Abstract
We previously identified a novel insect picorna-like virus, termed Kakugo virus (KV), from the brains of aggressive worker honeybees that had counterattacked a giant hornet. To survey the prevalence of KV in worker populations engaged in various labors, we quantified KV genomic RNA. KV was detected specifically from aggressive workers in some colonies, while it was also detected from other worker populations in other colonies where the amount of KV detected in the workers was relatively high, suggesting that KV can infect various worker populations in the honeybee colonies. To investigate whether the KV strains detected were identical, phylogenetic analysis was performed. There was less than a 2% difference in the RNA-dependent RNA polymerase (RdRp) sequences between KV strains from aggressive workers and those from other worker populations, suggesting that all of the viruses detected were virtually the same KV. We also found that some of the KV-infected colonies were parasitized by Varroa mites, and the sequences of the KV strains detected from the mites were the same as those detected from the workers of the same colonies, suggesting that the mites mediate KV prevalence in the honeybee colonies. KV strains had approximately 6% and 15% sequence differences in the RdRp region from deformed wing virus and Varroa destructor virus 1, respectively, suggesting that KV represents a viral strain closely related to, but distinct from, these two viruses.
The European honeybee (Apis mellifera L.) is a eusocial insect, and female adults differentiate into a reproductive caste (queen) and a sterile caste (workers) (33). Workers are devoted to the labors required to maintain colony activity rather than their own reproduction, and labors are divided according to age after eclosion. For example, young workers nurse the brood and the queen in the hive (nurse bees), middle-aged workers guard their colony at the hive entrance (guards), and older workers forage for nectar and pollen (foragers). These labors by sterile workers are altruistic, especially the attacking behavior of guards against their natural enemies, such as the giant hornet Vespa mandarinia japonica, which often results in the death of the worker after stinging behavior (9, 32). Little is known, however, of the molecular basis and evolutional process underlying these self-sacrificing behaviors.
We previously screened genes differentially expressed between highly aggressive guards that had attacked a giant hornet (Vespa mandarinia japonica) used as a decoy (attackers) and nonaggressive inside workers that had escaped from the decoy (escapers) using the differential-display method, to examine the molecular basis of their aggressive behavior in the brain. As a result, we identified a novel RNA termed Kakugo RNA (13). cDNA cloning of Kakugo RNA revealed that it shares structural characteristics with the insect picorna-like virus genome. Furthermore, inoculation experiments indicated that Kakugo RNA has infectivity in worker honeybees. Thus, we concluded that a novel insect picorna-like virus, termed Kakugo virus (KV), had infected the brains of the aggressive workers. These results suggested a possible relationship between KV infection and honeybee aggression (13). The causal association between KV infection and worker aggressive behaviors, however, remains unknown.
On the other hand, some groups recently reported sequences of two kinds of viruses that are very closely related to KV. One is the Varroa destructor virus 1 (VDV-1), identified from honeybee ectoparasitic mites (Varroa destructor), whose genome and polyprotein share 95% homology with those of KV (27). Another is deformed wing virus (DWV), which has been known as a pathogenic virus of the honeybee. The genomic sequence of DWV registered in the GenBank database shares 98% homology with that of KV (12, 13, 20). Although the pathogenicity of DWV infection is not as serious when it infects adult honeybees (23), DWV infection in larvae might cause wing deformity after eclosion and shorten the life span (1). However, bees with deformed wings were not identified among the attackers in our previous experiments (13). These findings suggest that KV, VDV-1, and DWV have different pathogenicities, though they share structural similarities.
In the present epidemiologic study, we examined the distribution of KV among various worker populations to analyze the relationship between KV infection and aggressive worker behaviors. We also compared the nucleotide sequences of the detected KV strains phylogenetically to examine whether they represent identical or different virus species.
MATERIALS AND METHODS
Collection of honeybees.
European honeybee A. mellifera L. Italian race colonies maintained at the Experimental Station for Medical Plant Studies (Kemigawa, Hanamigawa-ku, Chiba, Japan) or in the Department of Biological Sciences of the University of Tokyo (Hongo, Bunkyo-ku, Tokyo, Japan) were used. Attackers, nurse bees, foragers, and guards were collected based on their behaviors as reported previously (3, 13, 19). We also collected workers randomly from inside the hive (inside bees). In addition, aggressive bees, which we termed “reserves,” were collected from among those that scrambled from inside the hive to attack a giant hornet that was presented at the entrance of the hive when there were no guards. The bees were collected with tweezers and immediately anesthetized on ice. The brains were dissected from the heads under a microscope and stored at −80°C before use. The samples used in the present experiments were as follows: K3-F, mushroom bodies (MBs) (the higher center of the insect brain [15]) from 100 foragers collected from colony 3 at Kemigawa on 4 Novembere 2000 (note that the sample name, K3-F, is based on the location of the colony [Kemigawa], the colony number [3], and the worker population [foragers] from which the sample was derived); K3-N, MBs from 100 nurse bees collected from colony 3 at Kemigawa on 7 November 2000; K3-A, MBs from 102 attackers collected from colony 3 at Kemigawa on 8 November 2000; K3-E, MBs from 100 escapers collected from colony 3 at Kemigawa on 8 November 2000; H7-A, whole heads from 70 attackers collected from colony 7 at Hongo on 14 November 2000; H7-F, whole heads from 70 foragers collected from colony 7 at Hongo on 14 November 2000; K4-A, MBs from 100 attackers collected from colony 4 at Kemigawa on 4 August 2000; K4-I, MBs from 102 inside bees collected from colony 4 at Kemigawa on 4 August 2000; K5-G, whole heads from 70 guards collected from colony 5 at Kemigawa on 3 November 2004; K6-G, whole heads from 70 guards collected from colony 6 at Kemigawa on 3 November 2004; H8-R, whole heads from 70 reserve bees collected from colony 8 at Hongo on 1 December 2004; H9-R, whole heads from 70 reserve bees collected from colony 9 at Hongo on 1 December 2004; H10-R, whole heads from 70 reserve bees collected from colony 10 at Hongo on 1 December 2004; H12-A, antennal lobes (ALs) from 10 attackers against Vespa simillima xanthoptera collected from colony 12 at Hongo on 21 September 2005; and H12-E, ALs from 10 escapers from Vespa simillima xanthoptera collected from colony 12 at Hongo on 21 September 2005. To analyze the tissue distribution of KV, reserve bees were collected from colony 6, and the brain and other tissues of the head, thorax, and abdomen were dissected from each bee.
Isolation of total RNA.
Brain, head, or other tissue was homogenized in 800 μl TRIzol reagent (Invitrogen, Carlsbad, CA). To analyze the KV infection rate using reserve bees, 1/10 (80 μl) of each 10-head homogenate (each 800 μl) was combined as a “group” (total, 800 μl), and total RNA was extracted. The rest of each head homogenate (720 μl) was stored at −80°C for further analysis. KV infection was analyzed using total RNA isolated from whole heads of “groups” (n = 10 each) of foragers, nurse bees, or inside bees. To isolate total RNA from one brain, 5 μg of glycogen (Ambion, Austin, TX) was added as a carrier during RNA precipitation.
Quantification of KV genomic RNA.
Total RNA (0.5 to 1.0 μg) was treated with RNase-free DNase I and then reverse transcribed with or without Superscript II (Invitrogen) using an oligo(dT) primer. PCR was performed with a Light Cycler (Roche, Basel, Switzerland) and ExTaq DNA polymerase (Takara, Japan) using gene-specific primers (+8388 to +8415 and +8745 to +8765 for Kakugo cDNA and +2 to +21 and +376 to +395 for honeybee cytoplasmic actin cDNA) and fluorescent probes (+8654 to +8679 and +8681 to +8716 for Kakugo cDNA and +297 to +313 and +315 to +354 for actin cDNA) in a total volume of 20 μl (13). The PCR conditions used were as follows: 95°C for 1 s, 55°C for 10 s, and 72°C for 10 s. Kakugo and actin cDNA clones at known concentrations were used as standards. The relative Kakugo RNA content was estimated by normalization using the amount of actin mRNA. When the relative Kakugo RNA content was over 0.001, it was judged as KV infection, because the detection limit was around 0.001 in our experiments.
Collection of Varroa mites and quantification of KV genomic RNA.
Varroa mites that were attached to the honeybees or floating after the honeybees were soaked in ice-cold water were collected with tweezers. Total RNA was isolated from each mite using TRIzol, and then Kakugo RNA was quantified using quantitative reverse transcription (RT)-PCR. To normalize the amount of template, partial actin cDNA was subcloned from Varroa destructor and the actin mRNA was also quantified using specific primers (+1/+27 and +332/+353) and probes (+276/+304 and +306/+332).
Phylogenetic analysis.
Sequences corresponding to the RNA-dependent RNA polymerase (RdRp) region and the VP1 region were amplified using RT-PCR. For primers, sequences conserved between KV and DWV were used: +2528/+2547 and +4004/+4022 for VP1 of KV and +8755/+8783 and +9879/+9906 for RdRp of KV. PCR was performed using Thermococcus kodakaraensis DNA polymerase (TOYOBO, Japan) with the following program: 35 cycles of 94°C for 15s or 30 s, 55°C for 30 s, and 68°C for 1 min 30s. PCR products were purified using a Qiaquick PCR purification kit (QIAGEN, Tokyo, Japan) or precipitation in ethanol, and the nucleotide sequences of both strands were determined with a Dye-Terminator cycle-sequencing kit ver. 3.1 and an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). For sequencing, multiple primers were designed to correspond to the following regions: +2528/+2547, +2933/+2962, +3162/+3189, +3569/+3592, +3587/+3612, and +3917/+3936 for VP1 of KV and +8755/+8783, +9191/+9211, +9363/+9390, and +9760/+9788 for RdRp of KV. The site at which nucleotide signals overlapped was designated N. To examine the rate of mismatches during the PCR, a specific KV plasmid was used in every experiment as a control, and the analyzed sequences were always 100% identical between experiments.
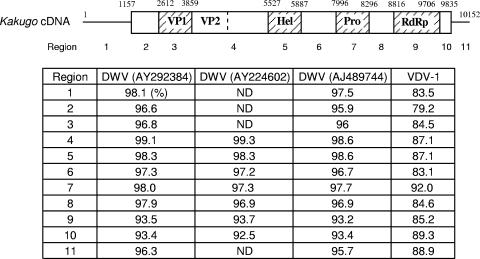
The nucleotide sequences in the coding region corresponding to VP1 (+2612/+3859) and RdRp (+8816/+9706) were used in the phylogenetic analysis with the neighbor-joining method using the Clustal W program (31). The statistical significance of branch order was estimated by performing 1,000 replications of bootstrap resampling of the original aligned nucleotide sequences. The genome information of DWV was based on the registered sequences in GenBank: AJ489744 (Rossi and Lanzi, Italy; sequence deposited in the database); AY292384 (DeMiranda et al., United States; sequence deposited in the database); and AY224602 (Gauthier et al., France; sequence deposited in the database).
Nucleotide sequence accession number.
The nucleotide sequence for Varroa actin cDNA reported in the present paper was submitted to the DDBJ/EMBL/GenBank data bank under accession number AB242568.
RESULTS
Analysis of KV prevalences among various worker populations.
To examine whether KV infects only attackers, we investigated the prevalences of KV among various worker populations. RNA was extracted from each worker population (n = 70 to 100 each) that displayed distinct behaviors in each colony, and the Kakugo RNA was quantified using quantitative RT-PCR. The relative Kakugo RNA contents were normalized to those of actin mRNA and compared.
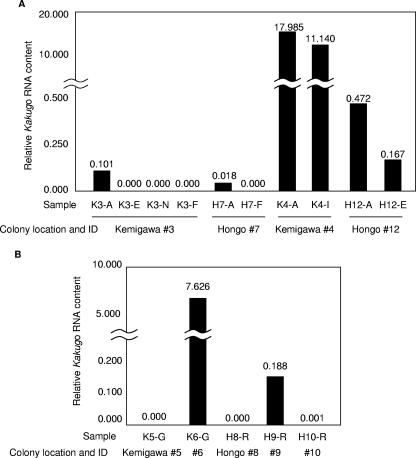
As reported previously, KV was detected in attackers, but not escapers, nurse bees, or foragers, in colony 3 (Kemigawa) (13). Quantitative RT-PCR of the Kakugo RNA of the same samples confirmed the previous results (Fig. 1A). Similarly, KV was detected in attackers, but not foragers, in colony 7 (Hongo). Colony 4 (Kemigawa) was so aggressive as a whole that the inside workers also participated in defensive attacks when the giant hornets were presented as decoys. Therefore, we were obliged to collect workers randomly from inside the hive (inside bees) instead of escapers. As a result, large amounts of KV were detected, not only in attackers, but also in inside bees (Fig. 1A). KV was also detected in both attackers and escapers in colony 12 (Hongo). These results indicate that attacker-specific KV infection occurs only under restricted colony conditions. The amounts of Kakugo RNA detected in colonies 4 and 12 were larger than those in colonies 3 and 7, particularly in colony 4, where it was 100 to 1,000 times greater (Fig. 1A). These results indicate that KV can be infectious to various worker populations and suggest that the attacker-specific KV infection occurs under some restricted conditions.
FIG. 1.
Comparison of the KV contents among various worker populations. (A) Seventy to 100 workers per sample were collected from four colonies according to their behavior as attackers (K3-A, H7-A, and K4-A), escapers (K3-E), nurse bees (K3-N), foragers (K3-F and H7-F), and inside workers (K4-I). In addition, 10 workers per sample were collected as attackers (H12-A) and escapers (H12-E) against Vespa simillima xanthoptera. The Kakugo RNA and actin mRNA were quantified using quantitative RT-PCR. The relative Kakugo RNA contents normalized to that of actin mRNA are indicated by the vertical bars and above the bars. (B) Seventy to 100 guards (G) or reserve bees (R) per sample were collected from five colonies (K5, K6, H8, H9, and H10; the samples are indicated as combinations, e.g., K5-G). The relative Kakugo RNA contents normalized to that of actin mRNA are indicated by the vertical bars and the numbers above the bars.
We next compared the amounts of KV in worker populations from colonies that were maintained at the same place and time. First, KV RNA was quantified using guards collected on 3 November 2004 from colonies 5 and 6 kept at Kemigawa. We examined the guards instead of attackers because we could not obtain giant hornets as decoys at that time. KV was detected in colony 6, but not colony 5 (Fig. 1B). Second, reserves were collected from three colonies (8 to 10) kept at Hongo on 1 December 2004 (see Materials and Methods). Since bee colonies do not have guards at the end of autumn in Japan, we collected aggressive bees from inside the hive (reserves; see Materials and Methods) using giant hornets as decoys. KV was detected in colony 9, but not in colonies 8 and 10 (Fig. 1B). These results indicated that the prevalences of KV are different among colonies, even when the seasons and locations are the same, suggesting that the KV infection rates and amounts of virus differ according to each colony condition.
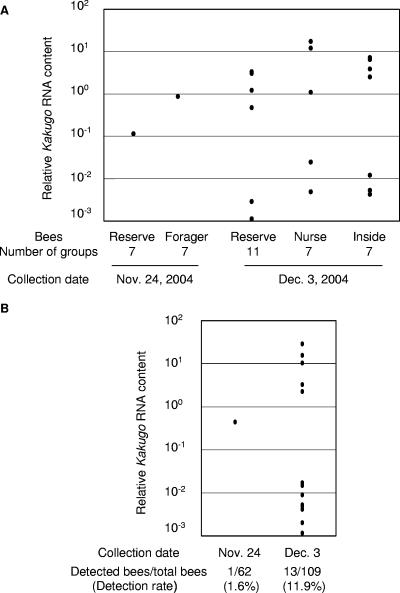
To test whether KV is epidemic in a colony, we collected reserves, as well as the other worker populations, twice within 9 days (24 November and 3 December 2004) from colony 6 and examined the change in the infection ratio and the amounts of KV during this period. We collected 7 to 11 groups of workers, each of which included 10 reserves or other worker populations (foragers for 24 November and nurse bees and inside bees for 3 December). The KV RNAs in these groups were quantified by quantitative RT-PCR. KV was detected in all populations tested, indicating that KV infected various worker populations in this colony. Interestingly, both the number of KV-positive groups and the greatest amount of KV RNA detected in these groups were higher than those in reserves and foragers collected on 24 November (Fig. 2A). To investigate these changes in each individual bee, the KV RNA was quantified using each reserve bee collected on 24 November and 3 December. The number of KV-positive bees increased from 1 to 13, and the KV infection ratio increased from 1.6% to 11.9% (Fig. 2B). Five bees collected on 3 December contained 10 to 100 times more KV than the bees collected on 24 November. These results suggested that KV is epidemic in the honeybee colonies.
FIG. 2.
Change in KV prevalence within a colony. (A) Reserve bees or other worker populations (foragers collected on 24 November and nurse bees and inside bees collected on 3 December) were collected from colony 6. Each individual bee was homogenized separately, aliquots of the homogenates from 10 samples were mixed as a group, and the Kakugo RNA and actin mRNA of each group were quantified using quantitative RT-PCR. The relative Kakugo RNA contents normalized to that of actin mRNA are indicated by dots. The populations of bees, numbers of groups examined, and collection dates are indicated below the graph. (B) The relative Kakugo RNA contents of each individual were quantified using samples in the groups from which Kakugo RNA was detected in panel A and are indicated by dots. The numbers of collected reserve bees and Kakugo-positive reserve bees and the detection rates are indicated below.
Detection of KV from Varroa mites.
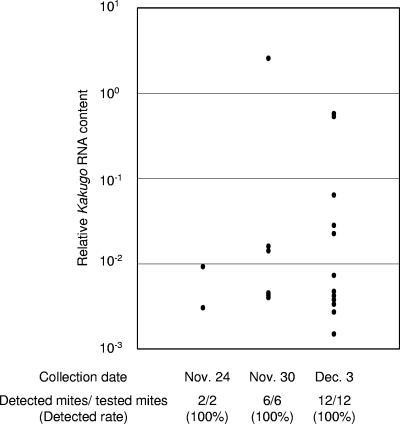
To determine the factors that affect KV prevalence in a colony, we observed colony 6 and noticed that the colony was parasitized with Varroa mites (Varroa destructor). These mites pierce the body wall of the honeybees between abdominal segments and feed on the hemolymph, and they often transmit various pathogenic viruses that cause colony collapse (1). Colony 12 was also infested with Varroa mites. To examine the possibility that Varroa mites mediate KV prevalence, we collected 2, 6, and 12 mites attached to the body walls of the workers collected on 24 November, 30 November, and 3 December from colony 6. All of these mites were attached to the nurse bees or inside bees. We quantified the Kakugo RNA in the mites by quantitative RT-PCR. KV was detected in all mites collected from the colony (Fig. 3), although the contents varied among individuals. No KV was detected, however, in any mites (n = 6) that were collected from the colonies where no KV was detected (data not shown). These results suggest that the mites mediated the KV prevalence in the honeybee colony.
FIG. 3.
Detection of KV in the Varroa mites that parasitized the honeybees. The Varroa mites that were attached to workers were collected on 24 November, 30 November, and 3 December, and the amounts of Kakugo RNA were quantified using quantitative RT-PCR. The relative Kakugo RNA contents normalized with that of Varroa destructor actin mRNA are indicated by dots. The numbers of collected mites and KV-positive mites and the detection rates are shown below.
Tissue distribution of Kakugo RNA.
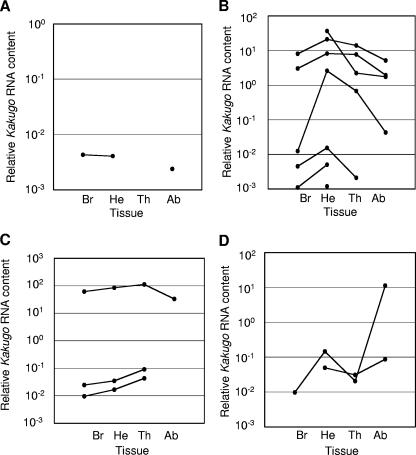
Some honeybee viruses that are transmitted by Varroa mites are distributed throughout various tissues in the honeybee (1, 29). Previously, we reported that KV was detected specifically in the brains of naturally infected attackers, whereas it was also detected in thoraxes and abdomens after artificial infection using the lysate of KV-infected workers (13). Here, we reexamined whether KV infects various body parts when it naturally infects the workers. Kakugo RNA was quantified by quantitative RT-PCR using total RNA from brains, heads without brains, thoraxes, and abdomens of 13 reserve bees that were collected from colony 6 on 3 December (Fig. 2B). KV was detected in all organs (Fig. 4), indicating that KV also infects various tissues when it naturally infects workers. Tissues with the greatest amounts of KV differed among individuals: one bee was infected with KV mostly in the brain, seven in heads without brains, two in the thorax, and three in the abdomen (Fig. 4), suggesting that the tissue distribution of KV might be affected by the physiological condition of workers.
FIG. 4.
Tissue distribution of KV in workers infected naturally with KV. The relative Kakugo RNA contents in the brains (Br), heads except brains (He), thoraxes (Th), and abdomens (Ab) of KV-positive reserve bees that were collected from colony 6 on 3 December 2004 (n = 13) were quantified using quantitative RT-PCR. The dots representing relative Kakugo RNA contents in each tissue from the same individual are connected by lines. When the amount of KV was below the detection limit of the quantitative RT-PCR in some of the tissue samples, the line was cut. (A to D) Examples showing that KV was detected mostly in the brain (A) (n = 1), head (B) (n = 7), thorax (C) (n = 3), and abdomen (D) (n = 2).
Phylogenetic analysis using sequences corresponding to the RdRp region.
The RT-PCR analysis described above led to the question of whether KVs detected from attackers and other various workers or Varroa mites were identical, because the sequences were not determined. In addition, because KV is very closely related to DWV and VDV-1, it was possible that these viruses were detected together. Furthermore, VDV-1 infects Varroa mites (27) and DWV may also infect mites, based on immunologic and RT-PCR methods (2, 25, 29, 30, 35). Therefore, we analyzed the sequences of the genomic RNAs of the viruses that we tentatively identified as KV in the above-mentioned experiments. We determined partial genomic sequences of viruses detected from worker and mite samples and compared them to each other, as well as with those of DWV and VDV-1. For sequencing, we used the RdRp region, because it had the lowest homologies (93.2 to 93.7%) among KV and three DWV strains (Fig. 5). The RdRp region was amplified from total RNA using RT-PCR and then purified. Direct sequencing was used, because it enabled us to analyze the major viral population in the samples and to exclude possible nucleotide mismatching during cloning procedures (11). To enable detection of both KV and DWV, even when DWV contaminated the samples, we designed primers for PCR and sequencing using sequences common to these two viruses.
FIG. 5.
Sequence identities between KV, DWV, and VDV-1 in various genomic regions. (Top) Structure of the Kakugo cDNA. The open and shaded boxes show open reading frames and domains of the Kakugo polyprotein, such as virion protein (VP), helicase (Hel), protease (Pro), and RdRp domains. The numbers above the cDNA indicate base positions corresponding to each domain from the 5′ terminus. Kakugo cDNA was divided into 11 regions: 1, 5′ untranslated region (UTR); 2, coding region for the N-terminal region; 3, VP1; 4, coding region between VP1 and Hel; 5, Hel; 6, coding region between Hel and Pro; 7, Pro; 8, coding region between Pro and RdRp; 9, RdRp; 10, coding region for the C terminus region; 11, 3′ UTR. (Bottom) Nucleotide sequence identities between the Kakugo RNA and the genomes of DWV and VDV-1 (27) in each region are indicated. Three DWV strains and one VDV-I strain, whose genome sequences are registered in GenBank, were used (the GenBank accession numbers for DWV are shown in parentheses). ND, not determined.
All of the viral sequences that were detected from worker populations used in our experiments had less than 2% sequence differences from that of the KV consensus genomic sequence that we reported previously (13) and approximately 6% and 15% sequence differences from those of DWV and VDV-1 (Fig. 6), strongly suggesting that all the viruses we detected using RT-PCR in the present study represented KV substrains and not DWV or VDV-1. Even when we analyzed KV genomic sequences from a single bee, there were some nucleotide positions with mixed sequencing signals (designated N in the sequences registered in GenBank) (Table 1), suggesting that there are KV quasispecies.
FIG. 6.
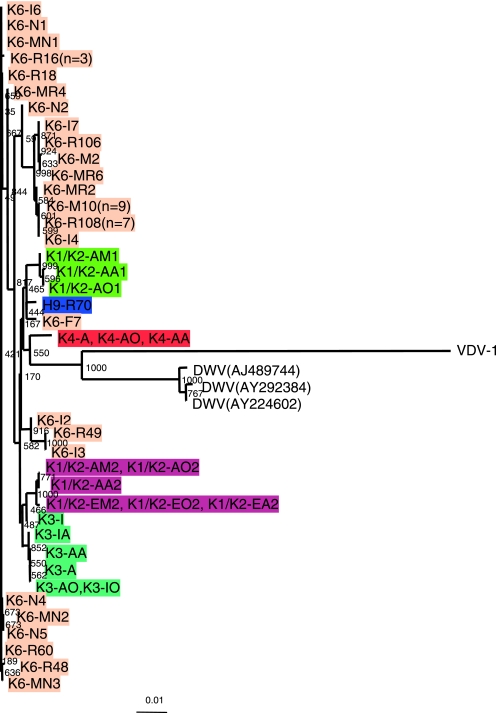
Phylogenetic analysis of KV in the RdRp region detected from various worker populations and Varroa mites. The phylogenetic tree was constructed with the nucleotide sequences encompassing motifs 1 to 8 of the RdRp domain (891 bp) using the neighbor-joining method. The samples obtained from the same colony are indicated by the same background color. The samples with light-orange backgrounds were derived from colony 6. The sample type is indicated by the first letter of each sample after the colony identification number: A, attacker; F, forager; E, escaper; R, reserve bee; N, nurse bee; I, inside bee; M, Varroa mite. (Note that the sample name, K3-FO, is based on the location of the colony [Kemigawa], the colony number [3], the worker population [foragers], and the tissue [optic lobes] from which the sample was derived.) The numbers at the branches show bootstrap values obtained after 1,000 replications of bootstrap sampling. The bar shows distances.
TABLE 1.
Sequence information for the RdRp and VP1 regions of KV substrains
| Sample | Yr | Colony IDa | Sample category and tissueb | Accession no.c
|
|
|---|---|---|---|---|---|
| RdRp | VP1 | ||||
| K1/K2-AM1 | 1999 | 1 and 2 in Kemigawa | MBs of attackers | AB242577 | AB245505 |
| K1/K2-AO1 | 1999 | 1 and 2 in Kemigawa | OLs of attackers | AB242576 | ND |
| K1/K2-AA1 | 1999 | 1 and 2 in Kemigawa | ALs of attackers | AB242575 | ND |
| K1/K2-AM2 | 1999 | 1 and 2 in Kemigawa | MBs of attackers | AB242578 | AB245504 |
| K1/K2-AO2 | 1999 | 1 and 2 in Kemigawa | OLs of attackers | AB242578 | ND |
| K1/K2-AA2 | 1999 | 1 and 2 in Kemigawa | ALs of attackers | AB242579 | ND |
| K1/K2-EM2 | 1999 | 1 and 2 in Kemigawa | MBs of escapers | AB242580 | ND |
| K1/K2-EO2 | 1999 | 1 and 2 in Kemigawa | OLs of escapers | AB242580 | ND |
| K1/K2-EA2 | 1999 | 1 and 2 in Kemigawa | ALs of escapers | AB242580 | ND |
| K3-A | 2000 | 3 in Kemigawa | MBs of attackers | AB242585 | AB245506 |
| K3-AO | 2000 | 3 in Kemigawa | OLs of attackers | AB242581 | ND |
| K3-AA | 2000 | 3 in Kemigawa | ALs of attackers | AB242583 | ND |
| K3-I | 2000 | 3 in Kemigawa | MBs of inside bees | AB242586 | ND |
| K3-IO | 2000 | 3 in Kemigawa | OLs of inside bees | AB242582 | ND |
| K3-IA | 2000 | 3 in Kemigawa | ALs of inside bees | AB242584 | ND |
| K4-A | 2001 | 4 in Kemigawa | MBs of attackers | AB242587 | AB245498 |
| K4-AO | 2001 | 4 in Kemigawa | OLs of attackers | AB242587 | ND |
| K4-AA | 2001 | 4 in Kemigawa | ALs of attackers | AB242587 | ND |
| H9-R70 | 2004 | 9 in Hongo | WHs of reserves | AB250361 | AB245502 |
| K6-F7 | 2004 | 6 in Kemigawa | WH of forager | AB250362 | ND |
| K6-R49 | 2004 | 6 in Kemigawa | WH of reserve | AB250363 | AB245500 |
| K6-I3 | 2004 | 6 in Kemigawa | WH of inside bee | AB250363 | AB245501 |
| K6-I2 | 2004 | 6 in Kemigawa | WH of inside bee | AB251935 | ND |
| K6-I4 | 2004 | 6 in Kemigawa | WH of inside bee | AB242594 | ND |
| K6-R108 | 2004 | 6 in Kemigawa | WH of reserve (n = 7) | AB242594 | AB245503 |
| K6-M10 | 2004 | 6 in Kemigawa | WB of mite (n = 9) | AB242594 | AB245503 |
| K6-MR2 | 2004 | 6 in Kemigawa | WB of mite | AB242592 | ND |
| K6-M2 | 2004 | 6 in Kemigawa | WB of mite | AB242589 | ND |
| K6-MR6 | 2004 | 6 in Kemigawa | WB of mite | AB242590 | ND |
| K6-I7 | 2004 | 6 in Kemigawa | WH of inside bee | AB242591 | ND |
| K6-R106 | 2004 | 6 in Kemigawa | WH of reserve | AB242591 | ND |
| K6-I7 | 2004 | 6 in Kemigawa | WH of inside bee | AB242591 | ND |
| K6-MR4 | 2004 | 6 in Kemigawa | WB of mite | AB242588 | ND |
| K6-N4 | 2004 | 6 in Kemigawa | WH of nurse bee | AB242570 | AB245499 |
| K6-N5 | 2004 | 6 in Kemigawa | WH of nurse bee | AB250360 | ND |
| K6-MN2 | 2004 | 6 in Kemigawa | WB of mite | AB242571 | ND |
| K6-R16 | 2004 | 6 in Kemigawa | WH of reserve (n = 3) | AB242573 | AB245499 |
| K6-R18 | 2004 | 6 in Kemigawa | WH of reserve | AB242574 | ND |
| K6-N1 | 2004 | 6 in Kemigawa | WH of nurse | AB242572 | ND |
| K6-I6 | 2004 | 6 in Kemigawa | WH of inside bee | AB242572 | ND |
| K6-R60 | 2004 | 6 in Kemigawa | WH of reserve | AB242572 | ND |
| K6-MN1 | 2004 | 6 in Kemigawa | WB of mite | AB242572 | AB245499 |
| K6-R48 | 2004 | 6 in Kemigawa | WH of reserve | AB242593 | ND |
| K6-MR8 | 2004 | 6 in Kemigawa | WB of mite | AB242593 | ND |
| K6-MN3 | 2004 | 6 in Kemigawa | WB of mite | AB242569 | ND |
Colony ID includes the colony number and colony location.
If identical sequences were found in several samples in the same condition, the number of samples (n) is indicated. OL, optic lobe; WH, whole head; WB, whole body.
ND; not determined.
To examine whether KV genomic sequences derived from attackers were different from those derived from other worker populations, phylogenetic analysis was performed using these sequences. There were instances in which identical KV genomic sequences were obtained from aggressive reserves, as well as other worker populations derived from colony 6: K6-R108 and K6-I4; K6-R106 and K6-I7; K6-R49 and K6-I3; and K6-R60, K6-I6, and K6-N1 (Fig. 6). These results indicate that a single KV substrain can infect both the aggressive reserves and other worker populations. The KV substrains obtained from different worker populations derived from the same colony tended to form a cluster in which the substrains had less than 1.6% sequence differences: K1/K2-AM2, K1/K2-AO2, K1/K2-AA2, K1/K2-EM2, K1/K2-EO2, and K1/K2-EA2, less than 0.2% difference; K3-I, K3-IA, K3-AA, K3-A, K3-AO, and K3-IO, less than 1.6% difference (Fig. 6). These results suggest that KV substrains diverged within these colonies. In contrast, KV substrains derived from the same colony were located in different clusters of the phylogenic tree: K6-F7, K6-I2, K6-R49, and K6-I3 in colony 6, which was infested with Varroa mites (Fig. 6). These results indicate that multiple KV substrains were epidemic simultaneously in colony 6.
To examine whether the viruses detected from the Varroa mites were identical with those from the worker populations where the mites were collected, the sequences of KV viruses detected from Varroa mites were determined. Many of the viral sequences from the Varroa mites were identical in both the RdRp and VP1 regions with those from worker populations (Fig. 6 and 7 and Table 1). Sequences from mites, K6-MN1 and K6-M10, were identical with those from the bees of the same colony, K6-R14 and K6-R108, respectively, indicating that the same KV substrains infected the honeybees and Varroa mites in these colonies and that KV can be transmitted between the honeybee and the Varroa mite.
FIG. 7.
Comparison of phylogenetic trees constructed using RdRp and VP1 regions. A representative sample was selected from each cluster of the phylogenetic tree shown in Fig. 6, and the phylogenetic tree was reconstructed with the nucleotide sequences encompassing the VP1 region (A) and motifs 1 to 8 of the RdRp domain (B) using the neighbor-joining method. Samples obtained from the same colony are indicated by the same background color. The samples with light-orange backgrounds were derived from colony 6. The sample type is indicated by the first letter of each sample after the colony identification number: A, attacker; R, reserve bee; N, nurse bee; I, inside bee; M, Varroa mite. The numbers at the branches show bootstrap values obtained after 1,000 replications of bootstrap sampling. The bar indicates distance.
Phylogenetic analysis using sequences corresponding to the VP1 region.
Due to genomic recombination, the sequence diversities in each region of the genomic RNA sometimes differ, even in the same picornavirus species (18, 21, 22, 26). Therefore, we performed phylogenetic analysis of KV substrains using the sequences corresponding to the VP1 region and compared the results with the above-mentioned results for the RdRp region. For this purpose, representative viruses were selected from each cluster of the phylogenetic tree constructed using the RdRp region, and their sequences corresponding to the VP1 region were determined. The difference among KV substrains was at most approximately 3% (Fig. 7B), which was more divergent than that estimated using sequences corresponding to the RdRp region (Fig. 7A and Table 1). The sequence difference between KV substrains and DWV also remained less than 3%, whereas that between KV and VDV-1 was approximately 15%. Together with the results with the RdRp region, these results suggest that KV is more closely related to DWV than VDV-1, confirming the previous reports (6).
DISCUSSION
Prevalence of KV among honeybee colonies and individual workers within colonies.
In the present study, we performed an epidemiologic study and found two colonies in which KV was detected only in the attackers and three colonies in which KV was detected in various worker populations (Fig. 1). These results, along with the phylogenetic analysis (Fig. 6), indicate that the same KV strain can infect not only attackers, but also various worker populations. The mechanism of KV progression and the reason that KV was infectious only to the attackers in colony 3 (Kemigawa) and colony 7 (Hongo) are unclear due to the limited data. Considering that the amount of KV in the attackers of the first two colonies was less than that in various worker populations of the other colonies and that KV can be epidemic in honeybee hives (Fig. 2), attacker-specific KV infection might represent its primary, or rather last, infection phase. Alternatively, it is also plausible that ecologic differences, such as location, season, and year, affect KV prevalence.
We observed that colony 4 (Kemigawa), which was heavily infected with KV, was so vigorous and aggressive that it was difficult to collect escapers. Colony 6, which was also infected with KV, survived for at least 2 months after KV detection (data not shown), suggesting that KV infection is not extremely lethal for honeybees. We previously speculated that the workers infected with KV might become more aggressive than noninfected workers (12). More studies are needed to examine this possibility, especially at the individual worker level. On the other hand, KV infection might not be essential for the intrinsic worker aggressive behaviors, because there were some colonies in which no significant KV was detected from any worker populations, including the attackers (data not shown).
Tissue distribution of KV in the worker honeybees.
We previously reported that KV was detectable in the mushroom bodies of the brains of the attackers but was scarcely detectable in the total heads, thoraxes, and abdomens (13). In the present study, KV was detected not only in the brains, but also in the heads without brains, thoraxes, and abdomens of the reserve bees that were confirmed to be infected with KV (Fig. 4). There was less than a 0.1% difference in the sequences corresponding to the RdRp region of KV substrains that were detected from each of their tissues (n = 3) (data not shown). It is possible that the tissue distributions of KV are different in attackers and reserves. On the other hand, it is also possible that our previous experimental design was not appropriate: we used brains from 100 attackers and heads, thoraxes, and abdomens from 5 attackers for KV detection by quantitative RT-PCR (13). Considering that the KV infection rate ranged from 1.6% to 11.9% even in the heavily infected reserve bees from colony 6 (Fig. 2B), it is possible that the 5 attackers did not include enough KV-infected bees whereas some of the former 100 attackers contained KV. As discussed below, KV might be introduced into the hemolymph of the bees from bites by Varroa mites. Therefore, it seems rational to conclude that KV can infect various organs/tissues, at least in the reserves. It is interesting that the tissue distributions were quite different among the infected bees. It is plausible that the physiological condition of the workers reflects the tissue distribution of KV, and this could reflect on the different pathogenicities proposed for KV, DWV, and VDV-1.
Prevalence of KV substrains among honeybee colonies and its possible infection route.
We also performed phylogenetic analysis of KV using single bees in a single colony, enabling us to observe the prevalence of KV as a wild insect virus at each substrain level. There were tendencies toward infection of a single colony with KV substrains that are very closely related to each other, forming a cluster in the phylogenetic tree and suggesting that KV diverges within a colony. This finding suggests that a colony infected with KV for a relatively long time can lead to the divergence of a single KV strain. The relation between KV and the honeybee might somehow be symbiotic. In contrast, multiple KV substrains were detected in colony 6, where the Varroa mite infection occurred. The fact that the same KV substrains were detected from both the workers and Varroa mites strongly suggests that the mites mediate KV prevalence in the colony and that the mites caused infection with multiple KV substrains in colony 6. Varroa mite infestation results in decreased honeybee immunity (14, 34), and this might further promote the KV epidemic in the colony.
We did not observe Varroa mite parasitism in colonies in which KV was detected only in attackers (colonies 3 and 7). Therefore, it is also possible that Varroa mite parasitism is not essential for KV infection in a honeybee colony, as reported for DWV (7). Recent studies have suggested that various bee viruses can be vertically transmitted (4, 5, 8, 10, 28), and this might also be the case for KV. It is likely that when Varroa mites parasitize a colony already infected with KV, they contribute to expanding the KV prevalence, as reported for DWV, sacbrood virus, and Kashmir bee virus infections (28, 29).
Relationship between KV, DWV, and VDV-1.
The full-length genomic sequences of DWV and VDV-1 were recently reported, and the results indicated that KV, DWV, and VDV-1 are closely related to each other. So far, different pathogenicities have been proposed for KV and DWV: association with aggressive worker behaviors and shrunken worker wings, respectively. In our experiments, we observed no attackers with shrunken wings. In addition, no DWV sequence was detected in the phylogenetic-tree analysis, although we used sequences that were common to KV and DWV as primers.
The tissue distribution of DWV was also different from that of KV in that DWV was not detected in the heads of healthy workers (35). Thus, it is likely that differences in their genomic sequences generate different pathogeneses and tissue distributions. The nucleotide sequence alternation was much higher in the RdRp region (approximately 6% between KV and DWV) than that in the VP1 region (less than 3%), and in the RdRp region, the alternation resulted in a 2-amino-acid-residue substitution between KV strains and DWV (data not shown). These results suggest that KV represents a virus strain distinct from DWV. Indeed, at least a 1% sequence difference, or substitution of a few amino acids, is enough to generate distinct toxicities in some kinds of viruses (16, 17, 24). It is also possible that KV and DWV represent a regional difference (e.g., KV in Japan and DWV in Europe) in what is essentially the same virus. Artificial infection experiments using infectious clones of these viruses will be needed to test these possibilities.
Acknowledgments
We thank Yutaka Orihara for permitting us to keep bee colonies in the Experimental Station for Medical Plant Studies, Graduate School of Pharmaceutical Sciences, at the University of Tokyo.
This work was supported by the Program for Promotion of Basic Research Activities for Innovative Bioscience (PROBRAIN) and Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture of Japan; Terumo Life Science Foundation; and the Naito Foundation. T.F. is the recipient of a Grant-in-Aid for young scientists from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print on 13 September 2006.
REFERENCES
- 1.Bailey, L., and B. V. Ball. 1991. Honey bee pathology. Academic Press Inc., San Diego, Calif.
- 2.Bowen-Walker, P. L., S. J. Martin, and A. Gunn. 1999. The transmission of deformed wing virus between honeybees (Apis mellifera L.) by the ectoparasitic mite Varroa jacobsoni Oud. J. Invertebrate Pathol. 73:101-106. [DOI] [PubMed] [Google Scholar]
- 3.Breed, M. D., T. A. Smith, and A. Torres. 1992. Role of guard honey bees (Hymenoptera: Apidae) in nestmate discrimination and replacement of removed guards. Ann. Entomol. Soc. Am. 85:633-637. [Google Scholar]
- 4.Chen, Y., J. Evans, and M. Feldlaufer. 2006. Horizontal and vertical transmission of viruses in the honey bee, Apis mellifera. J. Invertebrate Pathol. 92:152-159. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y., J. S. Pettis, and M. F. Feldlaufer. 2005. Detection of multiple viruses in queen of the honey bee Apis mellifera L. J. Invertebrate Pathol. 90:118-121. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Y., Y. Zhao, J. Hammond, H. Hsu, J. Evans, and M. Feldlaufer. 2004. Multiple virus infections in the honey bee and genome divergence of honey bee viruses. J. Invertebrate Pathol. 87:84-93. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y. P., J. A. Higgins, and M. F. Feldlaufer. 2005. Quantitative real-time reverse transcription-PCR analysis of deformed wing virus infection in the honeybee (Apis mellifera L.). Appl. Environ. Microbiol. 71:436-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y. P., J. S. Pettis, A. Collins, and M. F. Feldlaufer. 2006. Prevalence and transmission of honeybee viruses. Appl. Environ. Microbiol. 72:606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dade, H. A. 1962. Anatomy and dissection of the honeybee. International Bee Research Association, Cardiff, United Kingdom.
- 10.Fievet, J., D. Tentcheva, L. Gauthier, J. de Miranda, F. Cousserans, M. E. Colin, and M. Bergoin. 2006. Localization of deformed wing virus infection in queen and drone Apis mellifera L. Virol. J. 3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forns, X., J. Bukh, R. H. Purcell, and S. U. Emerson. 1997. How Escherichia coli can bias the results of molecular cloning: preferential selection of defective genomes of hepatitis C virus during the cloning procedure. Proc. Natl. Acad. Sci. USA 94:13909-13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiyuki, T., H. Takeuchi, M. Ono, S. Ohka, T. Sasaki, A. Nomoto, and T. Kubo. 2005. Kakugo virus from brains of aggressive worker honeybees. Adv. Virus Res. 65:1-27. [DOI] [PubMed] [Google Scholar]
- 13.Fujiyuki, T., H. Takeuchi, M. Ono, S. Ohka, T. Sasaki, A. Nomoto, and T. Kubo. 2004. Novel insect picorna-like virus identified in the brains of aggressive worker honeybees. J. Virol. 78:1093-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregory, P. G., J. D. Evans, T. Rinderer, and L. de Guzman. 2005. Conditional immune-gene suppression of honeybees parasitized by Varroa mites. J. Insect Sci. 5:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heisenberg, M. 1998. What do the mushroom bodies do for the insect brain? An introduction. Learning Memory 5:1-10. [PMC free article] [PubMed] [Google Scholar]
- 16.Kagiwada, S., Y. Yamaji, K. Komatsu, S. Takahashi, T. Mori, H. Hirata, M. Suzuki, M. Ugaki, and S. Namba. 2005. A single amino acid residue of RNA-dependent RNA polymerase in the Potato virus X genome determines the symptoms in Nicotiana plants. Virus Res. 110:177-182. [DOI] [PubMed] [Google Scholar]
- 17.Kanno, T., D. Mackay, G. Wilsden, and P. Kitching. 2001. Virulence of swine vesicular disease virus is determined at two amino acids in capsid protein VP1 and 2A protease. Virus Res. 80:101-107. [DOI] [PubMed] [Google Scholar]
- 18.Koenen, F., H. Vanderhallen, N. D. Dickinson, and N. J. Knowles. 1999. Phylogenetic analysis of European encephalomyocarditis viruses: comparison of two genomic regions. Arch. Virol. 144:893-903. [DOI] [PubMed] [Google Scholar]
- 19.Kubo, T., M. Sasaki, J. Nakamura, H. Sasagawa, K. Ohashi, H. Takeuchi, and S. Natori. 1996. Change in the expression of hypopharyngeal-gland proteins of the worker honeybees (Apis mellifera L.) with age and/or role. J. Biochem. 119:291-295. [DOI] [PubMed] [Google Scholar]
- 20.Lanzi, G., J. R. de Miranda, M. B. Boniotti, C. E. Cameron, A. Lavazza, L. Capucci, S. M. Camazine, and C. Rossi. 2006. Molecular and biological characterization of deformed wing virus of honeybees (Apis mellifera L.). J. Virol. 80:4998-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindberg, A. M., P. Andersson, C. Savolainen, M. N. Mulders, and T. Hovi. 2003. Evolution of the genome of Human enterovirus B: incongruence between phylogenies of the VP1 and 3CD regions indicates frequent recombination within the species. J. Gen. Virol. 84:1223-1235. [DOI] [PubMed] [Google Scholar]
- 22.Lukashev, A. N., V. A. Lashkevich, O. E. Ivanova, G. A. Koroleva, A. E. Hinkkanen, and J. Ilonen. 2003. Recombination in circulating enteroviruses. J. Virol. 77:10423-10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin, S. J. 2001. The role of Varroa and viral pathogens in the collapse of honeybee colonies: a modeling approach. J. Appl. Ecol. 38:1082-1093. [Google Scholar]
- 24.Nomoto, A., T. Omata, H. Toyoda, S. Kuge, H. Horie, Y. Kataoka, Y. Genba, Y. Nakano, and N. Imura. 1982. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc. Natl. Acad. Sci. USA 79:5793-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nordström, S. 2003. Distribution of deformed wing virus within honey bee (Apis mellifera) brood cells infested with the ectoparasitic mite Varroa destructor. Exp. Appl. Acarol. 29:293-302. [DOI] [PubMed] [Google Scholar]
- 26.Oberste, M. S., K. Maher, and M. A. Pallansch. 2004. Evidence for frequent recombination within species Human enterovirus B based on complete genomic sequences of all thirty-seven serotypes. J. Virol. 78:855-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ongus, J. R., D. Peters, J. M. Bonmatin, E. Bengsch, J. M. Vlak, and M. M. van Oers. 2004. Complete sequence of a picorna-like virus of the genus Iflavirus replicating in the mite Varroa destructor. J. Gen. Virol. 85:3747-3755. [DOI] [PubMed] [Google Scholar]
- 28.Shen, M., L. Cui, N. Ostiguy, and D. Cox-Foster. 2005. Intricate transmission routes and interactions between picorna-like viruses (Kashmir bee virus and sacbrood virus) with the honeybee host and the parasitic varroa mite. J. Gen. Virol. 86:2281-2289. [DOI] [PubMed] [Google Scholar]
- 29.Shen, M., X. Yang, D. Cox-Foster, and L. Cui. 2005. The role of varroa mites in infections of Kashmir bee virus (KBV) and deformed wing virus (DWV) in honey bees. Virology 342:141-149. [DOI] [PubMed] [Google Scholar]
- 30.Tentcheva, D., L. Gauthier, N. Zappulla, B. Dainat, F. Cousserans, M. E. Colin, and M. Bergoin. 2004. Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. Appl. Environ. Microbiol. 70:7185-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson, J. D., D. G. Higgins, and T. J. Gibsons. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson, E. O. 1975. Sociobiology: the new synthesis. Harvard University Press, Cambridge, Mass.
- 33.Winston, M. L. 1987. The biology of the honeybee. Harvard University Press, Cambridge, Mass.
- 34.Yang, X., and D. L. Cox-Foster. 2005. Impact of an ectoparasite on the immunity and pathology of an invertebrate: evidence for host immunosuppression and viral amplification. Proc. Natl. Acad. Sci. USA 102:7470-7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue, C., and E. Genersch. 2005. RT-PCR analysis of Deformed wing virus in honeybees (Apis mellifera) and mites (Varroa destructor). J. Gen. Virol. 86:3419-3424. [DOI] [PubMed] [Google Scholar]