathogens of humans, animals and plants. J. Antimicrob. Chemother. 46:848-849. [DOI] [PubMed] [Google Scholar]
- 279.Sundin, G. W., and C. L. Bender. 1996. Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals, and plants. Mol. Ecol. 5:133-143. [DOI] [PubMed] [Google Scholar]
- 280.Sundin, G. W., and C. L. Bender. 1993. Ecological and genetic analysis of copper and streptomycin resistance in Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 59:1018-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Sundin, G. W., and C. L. Bender. 1995. Expression of the strA-strB streptomycin resistance genes in Pseudomonas syringae and Xanthomonas campestris and characterization of IS6100 in X. campestris. Appl. Environ. Microbiol. 61:2891-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Suter, T. M., V. K. Viswanathan, and N. P. Cianciotto. 1997. Isolation of a gene encoding a novel spectinomycin phosphotransferase from Legionella pneumophila. Antimicrob. Agents Chemother. 41:1385-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Swenson, C. E., K. A. Stewart, J. L. Hammett, W. E. Fitzsimmons, and R. S. Ginsberg. 1990. Pharmacokinetics and in vivo activity of liposome-encapsulated gentamicin. Antimicrob. Agents Chemother. 34:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Taber, H. W., J. P. Mueller, P. F. Miller, and A. S. Arrow. 1987. Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 51:439-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Tadakuma, T., N. Ikewaki, T. Yasuda, M. Tsutsumi, S. Saito, and K. Saito. 1985. Treatment of experimental salmonellosis in mice with streptomycin entrapped in liposomes. Antimicrob. Agents Chemother. 28:28-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Tauch, A., S. Krieft, J. Kalinowski, and A. Puhler. 2000. The 51,409-bp R-plasmid pTP10 from the multiresistant clinical isolate Corynebacterium striatum M82B is composed of DNA segments initially identified in soil bacteria and in plant, animal, and human pathogens. Mol. Gen. Genet. 263:1-11. [DOI] [PubMed] [Google Scholar]
- 288.Taylor, D. E., W. Yan, L. K. Ng, E. K. Manavathu, and P. Courvalin. 1988. Genetic characterization of kanamycin resistance in Campylobacter coli. Ann. Inst. Pasteur Microbiol. 139:665-676. [DOI] [PubMed] [Google Scholar]
- 289.Tenorio, C., M. Zarazaga, C. Martinez, and C. Torres. 2001. Bifunctional enzyme 6′-N-aminoglycoside acetyltransferase-2"-O-aminoglycoside phosphotransferase in Lactobacillus and Pediococcus isolates of animal origin. J. Clin. Microbiol. 39:824-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Tenover, F. C., and P. M. Elvrum. 1988. Detection of two different kanamycin resistance genes in naturally occurring isolates of Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 32:1170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Tenover, F. C., T. Gilbert, and P. O'Hara. 1989. Nucleotide sequence of a novel kanamycin resistance gene, aphA-7, from Campylobacter jejuni and comparison to other kanamycin phosphotransferase genes. Plasmid 22.:52-58. [DOI] [PubMed] [Google Scholar]
- 292.Tenover, F. C., K. L. Phillips, T. Gilbert, P. Lockhart, P. J. O'Hara, and J. J. Plorde. 1989. Development of a DNA probe from the deoxyribonucleotide sequence of a 3-N-aminoglycoside acetyltransferase [AAC(3)-I] resistance gene. Antimicrob. Agents Chemother. 33:551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.The Aminoglycoside Resistance Study Groups. 1995. The most frequently occurring aminoglycoside resistance mechanisms-combined results of surveys in eight regions of the world. J. Chemother. 7(Suppl. 2):17-30. [PubMed] [Google Scholar]
- 294.Thompson, C. J., and G. S. Gray. 1983. Nucleotide sequence of a streptomycete aminoglycoside phosphotransferase gene and its relationship to phosphotransferases encoded by resistance plasmids. Proc. Natl. Acad. Sci. USA 80:5190-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Thompson, J., P. A. Skeggs, and E. Cundliffe. 1985. Methylation of 16S ribosomal RNA and resistance to the aminoglycoside antibiotics gentamicin and kanamycin determined by DNA from the gentamicin-producer. Micromonospora purpurea. Mol. Gen. Genet. 201:168-173. [DOI] [PubMed] [Google Scholar]
- 296.Thungapathra, M., Amita, K. K. Sinha, S. R. Chaudhuri, P. Garg, T. Ramamurthy, G. B. Nair, and A. Ghosh. 2002. Occurrence of antibiotic resistance gene cassettes aac(6′)-Ib, dfrA5, dfrA12, and ereA2 in class I integrons in non-O1, non-O139 Vibrio cholerae strains in India. Antimicrob. Agents Chemother. 46:2948-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Tolmasky, M. E., and J. H. Crosa. 1993. Genetic organization of antibiotic resistance genes (aac(6′)-Ib, aadA, and oxa9) in the multiresistance transposon Tn1331. Plasmid 29:31-40. [DOI] [PubMed] [Google Scholar]
- 298.Tosini, F., P. Visca, I. Luzzi, A. M. Dionisi, C. Pezzella, A. Petrucca, and A. Carattoli. 1998. Class 1 integron-borne multiple-antibiotic resistance carried by IncFI and IncL/M plasmids in Salmonella enterica serotype Typhimurium. Antimicrob. Agents Chemother. 42:3053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Trieu-Cuot, P., and P. Courvalin. 1983. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′,5"-aminoglycoside phosphotransferase type III. Gene 23:331-341. [DOI] [PubMed] [Google Scholar]
- 300.Tsai, S. F., M. J. Zervos, D. B. Clewell, S. M. Donabedian, D. F. Sahm, and J. W. Chow. 1998. A new high-level gentamicin resistance gene, aph(2")-Id, in Enterococcus spp. Antimicrob. Agents Chemother. 42:1229-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Ubukata, K., N. Yamashita, A. Gotoh, and M. Konno. 1984. Purification and characterization of aminoglycoside-modifying enzymes from Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob. Agents Chemother. 25:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Udo, E. E., and A. A. Dashti. 2000. Detection of genes encoding aminoglycoside-modifying enzymes in staphylococci by polymerase chain reaction and dot blot hybridization. Int. J. Antimicrob. Agents 13:273-279. [DOI] [PubMed] [Google Scholar]
- 303.Umezawa, H., Y. Nishimura, T. Tsuchiya, and S. Umezawa. 1972. Syntheses of 6′-N-methyl-kanamycin and 3′,4′-dideoxy-6′-N-methylkanamycin B active against resistant strains having 6′-N-acetylating enzymes. J. Antibiot. (Tokyo) 25:743-745. [DOI] [PubMed] [Google Scholar]
- 304.Reference deleted
- 305.Vanhoof, R., E. Hannecart-Pokorni, and J. Content. 1998. Nomenclature of genes encoding aminoglycoside-modifying enzymes. Antimicrob. Agents Chemother. 42:483.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Vicens, Q., and E. Westhof. 2001. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure (Cambridge) 9:647-658. [DOI] [PubMed] [Google Scholar]
- 307.Vliegenthart, J. S., P. A. Ketelaar-van Gaalen, and J. A. van de Klundert. 1991. Nucleotide sequence of the aacC3 gene, a gentamicin resistance determinant encoding aminoglycoside-(3)-N-acetyltransferase III expressed in Pseudomonas aeruginosa but not in Escherichia coli. Antimicrob. Agents Chemother. 35:892-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Vogtli, M., and R. Hutter. 1987. Characterisation of the hydroxystreptomycin phosphotransferase gene (sph) of Streptomyces glaucescens: nucleotide sequence and promoter analysis. Mol. Gen. Genet. 208:195-203. [DOI] [PubMed] [Google Scholar]
- 309.Watanakunakorn, C. 1986. Effects of inoculum size on the activity of carboxy- and ureido-penicillins and effects of combinations of ureido-penicillins with aminoglycosides against resistant Pseudomonas aeruginosa. J. Antimicrob. Chemother. 17:91-95. [DOI] [PubMed] [Google Scholar]
- 310.Watanakunakorn, C., and C. Glotzbecker. 1979. In vitro activity of carbenicillin, ticarcillin, aminoglycosides and combinations against Staphylococcus aureus. J. Antimicrob. Chemother. 5:151-158. [DOI] [PubMed] [Google Scholar]
- 311.Werner, G., B. Hildebrandt, and W. Witte. 2001. Aminoglycoside-streptothricin resistance gene cluster aadE-sat4-aphA-3 disseminated among multiresistant isolates of Enterococcus faecium. Antimicrob. Agents Chemother. 45:3267-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.White, P. A., C. J. McIver, and W. D. Rawlinson. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45:2658-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Wilson, D. N., G. Blaha, S. R. Connell, P. V. Ivanov, H. Jenke, U. Stelzl, Y. Teraoka, and K. H. Nierhaus. 2002. Protein synthesis at atomic resolution: mechanistics of translation in the light of highly resolved structures for the ribosome. Curr. Protein Peptide Sci. 3:1-53. [DOI] [PubMed] [Google Scholar]
- 315.Wimberly, B. T., D. E. Brodersen, W. M. Clemons, Jr., R. J. Morgan-Warren, A. P. Carter, C. Vonrhein, T. Hartsch, and V. Ramakrishnan. 2000. Structure of the 30S ribosomal subunit. Nature 407:327-339. [DOI] [PubMed] [Google Scholar]
- 316.Wohlleben, W., W. Arnold, L. Bissonnette, A. Pelletier, A. Tanguay, P. H. Roy, G. C. Gamboa, G. F. Barry, E. Aubert, J. Davies, et al. 1989. On the evolution of Tn21-like multiresistance transposons: sequence analysis of the gene (aacC1) for gentamicin acetyltransferase-3-I (AAC(3)-I), another member of the Tn21-based expression cassette. Mol. Gen. Genet. 217:202-208. [DOI] [PubMed] [Google Scholar]
- 317.Wright, G. D., A. M. Berghuis, and S. Mobashery. 1998. Aminoglycoside antibiotics. Structures, functions, and resistance, p. 27-69. In B. P. Rosen and S. Mobashery (ed.), Resolving the antibiotic paradox: progress in understanding drug resistance and development of new antibiotics. Kluwer Academic/Plenum Publishers, New York. N.Y.
- 318.Wright, G. D., and P. Ladak. 1997. Overexpression and characterization of the chromosomal aminoglycoside 6′-N-acetyltransferase from Enterococcus faecium. Antimicrob. Agents Chemother. 41:956-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Wrighton, C. J., and P. Strike. 1987. A pathway for the evolution of the plasmid NTP16 involving the novel kanamycin resistance transposon Tn4352. Plasmid 17:37-45. [DOI] [PubMed] [Google Scholar]
- 320.Yoshizawa, S., D. Fourmy, R. G. Eason, and J. D. Puglisi. 2002. Sequence-specific recognition of the major groove of RNA by deoxystreptamine. Biochemistry 41:6263-6270. [DOI] [PubMed] [Google Scholar]
- 321.Yoshizawa, S., D. Fourmy, and J. D. Puglisi. 1998. Structural origins of gentamicin antibiotic action. EMBO J. 17:6437-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.You, I., R. Kariyama, M. J. Zervos, H. Kumon, and J. W. Chow. 2000. In-vitro activity of arbekacin alone and in combination with vancomycin against gentamicin- and methicillin-resistant Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 36:37-41. [DOI] [PubMed] [Google Scholar]
- 323.Young, E. J. 1995. An overview of human brucellosis. Clin. Infect. Dis. 21:283-289. [DOI] [PubMed] [Google Scholar]
- 324.Zalacain, M., A. Gonzalez, M. C. Guerrero, R. J. Mattaliano, F. Malpartida, and A. Jimenez. 1986. Nucleotide sequence of the hygromycin B phosphotransferase gene from Streptomyces hygroscopicus. Nucleic Acids Res. 14:1565-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Zeng, S., C. Hu, H. Wei, Y. Lu, Y. Zhang, J. Yang, G. Yun, W. Zou, and B. Song. 1993. Intravitreal pharmacokinetics of liposome-encapsulated amikacin in a rabbit model. Ophthalmology 100:1640-1644. [DOI] [PubMed] [Google Scholar]
- 326.Zhanel, G. G., D. J. Hoban, and G. K. Harding. 1991. The postantibiotic effect: a review of in vitro and in vivo data. Ann. Pharmacother. 25:153-163. [DOI] [PubMed] [Google Scholar]