Abstract
We initially identified lens epithelium-derived growth factor/p75 (LEDGF/p75) as a binding partner of human immunodeficiency virus type 1 (HIV-1) integrase. To investigate the role of LEDGF/p75 in HIV replication and its potential as a new antiviral target, we stably overexpressed two different fragments containing the integrase binding domain (IBD) of LEDGF/p75 fused to enhanced green fluorescent protein (eGFP). HIV-1 replication was severely inhibited by overexpression of the eGFP-IBD fusion proteins, while no inhibition was observed in cell lines overexpressing the interaction-deficient D366A mutant. Quantitative PCR pinpointed the block to the integration step, whereas nuclear import was not affected. Competition of the IBD fusion proteins with endogenous LEDGF/p75 for binding to integrase led to a potent defect in HIV-1 replication in both HeLaP4- and MT-4-derived cell lines. A previously described diketo acid-resistant HIV-1 strain remained fully susceptible to inhibition, suggesting that this strategy will also work in patients who harbor strains resistant to the current experimental integrase inhibitors. These data support LEDGF/p75 as an important cofactor for HIV replication and provide proof of concept for the LEDGF/p75-integrase interaction as a novel target for treating HIV-1 infection.
Currently, more than 20 drugs that belong to four different classes are approved by the FDA for clinical treatment of human immunodeficiency virus type 1 (HIV-1) infection. However, the emergence of resistant strains, especially in multidrug-experienced patients, compromises antiretroviral regimens (20). To expand the antiretroviral armament, coreceptor blockers and integrase inhibitors have recently entered clinical trials. Here we investigate whether the interaction of HIV-1 integrase with its cellular binding partner, lens epithelium-derived growth factor/p75 (LEDGF/p75), could constitute a new target for antiretroviral therapy.
Although lentiviruses carry the proteins required for all steps of the replication cycle, some steps require additional cellular cofactors. Several candidate cofactors of HIV integration have been proposed (for recent reviews, see references 33 and 38). LEDGF/p75 was identified as an interaction partner of HIV-1 integrase by coimmunoprecipitation of nuclear extracts of cells overexpressing HIV-1 integrase from a synthetic gene (integrases) (5) and was found to stimulate DNA binding and integration in vitro (2, 5). The interaction of LEDGF/p75 with HIV-1 integrase was subsequently confirmed in two independent reports (13, 33). LEDGF/p75 binds to the integrases of HIV-1, HIV-2, the simian immunodeficiency virus SIVmac, and feline immunodeficiency virus (FIV) but not to Moloney murine leukemia virus, respiratory syncytial virus, or human T-cell leukemia virus type 2 integrase, proving the lentiviral specificity of the interaction (2, 22). Earlier studies described LEDGF/p75 as a protein copurifying with the transcriptional coactivator PC4 (15) and as a growth factor (32). Later studies showed that LEDGF/p75 is a weak coactivator of general transcription but plays a protective role against cellular stress (30). LEDGF/p75 consists of 530 amino acids (aa) and contains an N-terminal PWWP motif involved in chromatin binding (Fig. 1a) (16). Accordingly, the nuclear accumulation of HIV-1 integrase is apparently due to chromosomal tethering by LEDGF/p75 (24, 37). Mutations in the nuclear localization signal (NLS) of LEDGF/p75 induce its cytoplasmic accumulation after transient overexpression. However, stably expressed LEDGF/p75 containing a mutant NLS still accumulates in the nucleus due to chromosomal association (23, 37). To date, it is not clear whether this NLS is also responsible for targeting the HIV-1 preintegration complex to the nucleus.
FIG. 1.
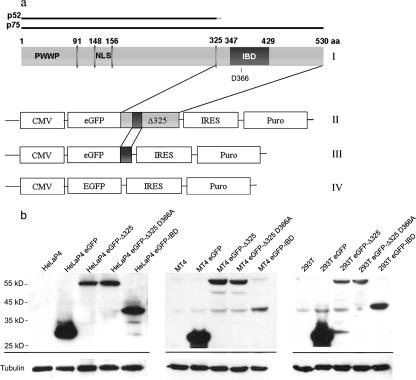
Selection of eGFP-IBD- and eGFP-Δ325-overexpressing cells. (a) Construction of lentiviral vectors encoding eGFP-Δ325 and eGFP-IBD. (I) Domain structure of LEDGF/p75. (II to IV) Schematic representations of the expression cassettes of pCombi eGFP-Δ325 IRES Puro, pCombi eGFP-IBD IRES Puro, and pCombi eGFP IRES Puro, respectively. (b) Western blot analysis of HeLaP4-, MT-4-, and 293T-derived cell lines. Equal amounts of protein extracted from each cell line were analyzed by Western blotting using an eGFP antibody. Equal loading was controlled by α-tubulin detection.
The minimal domain of LEDGF/p75 required for interaction with integrase (integrase binding domain [IBD]) was mapped to residues 347 to 429 (4, 37). In vitro experiments revealed that the IBD is necessary for interaction with integrase but not sufficient to stimulate strand transfer activity (4). By a yeast two-hybrid screen, the LEDGF/p75 interaction domain of HIV-1 integrase was mapped to the catalytic core, and several mutants defective for interaction with LEDGF/p75 were identified (13). Q168A mutant integrase does not interact with LEDGF/p75 but retains in vitro 3′ processing and strand transfer activity. A viral clone carrying the Q168A mutation was shown to be replication defective due to a block at the integration step. Although Llano et al. could show neither a reduction of vector transduction nor a reduction of HIV-1 replication in Jurkat cells depleted for LEDGF/p75 (22), transient and stable knockdown of LEDGF/p75 in HeLaP4 cells resulted in a two- to fivefold reduction in virus replication (36). The cause of this discrepancy is not clear but may be related to the nature of the cell lines used or the relative potency of the knockdown.
Here we report that stable overexpression of either of two C-terminal fragments of LEDGF/p75 containing the IBD inhibits HIV-1 replication and lentiviral vector transduction. An interaction-defective mutant version of the IBD had no effect on HIV-1 replication or vector transduction. Our results support a role for LEDGF/p75 in lentiviral integration and reveal the LEDGF/p75-integrase interaction as a promising new target for anti-HIV therapy.
MATERIALS AND METHODS
Plasmids.
To create the eukaryotic expression plasmid peGFP-IBD IRES Puro, the IRES Puro sequence was amplified by PCR with primers 5′ TGCTCTAGAGCTCTAGCCCAATTCCGC and 5′ TGCTCTAGATCAGGCACCGGGCTTGCGGG from pEF1 LEDGF back (22), digested with XbaI, and subcloned into pEGFP-C3 (Clontech) (peGFP IRES Puro). The IBD sequence (aa 347 to 429) was amplified by PCR from pCP6H75 (5), using primers 5′ CGGGAATTCGCCCTTGCGGCAGCCATATGTCAATGGATTCTCGA and 5′ CCGGGATCCTATACCAAGAACATGTT, digested with EcoRI and BamHI, and subcloned into peGFP-IRES Puro. To create peGFP-Δ325 IRES Puro, the IBD was removed by EcoRI and BamHI digestion and replaced by the Δ325 fragment (aa 325 to 530), which was amplified by PCR from pCP6H75 with primers 5′ GGGAATTCTAGAGCAGCAGAATAAAGATGAAG and 5′ CACGAGATCTGACTCGCGATTTCAAACCTGGAGACC. To construct the lentiviral transfer plasmids pCombi eGFP-IBD IRES Puro and pCombi eGFP-Δ325 IRES Puro, enhanced green fluorescent protein (eGFP) was removed from pCombi-eGFP (1) by XbaI-HpaI digestion and replaced by eGFP-IBD IRES Puro and eGFP-Δ325 IRES Puro, respectively. The D366A mutation was introduced into the Δ325 fragment of peGFP-Δ325 IRES Puro by using the method of Kirsch and Joly (19). The eGFP-Δ325 D366A IRES Puro fragment was subcloned into pCombi as described previously, resulting in pCombi eGFP-Δ325 D366A IRES Puro. To construct a fusion between monomeric red fluorescent protein (mRFP) and integrase (pmRFP-INs), eGFP was removed from peGFP-INs (24) by Eco47III-BglII digestion and replaced by mRFP amplified by PCR from CS-hrl-mRFP-ttk (kindly provided by S. S. Gambhir, Los Angeles, Calif.) (29) with primers 5′ TTCAGCGCTATGGCCTCCTCCGAGGACGTC and 5′ GAATTCAGATCTGGCGCCGGTGGAGTGGCGG. To construct a lentiviral vector encoding mRFP, eGFP was removed from pCombi-eGFP (1) by BamHI-SmaI digestion and replaced by mRFP cDNA. To construct a retroviral vector encoding an eGFP-firefly luciferase fusion protein (pLNC-GFP-Fluc), the Fluc coding region was removed from pCHMWS-Ffluc (C. Deroose et al., submitted for publication) by BamHI-XmaI digestion and cloned into the multiple cloning site of pEGFP-C1 (BD Biosciences, Erembodegem, Belgium) after BglII-XmaI digestion. The eGFP-Ffluc fragment was PCR amplified with the forward primer 5′ GGACCGGTGCAGTCGACGGTACCGC and the reverse primer 5′ GCATCGATGGTCCCGGGTTACACGG, digested with AgeI-ClaI, and cloned into pLNC-GFP (a kind gift from G. Towers, London, United Kingdom) after eGFP was removed by AgeI-ClaI digestion. To construct a lentiviral transfer plasmid with the eGFP-IBD IRES Puro or eGFP-Δ325 IRES Puro fragment under the control of a tetracycline-regulatable (Tet/off) promoter, eGFP was removed from pCCL.sin36.eGFP.PPT.Wpre.CMV.tTA-s2.tet (a kind gift from L. Naldini, Turino, Italy) by AgeI-HpaI digestion and replaced by the eGFP-IBD/Δ325 IRES Puro cassette (peGFP-IBD IRES Puro Tet/off and peGFP-Δ325 IRES Puro Tet/off, respectively). The IBD and Δ325 fragments were expressed in Escherichia coli as maltose binding protein (MBP) fusions. For construction of pMBP-IBD, the sequence corresponding to aa 347 to 429 was PCR amplified with primers 5′ TTCGGATCCATGGATTCTCGAC, containing a BamHI restriction site, and 5′ AGCCGTCGACTAAACCAAGAA, containing a SalI restriction site, using pCPnat75 (24) as a template. For construction of pMBP-Δ325, the sequence corresponding to aa 325 to 530 was PCR amplified with primers 5′ TTCAGAATTCCAGCAGAATAAAGAT, containing an EcoRI restriction site, and 5′ GCCAAGCTTTCAGTTATCTAGTGTAGT, containing a HindIII restriction site, using pCPnat75 as a template. Subsequent to amplification, the resulting products were digested with the respective restriction endonucleases and ligated into pMALTM-p2X (New England Biolabs Inc.). The D366A mutation was introduced into the Δ325 fragment of pMBP-Δ325 by using the method of Kirsch and Joly (19). The integrity of all plasmids was confirmed by sequencing.
Cell lines and cell culture.
HeLaP4 cells, obtained from the NIH reagent program, were grown in Dulbecco's modified Eagle's medium (Gibco BRL, Belgium) containing 10% fetal calf serum (FCS; International Medical, Belgium), 100 μg/ml streptomycin, 100 U/ml penicillin, and 0.5 mg/ml Geneticin (Gibco BRL) (HeLaP4 medium). MT-4 cells, a human T-cell line obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (contributed by Douglas Richman), were grown in RPMI 1640 medium (Gibco BRL) containing 10% FCS, 100 μg/ml streptomycin, 100 U/ml penicillin, and 2 mM l-glutamine (Gibco BRL) (MT-4 medium). 293T cells were provided by O. Danos (Evry, France) and were grown in Dulbecco's modified Eagle's medium containing 10% FCS, 100 μg/ml streptomycin, and 100 U/ml penicillin (293T medium). All cells were cultured at 37°C in a 5% CO2 humidified atmosphere.
Selection of eGFP-IBD- and eGFP-Δ325-expressing stable cell lines.
HeLaP4 and 293T cells were transduced with the lentiviral vector Combi eGFP-IBD IRES Puro, Combi eGFP-Δ325 IRES Puro, Combi eGFP-Δ325 D366A IRES Puro, or Combi eGFP-IRES Puro at a multiplicity of infection (MOI) of 1 to generate HeLaP4 eGFP-IBD, HeLaP4 eGFP-Δ325, HeLaP4 eGFP-Δ325 D366A, and HeLaP4 eGFP cells, respectively, or their 293T equivalents. Polyclonal cell lines were selected with 0.25 μg/ml puromycin. MT-4-derived cell lines were generated in a similar way, sorting the 10% most positive cells with FACSVantage (BD Biosciences) prior to selection. Cells (100,000) were retained after cell sorting to establish the polyclonal cell lines. To select HeLaP4 cell lines with the eGFP-IBD or -Δ325 IRES Puro cassette under control of the Tet/off promoter, HeLaP4 cells were transduced with the eGFP-IBD IRES Puro Tet/off or eGFP-Δ325 IRES Puro Tet/off lentiviral vector, and the 10% of cells with the most eGFP expression were sorted by FACSVantage. Monoclonal cell lines were selected with 0.25 μg/ml puromycin, and the clones with the highest eGFP expression levels, as determined by fluorescence-activated cell sorting (FACS) (FACSCalibur; BD Biosciences), were retained.
Protein purification, in vitro pull-down assay, and Western blotting.
Recombinant MBP-IBD, MBP-Δ325, and MBP-Δ325 D366A were expressed in BL21(DE3) cells (8). After transformation, the bacteria were grown to an optical density of 0.5 (MBP-IBD) or 0.6 (MBP-Δ325 and MBP-Δ325 D366A), and protein expression was induced by the addition of a final concentration of 0.5 mM (MBP-IBD) or 1 mM (MBP-Δ325) isopropyl-beta-d-thiogalactopyranoside. Following incubation at 37°C for 4 h, the bacteria were harvested and stored at −20°C until protein purification. For purification, the cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.2, 500 mM NaCl, 5 mM dithiothreitol, 1 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride). After complete lysis by ultrasonication, the supernatant was cleared by centrifugation, and recombinant proteins were bound to amylose resin (New England Biolabs Inc.). The resin was washed with 20 volumes of lysis buffer, and the MBP-tagged proteins were eluted with 10 1-ml fractions of lysis buffer supplemented with 10 mM maltose. The fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis for protein content, pooled, and concentrated by dialysis (overnight, 4°C) against storage buffer (50 mM Tris-HCl, pH 7.2, 500 mM NaCl, 50% [vol/vol] glycerol). Recombinant LEDGF/p75 and HIV-1 integrase containing a C-terminal His6 tag were purified as described previously for use in the in vitro pull-down assays (24).
Pulling down of LEDGF/p75 and MBP-IBD as well as MBP-Δ325 and MBP-Δ325 D366A by HIV-1 integrase was performed as described previously (24). Western blotting was performed as described previously (36).
Competition of integrase-Δ325 interaction by full-length LEDGF/p75.
To study the competition of full-length LEDGF/p75 and the Δ325 fragment, AlphaScreen technology (Perkin-Elmer, Benelux) was used. IN (0 or 100 nM) was preincubated in a 384-well plate for 30 min with various amounts of LEDGF/p75 at room temperature in assay buffer (25 mM Tris-HCl, pH 7.3, 150 mM NaCl, 1 mM MgCl2, 0.01% Tween 20, 0.1% bovine serum albumin). After preincubation, 0, 10, 30, or 100 nM MBP-Δ325 was added for cross-titration purposes. The new mixture was incubated for another 90 min at room temperature. For subsequent binding of the proteins to the beads, Ni chelate-coated acceptor beads and streptavidin-coated donor beads premixed with biotinylated anti-MBP antibody (Vector Laboratories) were added to a final concentration of 20 μg/ml. All dilutions were done in assay buffer, and if necessary, the final volume of the reaction was adjusted to 25 μl. In order to bind the anti-MBP antibody to the streptavidin-coated donor beads, the antibody was dialyzed overnight against assay buffer without bovine serum albumin, added to the beads at a final concentration of 50 nM, and incubated for 1 h at room temperature. After adding the beads to the binding assay, light exposure was omitted, and incubation was carried out for another 60 min before analyzing the interaction in an EnVision plate reader (Perkin-Elmer, Benelux).
Laser scanning microscopy.
HeLaP4, HeLaP4 eGFP, HeLaP4 eGFP-Δ325, HeLaP4 eGFP-Δ325 D366A, and HeLaP4 eGFP-IBD cells were transfected with pmRFP-INs and analyzed using an LSM510 unit (Carl Zeiss, Jena, Germany) as described previously (36). Immunofluorescence detection of endogenous LEDGF/p75 was performed as described previously (5).
Cellular FCCS analysis.
Cellular fluorescence cross-correlation spectroscopy (FCCS) measurements were performed on a commercial LSM510/ConfoCor2 system (Carl Zeiss, Jena, Germany). The 488-nm line of the Ar+ laser (Acousto-optical tunable filter, 0.1%; ∼ 25 μW) was used to excite eGFP, and the 543-nm line of the HeNe laser (AOTF, 7%; ∼70 μW) was used to excite mRFP1. The excitation light was reflected by a dichroic mirror (HFT 488/543) and focused through a type C Apochromat ×40/1.2-W objective lens. The fluorescence emission light was split by a second dichroic mirror (NFT 570) into two separate beam paths and passed through a 505- to 530-nm band-pass filter for eGFP fluorescence and a 600- to 650-nm band-pass filter for mRFP1 fluorescence. Each confocal pinhole diameter was set to 70 μm. After preparation of the cells in an eight-well chambered coverglass (Nunc A/S, Roskilde, Denmark), laser scanning microscopy was used to search for cells suitable for FCCS. The laser beam was focused at a selected spot, and FCCS measurement (10 times for 20 s each) was performed. Auto- and cross-correlation curves were evaluated by Levenberg-Marquardt nonlinear least-square fitting to a two-component model, using OriginPro 7.5 software (OriginLab, Northampton, MA). The relative cross-correlation was calculated from the following formula: CCrel. = Gcross(0)/Ggreen(0), with Gcross(0) and Ggreen(0) being the amplitudes of the cross and green autocorrelation curves, respectively.
Lentiviral and retroviral vector production.
The day before transfection, five million 293T cells were grown on an 8.5-cm-diameter cell culture dish in 5 ml Optimem (Gibco BRL) containing 2% FCS, 100 μg/ml streptomycin, and 100 U/ml penicillin. On day 2, the medium was replaced by 5 ml Optimem with 0% FCS, and cells were cotransfected with 10 μg of the packaging plasmid pCMVΔR8.91, 5 μg of the envelope plasmid pMDG (both provided by D. Trono, Geneva, Switzerland), and 20 μg of the respective transfer plasmid (11). For production of retroviral vectors, pCMVintron-gag-pol (a kind gift from G. Τowers, London, United Kingdom) was used as a packaging plasmid and pMDG was used as an envelope plasmid. On day 3, the medium was again replaced by 5 ml Optimem with 0% FCS, and on day 4, the supernatant was filtered through a 0.45-μm filter and concentrated 30-fold in a Vivaspin 15 50,000 MW column (Vivascience, Hannover, Germany) at 2,500 × g. Vector-containing supernatant was stored at −80°C.
Virus strains.
The molecular clone pNL4.3 was obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. HIV-1IIIb (28) and HIV-1L-708,906 were described previously (14). To generate vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped NL4.3 virus, five million 293T cells were plated 24 h prior to transfection with 20 μg pNL4.3 and 5 μg pMDG and further treated as described for lentiviral vector production, but the supernatant was not concentrated.
Lenti- and retroviral vector transduction.
To analyze the effects of eGFP-IBD and eGFP-Δ325 overexpression on lenti- or retroviral vector transduction, cells were transduced with serial dilutions of the Combi-mRFP or LNC-GFP FLuc vector, respectively. While HeLaP4 or 293T cells were seeded in a 96-well plate at 20,000 cells/well 1 day prior to transduction, the MT-4 cells were seeded on the day of transduction at 50,000 cells/well. At 4 hours posttransduction, the medium was replaced with fresh HeLaP4, 293T, or MT-4 medium. mRFP expression of the lentiviral vector was analyzed at 72 h posttransduction by FACS analysis. Firefly luciferase expression from the retroviral vector was measured with an IVIS system (Xenogen, Alameda, Calif.) 5 min after the addition of 150 ng/ml d-luciferin. Cells were imaged for between 1 and 5 min in the IVIS system, until the maximum signal was reached.
HIV-1 infection.
Prior to infection, HeLaP4 eGFP-IBD or -Δ325 Tet/off cells were incubated with or without 5 mg/ml doxycycline for at least 7 days. Infection of HeLaP4-derived cell lines was performed as described earlier (36). Infection of MT-4 cells was performed with 500,000 cells in 200 μl medium at different MOIs for 3 h. After 3 h, cells were washed twice with phosphate-buffered saline and resuspended in 5 ml MT-4 medium. HIV-1 replication was monitored by quantifying p24 antigen in the supernatant via an enzyme-linked immunosorbent assay (Alliance HIV-1 p24 ELISA kit; Perkin-Elmer, Zaventem, Belgium).
Real-time quantitative PCR analysis.
One day prior to infection, 2 × 105 293T cells were seeded into each well of a 24-well plate. Twenty-four hours later, cells were infected with VSV-G-pseudotyped HIV-1NL4.3 corresponding to 50,000 pg p24. As a negative control, the 293T cells were treated with 0.09 μg/ml zidovudine (AZT) during the experiment. DNA extraction and quantification of late reverse transcripts and two-long-terminal-repeat (2-LTR) circles were performed as described previously (39). The oligonucleotides used to quantify the late reverse transcripts primed in the HIV-1 gag gene and had the following sequences: forward primer, 5′ ATCAAGCAGCCATGCAAATGTT; reverse primer, 5′ CTGAAGGGTACTAGTAGTTCCTGCTATGTC; and probe, 5′ (6-carboxyfluorescein)-ACCATCAATGAGGAAGCTGCAGAATGGGA-(6- carboxytetramethylrhodamine). The primers and probe to quantify 2-LTR circles were identical to those used previously (39). To quantify the integration events, the infected 293T cells were passaged at least four times, and the total amount of HIV-1 DNA was quantified using the same primers and probe as those used for detection of the late reverse transcripts.
RESULTS
Selection of eGFP-IBD- and eGFP-Δ325-overexpressing cells.
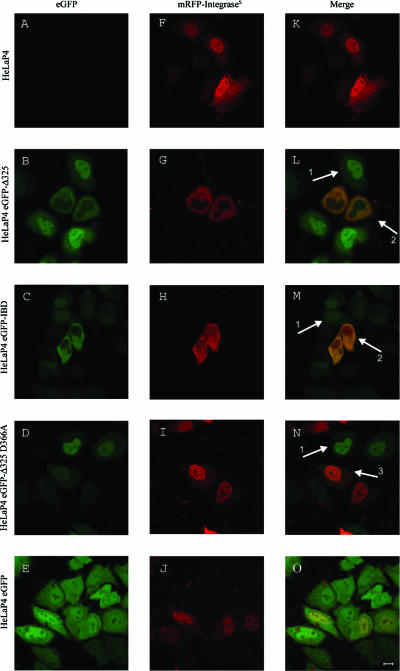
In an effort to inhibit HIV-1 replication by preventing interaction of HIV-1 integrase with LEDGF/p75, we generated two constructs encoding the IBD of LEDGF/p75 (4, 37). First, the IBD region (aa 347 to 429) of LEDGF/p75 was fused to the C terminus of eGFP (eGFP-IBD). The second construct comprised the C-terminal fragment (aa 325 to 530) of LEDGF/p75 (eGFP-Δ325) (Fig. 1a). Using lentiviral vector technology, HeLaP4-derived cell lines were generated to express the fusion proteins at high levels (HeLaP4 eGFP-IBD and HeLaP4 eGFP-Δ325). The following two control cell lines were established in parallel: HeLaP4 eGFP cells, expressing eGFP without a fusion, and HeLaP4 eGFP-Δ325 D366A cells, expressing the D366A mutant of eGFP-Δ325, which is known to be defective for interaction with HIV-1 integrase (7). The viability of all selected cell lines was similar to that of the parental HeLaP4 cell line, as determined by growth kinetics (data not shown). To exclude selection bias, CD4 and CXCR4 coreceptor expression was monitored in the HeLaP4-derived cell lines by FACS analysis. All cell lines were >99% positive for CD4 and CXCR4 expression. In a similar way, 293T- and MT-4-derived cell lines were generated. For the MT-4-derived cells, the 10% most-positive cells were sorted by FACS. Expression levels of the fusion proteins were verified by Western blotting (Fig. 1b). Confocal microscopy with the HeLaP4-derived cell lines revealed the localization of eGFP-IBD, eGFP-Δ325, and eGFP-Δ325 D366A fusion proteins to the nucleus (Fig. 2L, M, and N, arrows 1), unlike wild-type eGFP, which was present throughout the cell (Fig. 2O). As expected, a transiently expressed mRFP-integrases fusion protein was localized in the nuclei of wild-type HeLaP4 cells (Fig. 2F) (6, 10, 12, 21, 26, 27). Upon coexpression of mRFP-integrases and eGFP-IBD/eGFP-Δ325, however, a dramatic relocalization of each fusion protein from the nucleus to the cytoplasm was observed (Fig. 2L and M, arrows 2). In contrast, in the HeLaP4 eGFP-Δ325 D366A control cells expressing interaction-defective IBD, the fusion proteins retained their nuclear localization (Fig. 2N, arrow 3).
FIG. 2.
Relocalization of mRFP-integrases upon coexpression of eGFP-Δ325 or eGFP-IBD. HeLaP4, HeLaP4 eGFP-Δ325, HeLaP4 eGFP-IBD, HeLaP4 eGFP-Δ325 D366A, and HeLaP4 eGFP cells were transfected with pmRFP-INs 24 h before laser scanning microscopy. (A to E) eGFP expression; (F to J) mRFP-integrases expression; (K to O), merge. Bar, 10 μm. Arrows 1 represent nuclear localization of the overexpressed fragments, arrows 2 represent cytoplasmic tethering upon coexpression of integrase and eGFP-Δ325 or eGFP-IBD, and arrow 3 shows nuclear localization of coexpressed integrase and eGFP-Δ325 D366A.
Both the IBD and Δ325 fragments bind to integrase in vitro and in vivo.
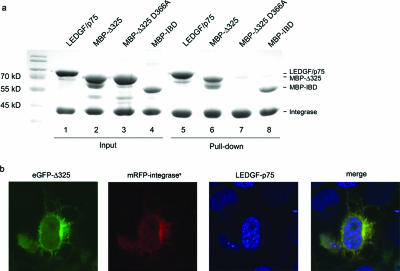
The confocal imaging data suggested a direct interaction of the IBD and Δ325 fragments with HIV-1 integrase. To verify this observation, an in vitro pull-down assay using His6-tagged integrase was performed as described previously (Fig. 3a) (24). Full-length LEDGF/p75 was readily recovered in complex with His6-tagged integrase (Fig. 3a, lane 5). The MBP-Δ325 and MBP-IBD fusions were pulled down as well (Fig. 3a, lanes 6 and 8). No pulling down was observed with MBP-Δ325 D366A (Fig. 3a, lane 7) or in the absence of HIV-1 integrase (data not shown). To confirm this interaction in vivo, FCCS was performed with the HeLaP4-derived cell lines (Table 1). This method allows the detection of interactions of two fluorescently labeled proteins in vivo by measuring simultaneous diffusion. HeLaP4 eGFP-Δ325, HeLaP4 eGFP-Δ325 D366A, and HeLaP4 eGFP-IBD cells were transfected with pmRFP-integrases. As a negative control, cells were transfected with an mRFP expression plasmid. As a positive control, HeLaP4 cells were transfected with a plasmid encoding an eGFP-mRFP fusion protein. Compared to the controls, cross-correlation of eGFP-Δ325 and eGFP-IBD with mRFP-integrases was detected, indicating that both proteins interact in vivo (Table 1). Cross-correlation was not observed in the HeLaP4 eGFP-Δ325 D366A cells. If eGFP-Δ325 competes with endogenous LEDGF/p75 for binding to integrase, then endogenous LEDGF/p75 should remain in the nucleus due to its nuclear localization signal, whereas both eGFP-Δ325 and integrase should migrate to the cytoplasm. As shown in Fig. 3b, the nuclear localization of endogenous LEDGF/p75 was indeed confirmed for HeLaP4 eGFP-Δ325 cells transfected with mRFP-integrases.
FIG. 3.
In vitro interaction of deletion mutants with integrase. (a) His6 tag-mediated pull-down of MBP-Δ325, MBP-Δ325 D366A, and MBP-IBD was carried out. Recombinant LEDGF/p75, MBP-Δ325, MBP-Δ325 D366A, and MBP-IBD were incubated with His6-tagged HIV-1 integrase, and complexes were recovered using Ni2+-nitrilotriacetic acid agarose beads. Proteins were separated via 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected using Coomassie blue staining. The relative positions of the proteins are indicated on the right. Molecular size markers are indicated on the left. Lanes 1 to 4 show input quantities of each protein; lanes 5 to 8 show the recovered proteins after pull-down. (b) HeLaP4 eGFP-Δ325 cells were transfected with pmRFP-INs 24 h before fixation and permeabilization. Endogenous LEDGF/p75 was stained with a combination of monoclonal anti-LEDGF/p75 and Alexa 633-conjugated anti-mouse antibodies. Cells were analyzed using laser scanning microscopy.
TABLE 1.
Simultaneous diffusion of HIV-1 integrase and LEDGF/p75 deletion mutantsa
| Cell line + integrase or control | Relative cross-correlation |
|---|---|
| HeLaP4 eGFP-Δ325 + mRFP | 0.140 ± 0.003 |
| HeLaP4 eGFP-Δ325 + mRFP-integrases | 0.600 ± 0.004 |
| HeLaP4 eGFP-IBD + mRFP-integrases | 0.467 ± 0.004 |
| HeLaP4 eGFP-Δ325 D366A + mRFP-integrases | 0.184 ± 0.015 |
| HeLaP4 + mRFP-eGFP | 0.468 ± 0.006 |
HeLaP4 eGFP-Δ325, HeLaP4 eGFP-Δ325 D366A, and HeLaP4 eGFP-IBD cells were transfected with an mRFP-integrases expression plasmid. As a negative control, cells were transfected with an mRFP expression plasmid. As a positive control, HeLaP4 cells were transfected with a plasmid encoding an eGFP-mRFP fusion protein. Interaction between a LEDGF/p75 deletion mutant and mRFP-integrases was detected by measuring the simultaneous diffusion of both proteins using fluorescence cross-correlation spectroscopy and was quantified as the relative extent of cross-correlation (data are average numbers ± SD for 10 measurements).
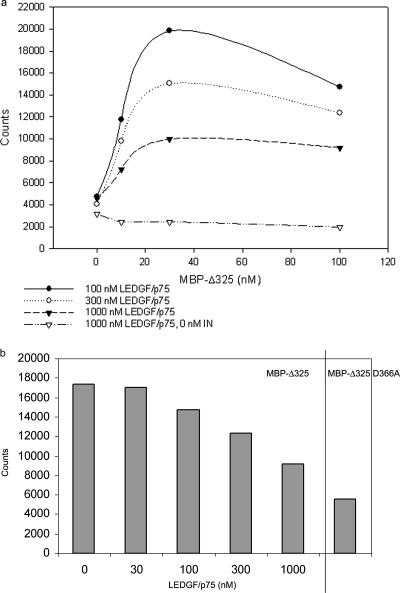
To investigate in molecular detail whether LEDGF/p75 and the deletion mutants compete directly for binding to integrase, AlphaScreen technology was used (35). In this assay, His6-tagged integrase was preincubated with various amounts of LEDGF/p75. Different concentrations of MBP-Δ325 were subsequently added. Ni chelate-coated acceptor beads and anti-MBP-coated donor beads were added to bind integrase and MBP-Δ325, respectively. Binding of the molecules on the beads leads to energy transfer from one bead to the other, ultimately producing a fluorescent signal (Fig. 4a). The fluorescent signal increased with increasing MBP-Δ325 concentrations (from 0.01 to 300 nM) until a plateau was reached (data not shown), indicating saturation of the integrase-MBP-Δ325 interaction. Representative interaction spectra are shown in Fig. 4a. On the other hand, when a larger amount of LEDGF/p75 was preincubated with integrase, the fluorescent signal decreased, from 17,000 to 9,000 counts (100 nM MBP-Δ325) (Fig. 4a and b). These results clearly indicate that LEDGF/p75 and MBP-Δ325 compete for binding to HIV-1 integrase.
FIG. 4.
Competition of integrase-Δ325 interaction by full-length LEDGF/p75. (a) His6-tagged integrase (100 nM) was preincubated with various amounts of LEDGF/p75. As a negative control, 0 nM integrase was preincubated with 1,000 nM LEDGF/p75. Increasing amounts of MBP-Δ325 were subsequently added for cross-titration purposes. The integrase-MBP-Δ325 interaction was measured by the AlphaScreen method. Ni chelate-coated acceptor beads and anti-MBP-coated donor beads were added to bind integrase and MBP-Δ325, respectively. (b) Effect of LEDGF/p75 on integrase-MBP-Δ325 interaction. Integrase was preincubated with different amounts of LEDGF/p75. MBP-Δ325 (100 nM) was subsequently added. As a negative control, 100 nM MBP-Δ325 D366A was used. The interaction between integrase and MBP-Δ325 or MBP-Δ325 D366A was measured using the AlphaScreen method. Experiments a and b were performed three times, and results of a representative experiment are shown.
Lentiviral but not retroviral vector transduction is strongly reduced by overexpression of eGFP-Δ325 or eGFP-IBD.
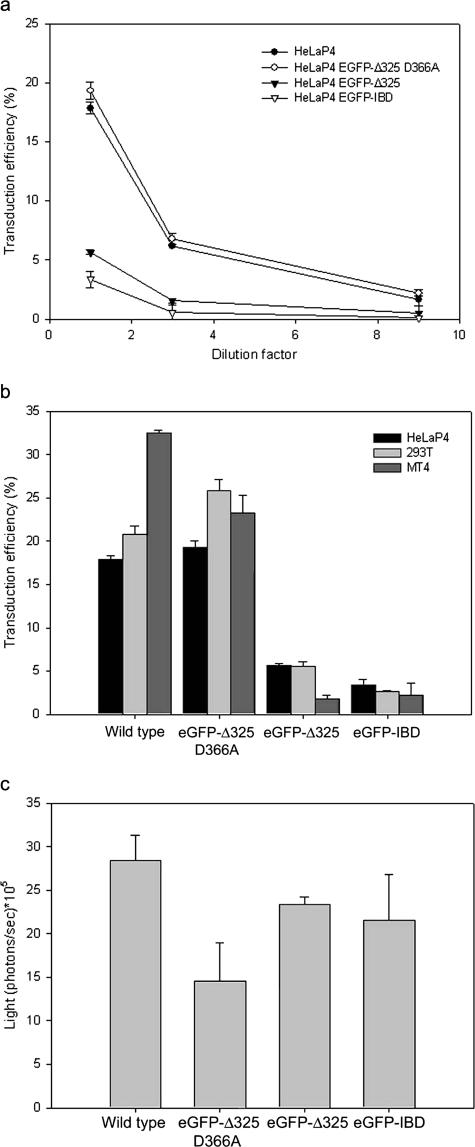
Next, we transduced HeLaP4, HeLaP4 eGFP-Δ325, HeLaP4 eGFP-Δ325 D366A, and HeLaP4 eGFP-IBD cells with a second-generation VSV-G-pseudotyped lentiviral vector (42) encoding mRFP (Fig. 5a). Transduction efficiency was measured by FACS at 72 h posttransduction. For different vector MOIs, the transduction efficiencies in HeLaP4 eGFP-Δ325 and HeLaP4 eGFP-IBD cells were three times lower than those in HeLaP4 and HeLaP4 eGFP-Δ325 D366A cells. To rule out cell type dependence, the MT-4- and 293T-derived cell lines were transduced in parallel. As observed for the HeLaP4-derived cell lines, a potent inhibition of vector transduction was observed in the presence of eGFP-Δ325 or eGFP-IBD (Fig. 5b). The inhibition was most pronounced in the MT-4-based cell lines (10- to 15-fold).
FIG. 5.
Effects of eGFP-Δ325 and eGFP-IBD overexpression on lentiviral and retroviral vector transduction. (a) Effects on lentiviral vector transduction of HeLaP4-derived cell lines. HeLaP4, HeLaP4 eGFP-Δ325 D366A, HeLaP4 eGFP-Δ325, and HeLaP4 eGFP-IBD cells were transduced with a threefold dilution series of an HIV-1-derived vector encoding mRFP. After 72 h, the percentage of mRFP-positive cells was determined by FACS analysis. (b) Effects on lentiviral vector transduction of HeLaP4-, 293T-, and MT-4-derived cell lines. Wild-type HeLaP4, 293T, and MT-4 cells and their derived cell lines overexpressing eGFP-Δ325 D366A, eGFP-Δ325, and eGFP-IBD were transduced with an mRFP-expressing HIV-1-derived vector at an MOI of 0.2, as determined in wild-type HeLaP4 cells. After 72 h, the percentage of mRFP-expressing cells was determined by FACS analysis. (c) Effects on retroviral vector transduction of HeLaP4-derived cell lines. HeLaP4, HeLaP4 eGFP-Δ325 D366A, HeLaP4 eGFP-Δ325, and HeLaP4 eGFP-IBD cells were transduced with a Moloney murine leukemia virus-derived retroviral vector encoding firefly luciferase. After 72 h, d-luciferin was added to the cell culture medium and incubated for 5 min. Bioluminescence was measured using an IVIS 100 system. In all panels, average numbers ± standard deviations (SD) for duplicate experiments are shown.
Next, the HeLaP4-derived cell lines were transduced with a Moloney murine leukemia virus-derived retroviral vector encoding eGFP-luciferase. No reduction in luciferase expression was observed in HeLaP4 cells overexpressing eGFP-Δ325 or eGFP-IBD in comparison with control cell lines (Fig. 5c). Identical results were obtained for the 293T-derived cell lines (data not shown).
HIV-1 replication is inhibited by overexpression of eGFP-Δ325 or eGFP-IBD.
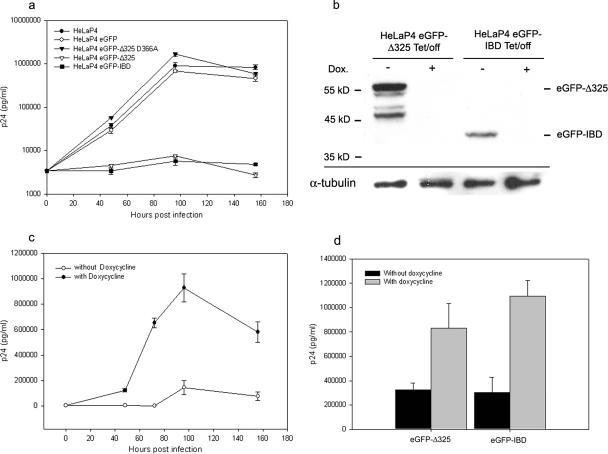
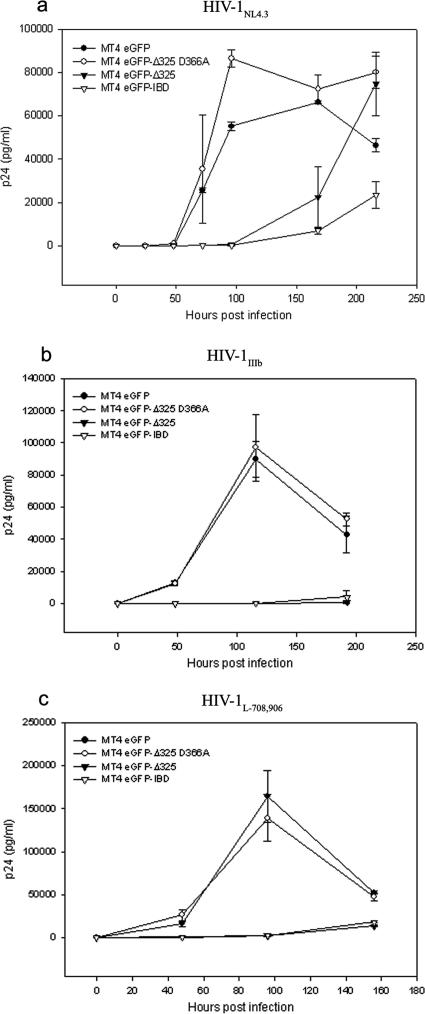
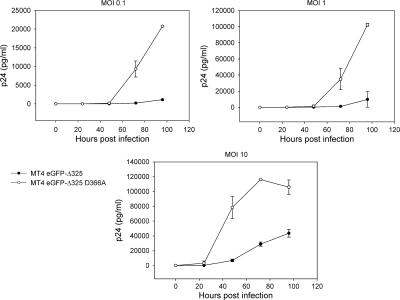
To investigate the effects of eGFP-Δ325 and eGFP-IBD overexpression on multiple-round HIV-1 replication, HeLaP4-derived cells were infected with HIV-1NL4.3 at an MOI of 0.01 (Fig. 6a). Viral replication was followed by harvesting of the supernatant and measurement of the p24 concentration. HIV replication was readily detected in HeLaP4, HeLaP4 eGFP-Δ325 D366A, and HeLaP4 eGFP cells. In HeLaP4 eGFP-Δ325 and HeLaP4 eGFP-IBD cells, however, no viral replication could be detected for 156 h after infection. Even at an MOI of 0.1 or after passaging the infected cells for more than 2 weeks, no viral breakthrough was detected (data not shown). To exclude any possible selection bias, we engineered monoclonal HeLaP4 cell lines expressing eGFP-IBD and eGFP-Δ325 from a tetracycline-dependent promoter (Tet/off) (40). FACS analysis (data not shown) and Western blotting revealed that high-level expression was shut off in the presence of 5 mg/ml doxycycline (Fig. 6b). Infection of HeLaP4 eGFP-IBD Tet/off cells with HIV-1NL4.3 at an MOI of 0.01 resulted in a clear inhibition of replication in the absence of doxycycline, whereas the addition of doxycycline rescued HIV-1 replication (Fig. 6c). Identical results were obtained when eGFP-Δ325 was transcribed from a Tet/off promoter (Fig. 6d). Because HeLaP4 cells are not physiological host cells for HIV, the MT-4-based T-cell lines were infected with HIV-1NL4.3 at an MOI of 0.1, and p24 accumulation in the supernatant was monitored (Fig. 7a). While HIV-1 replication was readily detected in the MT-4 eGFP and MT-4 eGFP-Δ325 D366A mutant cell lines, HIV-1 replication was inhibited >20-fold at 4 days postinfection in MT-4 eGFP-Δ325 and MT-4 eGFP-IBD cells. Viral breakthrough was delayed 4 days in these cell lines. To exclude virus strain specificity, the MT-4-derived cell lines were also infected with HIV-1IIIB and HIV-1L-708,906 at an MOI of 0.1 (Fig. 7b and c). HIV-1L-708,906 is a virus strain that is resistant to the integrase inhibitor L-708,906 (14). Again, a strong inhibition of replication of both viral strains was observed in both MT-4 eGFP-Δ325 and MT-4 eGFP-IBD cells. To further investigate viral breakthrough, MT-4 eGFP-Δ325 and MT-4 eGFP-Δ325 D366A cells were infected with HIV-1NL4.3 at different MOIs (Fig. 8). At an MOI of 10, viral breakthrough was delayed only 24 h, but viral replication was still threefold lower at 4 days postinfection.
FIG. 6.
Effects of eGFP-Δ325 and eGFP-IBD overexpression on HIV-1 replication in HeLaP4 cells. (a) HeLaP4, HeLaP4 eGFP-Δ325 D366A, HeLaP4 eGFP Δ325, HeLaP4 eGFP-IBD, and HeLaP4 eGFP cells were infected with HIV-1NL4.3 at an MOI of 0.01. HIV-1 replication was monitored by measurement of p24 in the supernatant at different times postinfection. The experiment was performed in quadruplicate, and average values ± SD are shown. (b) Western blot analysis of the monoclonal HeLaP4 eGFP-Δ325 Tet/off and HeLaP4 eGFP-IBD Tet/off cell lines. One week prior to Western blotting, cells were grown in the presence (+) or absence (−) of 5 mg/ml doxycycline. Equal amounts of protein extracted from each cell line were analyzed by Western blotting using an eGFP antibody. Equal loading was controlled by α-tubulin detection. (c) HeLaP4 Tet/off eGFP-IBD cells were cultured in the presence or absence of 5 mg/ml doxycycline for 7 days before infection with HIV-1NL4.3 at an MOI of 0.01. HIV-1 replication was monitored by measurement of p24 in the supernatant at different times postinfection. (d) HeLaP4 eGFP-Δ325 Tet/off and HeLaP4 eGFP-IBD Tet/off cells were cultured in the presence or absence of 5 mg/ml doxycycline for 7 days before infection with HIV-1NL4.3 at an MOI of 0.01. At 96 h postinfection, the p24 concentration in the supernatant was measured. Average numbers of p24 ± SD for duplicate experiments were determined.
FIG. 7.
Effects of eGFP-Δ325 and eGFP-IBD overexpression on HIV-1 replication in MT-4 cells. (a) MT-4 eGFP-Δ325 D366A, MT-4 eGFP Δ325, MT-4 eGFP-IBD, and MT-4 eGFP cells were infected with HIV-1NL4.3 (a) HIV-1IIIb (b), or HIV-1L-708,906 (c) at an MOI of 0.1. HIV-1 replication was monitored by measurement of p24 in the supernatant at different times postinfection. Average p24 values ± SD for triplicate experiments are shown.
FIG. 8.
Effect of MOI on viral replication in MT-4 eGFP-Δ325 and MT-4 eGFP-Δ325 D366A cells. MT-4 eGFP-Δ325 D366A and MT-4 eGFP Δ325 cells were infected with HIV-1NL4.3 at an MOI of 0.01, 0.1, or 1. HIV-1 replication was monitored by measurement of p24 in the supernatant at different times postinfection. Average p24 values ± SD for triplicate experiments are shown.
HIV-1 replication in eGFP-Δ325 and eGFP-IBD cells is blocked at the integration step.
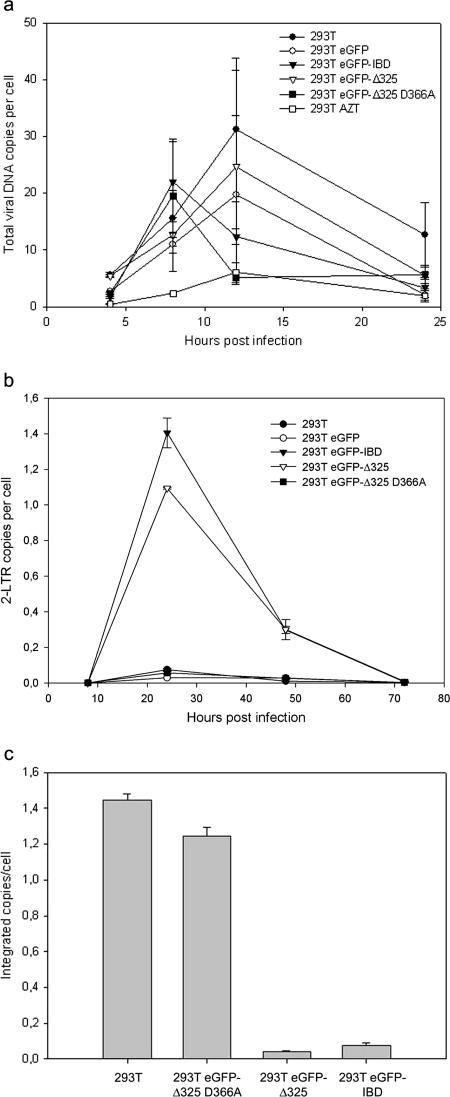
The manifest phenotype in HIV-1 replication experiments raised the question of whether the block in replication occurred during early or late steps of the viral life cycle. First, the effects of eGFP-Δ325 and eGFP-IBD expression on the late steps of viral replication were analyzed in the 293T-derived cell lines. 293T, 293T eGFP-Δ325, 293T eGFP-Δ325 D366A, 293T eGFP-IBD, and 293T eGFP cells were transfected with the molecular clone pNL4.3, and the virus was harvested at 48 h posttransfection. These viral stocks were analyzed for p24 content and titrated in MT-4 cells. No significant differences in p24 content or infectious titer were observed among the different cell lines (data not shown), ruling out an effect on virus production or replication fitness. In the next step, the formation of total HIV-1 DNA and 2-LTR circles was monitored by real-time quantitative PCR (Fig. 9). To ensure single-round infections, the 293T, 293T eGFP-Δ325, 293T eGFP-Δ325 D366A, 293T eGFP-IBD, and 293T eGFP cell lines were infected with a VSV-G-pseudotyped HIV-1NL4.3 virus. Plasmid contamination of DNase-treated vector stocks was controlled by including cells treated with AZT, inhibiting de novo viral DNA synthesis (Fig. 9a). DNA was extracted at several time points after infection, and the total HIV-1 DNA was measured by quantitative PCR using primers specific for the gag gene, not priming on to the lentiviral vector used to establish the cell lines. As shown in Fig. 9a, the accumulation of viral DNA at early time points was not affected upon expression of eGFP-IBD or eGFP-Δ325. In contrast, the number of 2-LTR circles increased 10-fold in the 293T eGFP-IBD and 293T eGFP-Δ325 cells (Fig. 9b). 2-LTR circles are formed in the nucleus and are an indirect indication of nuclear import. As a consequence, the presence of eGFP-Δ325 or eGFP-IBD obviously did not inhibit reverse transcription or nuclear import. The integration events could not be measured by real-time quantitative Alu PCR, which makes use of an LTR primer that also anneals to the lentiviral vector sequences present in the cell lines. Therefore, infected cells were passaged at least four times to dilute nonintegrated DNA, ensuring that only integrated copies of viral DNA remained. The absence of 2-LTR circles at 72 h postinfection (Fig. 9b) is indicative of the loss of nonintegrated DNA. Measuring the total HIV-1 DNA after four passages revealed a 20-fold reduction in the number of proviruses in the 293T eGFP-Δ325 and eGFP-IBD cell lines in comparison to the control cell lines (Fig. 9c).
FIG. 9.
eGFP-Δ325 and eGFP-IBD overexpression impairs HIV-1 integration. 293T, 293T eGFP, 293T eGFP-IBD, 293T eGFP-Δ325, and 293T eGFP-Δ325 D366A cells were infected with VSV-G-pseudotyped HIV-1NL4.3. To exclude plasmid contamination of vector stocks, cells were treated in a parallel experiment with 0.09 μg/ml AZT. DNAs were extracted at different times postinfection, and the amounts of total viral DNA (a) and 2-LTR circles (b) were determined by real-time quantitative PCR. To quantify integrated DNA (c), infected cells were passaged four times, and the total viral DNA was measured. All quantifications were performed in duplicate, and averages ± SD are shown.
DISCUSSION
Since LEDGF/p75 was discovered as an interaction partner of HIV-1 integrase (5), many studies have addressed its putative role in HIV-1 replication. Using RNA interference-mediated knockdown, a modest inhibition of HIV-1 replication was shown in HeLaP4 cells depleted of LEDGF/p75 (36). These results are at odds with the wild-type levels of HIV replication seen in LEDGF/p75-depleted Jurkat cells (22).
We hypothesized that on the condition that LEDGF/p75 plays a role as a DNA tethering factor in HIV-1 replication, expression of the C-terminal domain of LEDGF/p75 lacking the domains known to interact with chromatin (e.g., the PWWP and AT-hook domains) (31, 34) would compete with endogenous LEDGF/p75 for its interaction with integrase and thereby inhibit the function of LEDGF/p75. Overexpression of full-length LEDGF/p75 did not result in such a phenotype (36; data not shown). Here we demonstrate unequivocally that HIV-1 vector transduction and virus replication are indeed inhibited dramatically upon overexpression of the C-terminal fragment of LEDGF/p75 (Δ325) (aa 325 to 530) or the IBD (aa 347 to 429) due to a block of integration. Although we cannot formally exclude an indirect effect of the fusion protein, inhibition of virus replication was not observed upon overexpression of the eGFP-Δ325 D366A fragment, containing a single mutation that abolishes the interaction with IN (7). This suggests that the observed inhibition in viral replication and vector transduction is highly specific and indeed based on direct interaction of the overexpressed fragments (eGFP-IBD and eGFP-Δ325) with IN. The direct interaction of integrase and the eGFP-Δ325 and eGFP-IBD fragments was confirmed by in vivo fluorescence cross-correlation spectroscopy (Table 1). In addition to the eGFP fusions, cell lines overexpressing hemagglutinin-tagged Δ325 or IBD were made as well (data not shown). Inhibition of HIV-1 replication was also observed in these cell lines, although at a lower level, probably due to lower expression levels or incorrect folding of the proteins. This excludes the possibility of nonspecific inhibition by the eGFP moiety of the C-terminal fragments.
For the HeLaP4-derived cell lines, viral breakthrough could never be observed in the presence of eGFP-Δ325 or eGFP-IBD. In the T-cell line MT-4, viral breakthrough was severely delayed (Fig. 7 and 8). To exclude artifacts due to cell line selection, we also analyzed the inhibition of HIV-1 replication in HeLaP4 cells expressing eGFP-IBD or eGFP-Δ325 from a tetracycline-regulatable promoter (Fig. 6c and d). In these cell lines, inhibition of viral replication was clearly dependent on the presence of eGFP-IBD or eGFP-Δ325; upon doxycycline-mediated inhibition of eGFP-IBD or eGFP-Δ325 expression, HIV-1 replication was restored.
We previously analyzed the effect of LEDGF/p75 reduction on lentiviral vector transduction efficiency (36). At most, a 20% inhibition of transduction was observed in a monoclonal HeLaP4-derived cell line, with LEDGF/p75 levels reduced to close to the detection limit. Most probably, knockdown of the abundant LEDGF/p75 protein was not capable of reducing this cofactor below the threshold required for integration, although we could not exclude the possibility that lentiviral vector integration was less dependent on the presence of LEDGF/p75 than HIV-1 replication. The results presented here prove that LEDGF/p75 is also a cofactor for lentiviral vector transduction. Overexpression of eGFP-IBD or eGFP-Δ325 clearly reduced lentiviral vector transduction 3- to 15-fold, depending on the cell line used (Fig. 5a and b), whereas transduction with a retroviral vector was not inhibited at all (Fig. 5c). The lentiviral specificity of the phenotype described is in agreement with the established specificity of LEDGF/p75 for interactions with lentiviral integrases (2, 22).
In the HeLaP4, 293T, and MT-4 cell lines, we overexpressed Δ325 or IBD fused to eGFP to increase protein stability and for visualization by confocal microcopy. Interestingly, both fusion proteins localized to the nuclei of HeLaP4 cells, although the NLS of LEDGF/p75 (aa 148 to 156) is not present in these fragments (Fig. 2) (23, 37). Most probably, these fragments still bind DNA, as proposed previously (31), or are directed to the chromatin through interactions with other cellular proteins (25). Still, this residual nuclear localization is not sufficient to rescue viral replication. The D366A mutation renders the IBD defective for interaction with HIV-1 integrase (7). Interestingly, introduction of the D366A mutation into eGFP-Δ325 did not alter the cellular localization of this domain (Fig. 2N).
The nuclear localization of HIV-1 integrase has been the focus of many investigations (6, 10, 12, 21, 26, 27). According to Maertens et al. and Vanegas et al., transient coexpression of HIV-1 integrase and NLS-deficient LEDGF/p75 redirects both proteins to the cytoplasm (23, 37). However, in cell lines stably expressing NLS-deficient LEDGF/p75, both proteins are again directed to the nucleus due to cell division and subsequent tethering of LEDGF/p75 to the chromatin (37). In contrast, upon stable overexpression of eGFP-IBD or eGFP-Δ325, the fusion protein accumulates in the cytoplasm in the presence of mRFP-integrases (Fig. 2), while endogenous LEDGF/p75 retains its nuclear localization (Fig. 3b). This may be explained by the fact that the residual chromatin binding activity of the C-terminal fragments is masked by binding to integrase. By confocal microscopy, a clear correlation was observed between the amounts of eGFP-IBD or eGFP-Δ325 and mRFP-integrases expressed and their cellular distributions (data not shown). At a high mRFP-integrases-to-eGFP-Δ325 ratio, all eGFP-Δ325 was found in the cytoplasm, while part of the mRFP-integrases was tethered to the nucleus. It is likely that the truncated fragments and the endogenous LEDGF/p75 compete for binding with HIV integrase. When eGFP-Δ325 is titrated out by an excess of integrase, part of the integrase proteins accumulate in the nucleus through interaction with endogenous LEDGF/p75. The opposite was seen when integrase was titrated out by eGFP-Δ325.
Although our results show a physical interaction of IBD and Δ325 proteins with integrase in the cytoplasm, one has to be careful in interpretation since overexpressed integrase does not always predict the behavior of integrase during HIV-1 infection. Therefore, we investigated the mechanism of action of IBD by using quantitative PCR. Quantitative PCR analysis of 293T-derived cells infected with VSV-G-pseudotyped HIV-1 pinpointed the replication block in eGFP-IBD- and eGFP-Δ325-overexpressing cells to the integration step (Fig. 9c), whereas reverse transcription and nuclear import were not inhibited. On the contrary, a clear increase in 2-LTR circle formation was observed in the eGFP-IBD- and eGFP-Δ325-overexpressing cells (Fig. 9a and b), consistent with an inhibition of the DNA strand transfer reaction (17, 39). The growth kinetics of cells and overall eGFP expression remained unchanged during passaging, excluding the possibility that the observed integration defect was due to selective killing of eGFP-IBD- or eGFP-Δ325-overexpressing cells. Although eGFP-IBD and eGFP-Δ325 cells are capable of relocalizing mRFP-integrases from the nucleus to the cytoplasm (Fig. 2), the nuclear import of the HIV-1 preinitiation complex (PIC) was not inhibited. The most plausible explanation for the replication block is the inability of the PIC, lacking functional LEDGF/p75, to dock the preintegration complex to the chromatin, preventing viral integration. Nonintegrated viral DNA may then accumulate as 2-LTR (and 1-LTR) circles. In favor of a tethering role for LEDGF/p75, Ciuffi et al. recently showed that HIV-1 integration occurs less frequently in transcription units in LEDGF/p75-depleted cells (9). Nevertheless, at this stage we cannot completely rule out the possibility of a disturbed conformation of the PIC hampering integration. Inhibition by overexpressed IBD of the interaction of another cofactor with IN cannot be excluded either, but this would imply overlapping binding sites on integrase. The possibility of blocking HIV-1 replication by overexpressing part of a cellular protein has been described before (41). Yung and coworkers showed that overexpression of the S6 integrase binding domain of integrase interactor 1 (INI1) efficiently inhibited the production of viral particles. INI1 is part of the SWI/SNF complex involved in chromatin remodeling and interacts with HIV-1 integrase (18, 41). Whereas in the case of INI1 it has remained unclear whether the inhibition of virus production by the S6 fragment relates to the role of INI1 during HIV replication, in the case of LEDGF/p75 all evidence points to a role of LEDGF/p75 in integration.
Our data reveal that overexpression of the truncation mutants can compete with endogenous LEDGF/p75 and thereby inhibit virus replication. A true transdominant should not lose its phenotype when LEDGF/p75 is present in excess of the IBD. Although our Western blots suggest that the overexpressed fragments are present in excess of endogenous LEDGF/p75, the observed potency may hint at an apparent transdominant phenotype. Since it is possible that recruitment of LEDGF/p75 versus the IBD already occurs in the cytoplasmic PIC (22), cytoplasmic ratios of both proteins ought to be compared. The fact that the inhibition by overexpression of the deletion mutants can be overcome by a single point mutation (D366A), together with the recently described crystal structure of the IBD in complex with the integrase core domain (3), supports a very defined protein-protein interface. Our data highlight the LEDGF/p75-integrase interaction as a novel antiviral target for the development of small-molecule inhibitors. In this regard, it is interesting that no cross-resistance was observed with a diketo acid-resistant strain (Fig. 7c) (14). Like INI1, LEDGF/p75 is a cellular protein, and as a consequence, it is very unlikely that an immunogenic response will occur upon overexpression of LEDGF/p75 fragments. IBD-based gene therapeutic approaches against AIDS may thus be envisaged as well. Thus, far, no effect on cell viability was seen by overexpressing the IBD.
.
Acknowledgments
We acknowledge M. Michiels, A. Nijs, and B. Van Remoortel for excellent technical assistance, B. Van Maele for a critical reading of the manuscript, and A. Hantson for the luciferase-expressing retroviral vector. We appreciate the help of V. Van Duppen from the Hematology Division, KULeuven, with cell sorting. We thank the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, for providing different cell lines and the HIV pNL4.3 molecular clone.
Work at KULeuven was supported by the European Commission (LSHB-CT-2003-503480) (TRIoH project) and the SBO program of the Flemish Institute Supporting Scientific-Technological Research in Industry (IWT). J.D.R. and L.V. are funded by grants from the IWT.
Footnotes
Published ahead of print on 20 September 2006.
REFERENCES
- 1.Baekelandt, V., A. Claeys, K. Eggermont, E. Lauwers, B. De Strooper, B. Nuttin, and Z. Debyser. 2002. Characterization of lentiviral vector-mediated gene transfer in adult mouse brain. Hum. Gene Ther. 13:841-853. [DOI] [PubMed] [Google Scholar]
- 2.Busschots, K., J. Vercammen, S. Emiliani, R. Benarous, Y. Engelborghs, F. Christ, and Z. Debyser. 2005. The interaction of LEDGF/p75 with integrase is lentivirus-specific and promotes DNA binding. J. Biol. Chem. 280:17841-17847. [DOI] [PubMed] [Google Scholar]
- 3.Cherepanov, P., A. L. Ambrosio, S. Rahman, T. Ellenberger, and A. Engelman. 2005. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc. Natl. Acad. Sci. USA 102:17308-17313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherepanov, P., E. Devroe, P. A. Silver, and A. Engelman. 2004. Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J. Biol. Chem. 279:48883-48892. [DOI] [PubMed] [Google Scholar]
- 5.Cherepanov, P., G. Maertens, P. Proost, B. Devreese, J. Van Beeumen, Y. Engelborghs, E. De Clercq, and Z. Debyser. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278:372-381. [DOI] [PubMed] [Google Scholar]
- 6.Cherepanov, P., W. Pluymers, A. Claeys, P. Proost, E. De Clercq, and Z. Debyser. 2000. High-level expression of active HIV-1 integrase from a synthetic gene in human cells. FASEB J. 14:1389-1399. [DOI] [PubMed] [Google Scholar]
- 7.Cherepanov, P., Z. Y. Sun, S. Rahman, G. Maertens, G. Wagner, and A. Engelman. 2005. Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75. Nat. Struct. Mol. Biol. 12:526-532. [DOI] [PubMed] [Google Scholar]
- 8.Cherepanov, P., D. Surratt, J. Toelen, W. Pluymers, J. Griffith, E. De Clercq, and Z. Debyser. 1999. Activity of recombinant HIV-1 integrase on mini-HIV DNA. Nucleic Acids Res. 27:2202-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciuffi, A., M. Llano, E. Poeschla, C. Hoffmann, J. Leipzig, P. Shinn, J. R. Ecker, and F. Bushman. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11:1287-1289. [DOI] [PubMed] [Google Scholar]
- 10.Depienne, C., P. Roques, C. Creminon, L. Fritsch, R. Casseron, D. Dormont, C. Dargemont, and S. Benichou. 2000. Cellular distribution and karyophilic properties of matrix, integrase, and Vpr proteins from the human and simian immunodeficiency viruses. Exp. Cell Res. 260:387-395. [DOI] [PubMed] [Google Scholar]
- 11.De Rijck, J., B. Van Maele, and Z. Debyser. 2005. Positional effects of the central DNA flap in HIV-1-derived lentiviral vectors. Biochem. Biophys. Res. Commun. 328:987-994. [DOI] [PubMed] [Google Scholar]
- 12.Devroe, E., A. Engelman, and P. A. Silver. 2003. Intracellular transport of human immunodeficiency virus type 1 integrase. J. Cell Sci. 116:4401-4408. [DOI] [PubMed] [Google Scholar]
- 13.Emiliani, S., A. Mousnier, K. Busschots, M. Maroun, B. Van Maele, D. Tempe, L. Vandekerckhove, F. Moisant, L. Ben-Slama, M. Witvrouw, F. Christ, J. C. Rain, C. Dargemont, Z. Debyser, and R. Benarous. 2005. Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 280:25517-25523. [DOI] [PubMed] [Google Scholar]
- 14.Fikkert, V., B. Van Maele, J. Vercammen, A. Hantson, B. Van Remoortel, M. Michiels, C. Gurnari, C. Pannecouque, M. De Maeyer, Y. Engelborghs, E. De Clercq, Z. Debyser, and M. Witvrouw. 2003. Development of resistance against diketo derivatives of human immunodeficiency virus type 1 by progressive accumulation of integrase mutations. J. Virol. 77:11459-11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge, H., Y. Si, and R. G. Roeder. 1998. Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J. 17:6723-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge, Y. Z., M. T. Pu, H. Gowher, H. P. Wu, J. P. Ding, A. Jeltsch, and G. L. Xu. 2004. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 279:25447-25454. [DOI] [PubMed] [Google Scholar]
- 17.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646-650. [DOI] [PubMed] [Google Scholar]
- 18.Kalpana, G. V., S. Marmon, W. Wang, G. R. Crabtree, and S. P. Goff. 1994. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 266:2002-2006. [DOI] [PubMed] [Google Scholar]
- 19.Kirsch, R. D., and E. Joly. 1998. An improved PCR-mutagenesis strategy for two-site mutagenesis or sequence swapping between related genes. Nucleic Acids Res. 26:1848-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lalezari, J. P., K. Henry, M. O'Hearn, J. S. Montaner, P. J. Piliero, B. Trottier, S. Walmsley, C. Cohen, D. R. Kuritzkes, J. J. Eron, Jr., J. Chung, R. DeMasi, L. Donatacci, C. Drobnes, J. Delehanty, and M. Salgo. 2003. Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N. Engl. J. Med. 348:2175-2185. [DOI] [PubMed] [Google Scholar]
- 21.Limón, A., E. Devroe, R. Lu, H. Z. Ghory, P. A. Silver, and A. Engelman. 2002. Nuclear localization of human immunodeficiency virus type 1 preintegration complexes (PICs): V165A and R166A are pleiotropic integrase mutants primarily defective for integration, not PIC nuclear import. J. Virol. 76:10598-10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Llano, M., M. Vanegas, O. Fregoso, D. Saenz, S. Chung, M. Peretz, and E. M. Poeschla. 2004. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J. Virol. 78:9524-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maertens, G., P. Cherepanov, Z. Debyser, Y. Engelborghs, and A. Engelman. 2004. Identification and characterization of a functional nuclear localization signal in the HIV-1 integrase interactor LEDGF/p75. J. Biol. Chem. 279:33421-33429. [DOI] [PubMed] [Google Scholar]
- 24.Maertens, G., P. Cherepanov, W. Pluymers, K. Busschots, E. De Clercq, Z. Debyser, and Y. Engelborghs. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278:33528-33539. [DOI] [PubMed] [Google Scholar]
- 25.Maertens, G. N., P. Cherepanov, and A. Engelman. 2006. Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J. Cell Sci. 119:2563-2571. [DOI] [PubMed] [Google Scholar]
- 26.Petit, C., O. Schwartz, and F. Mammano. 1999. Oligomerization within virions and subcellular localization of human immunodeficiency virus type 1 integrase. J. Virol. 73:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pluymers, W., P. Cherepanov, D. Schols, E. De Clercq, and Z. Debyser. 1999. Nuclear localization of human immunodeficiency virus type 1 integrase expressed as a fusion protein with green fluorescent protein. Virology 258:327-332. [DOI] [PubMed] [Google Scholar]
- 28.Popovic, M., M. G. Sarngadharan, E. Read, and R. C. Gallo. 1984. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science 224:497-500. [DOI] [PubMed] [Google Scholar]
- 29.Ray, P., A. De, J. J. Min, R. Y. Tsien, and S. S. Gambhir. 2004. Imaging tri-fusion multimodality reporter gene expression in living subjects. Cancer Res. 64:1323-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma, P., D. P. Singh, N. Fatma, L. T. Chylack, Jr., and T. Shinohara. 2000. Activation of LEDGF gene by thermal—and oxidative—stresses. Biochem. Biophys. Res. Commun. 276:1320-1324. [DOI] [PubMed] [Google Scholar]
- 31.Singh, D. P., E. Kubo, Y. Takamura, T. Shinohara, A. Kumar, L. T. Chylack, Jr., and N. Fatma. 2006. DNA binding domains and nuclear localization signal of LEDGF: contribution of two helix-turn-helix (HTH)-like domains and a stretch of 58 amino acids of the N-terminal to the trans-activation potential of LEDGF. J. Mol. Biol. 355:379-394. [DOI] [PubMed] [Google Scholar]
- 32.Singh, D. P., N. Ohguro, T. Kikuchi, T. Sueno, V. N. Reddy, K. Yuge, L. T. Chylack, Jr., T. Shinohara, N. Fatma, and A. Kimura. 2000. Lens epithelium-derived growth factor: effects on growth and survival of lens epithelial cells, keratinocytes, and fibroblasts. Biochem. Biophys. Res. Commun. 267:373-381. [DOI] [PubMed] [Google Scholar]
- 33.Turlure, F., E. Devroe, P. A. Silver, and A. Engelman. 2004. Human cell proteins and human immunodeficiency virus DNA integration. Front. Biosci. 9:3187-3208. [DOI] [PubMed] [Google Scholar]
- 34.Turlure, F., G. Maertens, S. Rahman, P. Cherepanov, and A. Engelman. 2006. A tripartite DNA-binding element, comprised of the nuclear localization signal and two AT-hook motifs, mediates the association of LEDGF/p75 with chromatin in vivo. Nucleic Acids Res. 34:1663-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ullman, E. F., H. Kirakossian, S. Singh, Z. P. Wu, B. R. Irvin, J. S. Pease, A. C. Switchenko, J. D. Irvine, A. Dafforn, C. N. Skold, et al. 1994. Luminescent oxygen channeling immunoassay: measurement of particle binding kinetics by chemiluminescence. Proc. Natl. Acad. Sci. USA 91:5426-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandekerckhove, L., F. Christ, B. Van Maele, J. De Rijck, R. Gijsbers, C. Van den Haute, M. Witvrouw, and Z. Debyser. 2006. Transient and stable knock-down of the integrase co-factor LEDGF/p75 reveals its role in the replication cycle of human immunodeficiency virus. J. Virol. 80:1886-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanegas, M., M. Llano, S. Delgado, D. Thompson, M. Peretz, and E. Poeschla. 2005. Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J. Cell Sci. 118:1733-1743. [DOI] [PubMed] [Google Scholar]
- 38.Van Maele, B., K. Busschots, L. Vandekerckhove, F. Christ, and Z. Debyser. 2006. Cellular cofactors of HIV-1 integration. Trends Biochem. Sci. 31:98-105. [DOI] [PubMed] [Google Scholar]
- 39.Van Maele, B., J. De Rijck, E. De Clercq, and Z. Debyser. 2003. Impact of the central polypurine tract on the kinetics of human immunodeficiency virus type 1 vector transduction. J. Virol. 77:4685-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vigna, E., M. Amendola, F. Benedicenti, A. D. Simmons, A. Follenzi, and L. Naldini. 2005. Efficient Tet-dependent expression of human factor IX in vivo by a new self-regulating lentiviral vector. Mol. Ther. 11:763-775. [DOI] [PubMed] [Google Scholar]
- 41.Yung, E., M. Sorin, A. Pal, E. Craig, A. Morozov, O. Delattre, J. Kappes, D. Ott, and G. V. Kalpana. 2001. Inhibition of HIV-1 virion production by a transdominant mutant of integrase interactor 1. Nat. Med. 7:920-926. [DOI] [PubMed] [Google Scholar]
- 42.Zufferey, R., D. Nagy, R. J. Mandel, L. Naldini, and D. Trono. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871-875. [DOI] [PubMed] [Google Scholar]