Abstract
Following infection, the physical state of linear herpes simplex virus (HSV) genomes may change into an “endless” or circular form. In this study, using Southern blot analysis of the HSV genome, we provide evidence that immediate-early protein ICP4 is involved in the process of converting the linear HSV-1 ICP4-deleted mutant strain d120 genome into its endless form. Under conditions where de novo viral DNA synthesis was inhibited, the genome of the ICP4 deletion mutant d120 failed to assume an endless conformation following infection of Vero cells (compared with the ability of wild-type strain KOS). This defect was reversed in the Vero-derived cell line E5, which produces the ICP4 protein, suggesting that ICP4 is necessary and sufficient to complement the d120 defect. When ICP4 protein was provided by the replication-defective DNA polymerase mutant HP66, the genomes of mutant d120 could assume an endless conformation in Vero cells. Western blot analysis using antibody specific to the ICP4 protein showed that although the d120 virions contained ICP4 protein, the majority of that ICP4 protein was in a 40-kDa truncated form, with only a small fraction present as a full-length 175-kDa protein. When expression of ICP4 protein from E5 cells was inhibited by cycloheximide, the d120 virion-associated ICP4 protein was unable to mediate endless formation after infection of E5 cells. Collectively, these data suggest that ICP4 protein has an important role in mediating the endless formation of the HSV-1 genome upon infection and that this function can be provided in trans.
During the life cycle of herpes simplex virus (HSV), the viral genome exists in several different physical states, which may influence the outcome of infection. Following entry into a cell, the virion linear double-stranded herpes simplex virus type 1 (HSV-1) genome undergoes a change in physical state in which it assumes a nonlinear, presumably circular form that has been referred to as “endless” (12, 20, 31). Consistent with the nonlinear, circular form of the input genomes, Strang and Stow (28) demonstrated that the circularization of the HSV genome occurs early in lytic infection. This structural change occurs in nonneuronal and neuronal-like cells (29). The circular form predominates in latently infected cells in humans and experimental animals (9, 18, 22, 26). However, the kinetics of the state change is not known.
The cellular and viral factors involved in this transformation are not well understood, but the transformation from linear to circular form has generally been assumed to be essential for replication (12, 20) until the recent challenge by Jackson and Deluca (14). These investigators assert that the circular form of the viral genome is not the template for DNA replication but is the structure that is preferred for the establishment of latency.
HSV-1 ICP4 is a major regulatory protein that is synthesized immediately after the infection of permissive cells in tissue culture and is essential for the virus life cycle (6, 15, 24, 33). Although not all modifications of the ICP4 protein have been fully characterized, it is modified by phosphorylation (3, 23) and nucleotidylation (4, 19). Both positive and negative regulation of viral functions are achieved by binding of the ICP4 protein to different consensus and nonconsensus sites throughout the genome. These interactions are regulated by the degree of phosphorylation of different sites on the protein (1, 34). In addition, the ICP4 protein has been suggested to be involved in the initial stage of formation of viral DNA replication complexes (2, 30), which appear to occur adjacent to ND10 sites at prereplicative domains (10, 11, 17, 27, 32). This suggests an important role of the ICP4 protein in initiating viral DNA replication, possibly by mediating the maturation of prereplicative domains.
In the course of studying the stability of the herpes simplex virus type 1 viral genome in neuronal-like cell cultures, we observed a surprising property associated with ICP4-defective virus: the genome of ICP4 deletion mutant d120 remained linear up to 30 days postinfection (Y.-H. Su, A. Ng, and T. M. Block, unpublished observation). To explore the possibility that the ICP4 protein is necessary for viral genome circularization, we studied the genomic structure of an ICP4-deleted mutant, d120, in Vero cells and its Vero-derived complementary E5 cells (from which mutant d120 virus stock was prepared).
Here we report that the ICP4-deleted mutant d120 genome failed to assume an endless state upon infection in Vero cells. The inability of the d120 mutant genome to form the endless state can be rescued by infecting (i) Vero-derived ICP4 expressing E5 cells or (ii) Vero cells preinfected with an HSV-1 DNA polymerase mutant, HP66. Furthermore, the d120 virion-associated ICP4 protein was found to be mainly in a truncated 40-kDa form and thus failed to mediate the endless structure formation of the d120 genome in the infected E5 cells when de novo protein synthesis was inhibited. Our results strongly suggest that the failure of the d120 genome to form endless genomes upon infection of Vero cells is due to its lack of functional ICP4 protein.
MATERIALS AND METHODS
Virus and cells.
HSV-1 wild-type strain KOS was prepared in Vero cells, and HSV-1 mutant HP66, containing a 2.3-kb deletion in the locus of DNA polymerase (16), kindly provided by Don Coen (Harvard Medical School, Boston, MA), was prepared in the complementing cell line polB3 (13) (kindly provided by Charles Hwang, SUNY Health Science Center, Syracuse, NY). HSV-1 mutant d120, containing a 4.1-kb deletion in both copies of the ICP4 gene (8), and the complementary cell line E5 (7) were kindly provided by Priscilla Schaffer (Harvard Medical School, Boston, MA). The stock of mutant d120 virus was prepared in E5 cells. Every stock of d120 virus was examined for its infectivity on Vero cells. This was done by infecting 106 PFU of viruses from the stock in Vero monolayer, harvesting the culture medium, and then determining the existence of any infectious progeny by plaque assay on Vero cells.
Nuclear DNA isolation.
To isolate nuclear DNA, cells were scraped into medium and collected by centrifugation at 1,000 rpm for 10 min at 4°C. The cell pellet was resuspended into lysis buffer (1 mM CaCl2, 60 mM KCl, 15 mM NaCl, 3 mM MgCl2, 10 mM Tris, pH 7.5, 5% sucrose) containing 0.5% NP-40, homogenized with a Dounce homogenizer, and washed in nuclei lysis buffer containing 0.1% sodium deoxycholate. The nuclei were then collected by centrifugation at 2,000 rpm for 10 min at 4°C. The pelleted nuclei were subjected to DNA isolation by sodium dodecyl sulfate (SDS)/proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation.
Virion DNA preparation.
The method to isolate HSV virion DNA was described previously (29). Briefly, HSV-1-infected CV-1 cells were scraped into medium and collected by centrifugation at 1,500 rpm for 10 min at 4°C. The cells were lysed by sonication at 40% power (Heat System Ultrasonicator, Farmington, NY) for 1 min. Cell debris was separated from virions by centrifugation at 2,000 rpm for 15 min at 4°C. HSV-1 virions were pelleted by centrifugation at 28,000 rpm using an SW41 rotor for 1 h at 10°C. Isolated virions were then lysed in virion lysis buffer (0.25% Triton X-100, 10 mM EDTA, 10 mM Tris, pH 8.0), and viral capsids were isolated by centrifugation through a 20% sucrose cushion at 28,000 rpm using an SW55 rotor for 50 min at 4°C. DNA was isolated by SDS/proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation.
DNA structure analysis by Southern blot hybridization.
To determine the structure of HSV-1 DNA following infection, Vero or E5 cells were infected with wild-type KOS or the ICP4-deleted mutant d120 at a multiplicity of infection (MOI) of 5 in the presence or absence of the DNA replication inhibitor phosphonoacetic acid (PAA; 400 μg/ml) or protein synthesis inhibitor cycloheximide (CHX; 50 μg/ml). Three hours postinfection (hpi), nuclear DNA was isolated as mentioned above. To control for the effectiveness of PAA on viral DNA replication inhibition, KOS DNA in infected Vero cells after 8 hpi was determined in the presence of PAA and showed no detectable increase in the amount of HSV-1 KOS DNA after 8 hpi compared to the input DNA.
Infected or uninfected nuclear DNA was digested with the restriction endonuclease BamHI. The digested DNA was resolved on a 1% agarose gel and transferred to a nylon membrane by capillary transfer. The probes, BamHI B and PS fragments, were isolated from cloned plasmids pRB112 (21) and pBamSP (5), respectively. The probes were prepared by labeling with [α-32P]dCTP using the random primer labeling kit (Radprime DNA labeling system; Invitrogen, Carlsbad, CA) per the manufacturer's specification. The membrane was hybridized with the selected 32P-radiolabeled probe of interest. The autoradiographic image was generated and quantified by a PhosphorImager (Bio-Rad Laboratories, Inc., Hercules, CA).
Purification of virions and preparation of infected cell crude lysates for Western blot analysis.
To prepare the virions, Vero cells were infected with HSV-1 wild-type strain KOS, and the ICP4-expressing E5 cells were infected with the HSV-1 ICP4-deleted mutant strain d120, at an MOI of 5. After 1 h of absorption at 37°C, maintenance medium (Dulbecco's modified Eagle medium) containing 5% newborn calf serum was added. All incubations were carried out at 37°C. At 48 hpi, infected cells were harvested and pelleted at 1,500 rpm at 4°C for 10 min. The cell pellet was lysed by addition of the lysis buffer (0.25% Triton X-100, 10 mM EDTA, and 10 mM Tris, pH 8.0). Some of the lysate was stored at −20°C as infected cell crude lysate.
For virion purification, the cell debris was removed by centrifugation at 1,500 rpm. The virions were pelleted from the supernatant by centrifugation at 30,000 × g for 1 h at 4°C using a Sorvall SS34 rotor and washed once with 0.1% sodium deoxycholate in TNE (0.01 M Tris, 0.14 M NaCl, and 5 mM EDTA, pH 7.5) to strip off proteins that associated outside of the viral envelope. The virion pellet was first suspended in TNE, layered on top of 20% (wt/vol) sucrose, and centrifuged overnight at 50,000 rpm in a Beckman ultracentrifuge in an SW55 rotor to pellet the virions. The 20% sucrose pelleted virions were resuspended in TNE and layered onto a 20% to 60% (wt/wt) sucrose gradient, followed by centrifugation at 50,000 rpm for 20 h at 4°C in an SW55 rotor. The virion band which was found at the 45% to 50% sucrose position in the gradient was collected. The fractions above and below the virion band were also collected and concentrated using a microconcentator-10 (Micron, Danvers, MA) according to the manufacturer's specification for Western blot analysis.
Western blot analysis and antibodies.
The mouse anti-ICP4 (H1101) monoclonal antibody was purchased from The Goodwin Institute for Cancer Research Inc. (Plantation, FL). Mouse monoclonal anti-glycoprotein D (gD) (HA025) and anti-ICP27 (P1119) were purchased from Virusys Corp. (Sykesville, MD). Rabbit polyclonal anti-VP16 (V4388) was obtained from Sigma-Aldrich (Saint Louis, MO). Mouse monoclonal anti-actin antibody (MAB1501) was purchased from Chemicon Corp. (Temecula, CA).
Protein samples prepared for Western blot analysis were electrophoretically separated in a 9% SDS-polyacrimide gel under denaturing and reducing conditions. The gel was transferred to a polyvinylidene difluoride membrane manufactured by Amersham Biosciences (Piscataway, NJ) at 0.03 Å at 4°C overnight. The HSV-1 ICP4, VP16, ICP27, and gD proteins and β-actin were detected, respectively, with specific antibodies at the following dilutions: anti-ICP4 antibody, 1:5,000; anti-VP16 antibody, 1:400; anti-actin antibody, 1:2,000; anti-gD antibody, 1:10,000; and anti-ICP27 antibody, 1:200. The desired protein-antibody complexes were visualized by using the appropriate species-specific secondary antibodies, followed by using the SuperSignal Western Chemiluminescent Detection tool according to the manufacturer's instructions (Pierce, Rockford, IL).
RESULTS
Failure of the HSV-1 strain KOS ICP4 mutant d120 genome to assume an endless structure in Vero cells after infection.
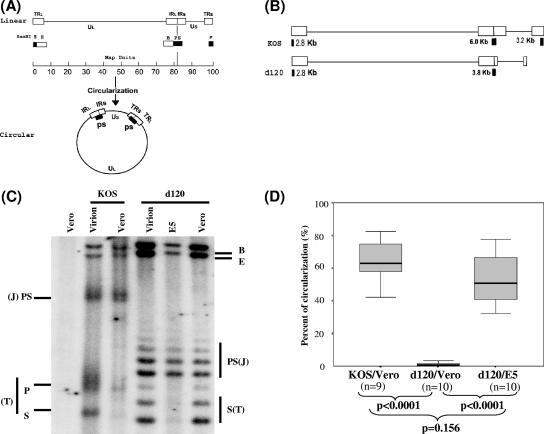
To determine if the ICP4 protein is involved in a genomic structural change after infection, the ICP4-deleted mutant virus, d120, was used to infect Vero cells as well as the Vero-derived ICP4-expressing E5 cells at an MOI of 5. Mutant d120, as described below, is derived from strain KOS and contains a 4.1-kb deletion in both ICP4 loci (8), as illustrated in Fig. 1A and B, which also shows two conformations of the viral genome: linear and circular.
FIG. 1.
Southern blot analysis of HSV-1 DNA structures of wild-type strain KOS and ICP4-deleted mutant d120. (A) Maps of the HSV-1 genome as linear and circular forms. The linear map of the HSV-1 genome is divided into unique long (UL) and unique short (US) segments with open boxes (TRL and TRS indicate long and short terminal repeats, respectively; IRL and IRS indicate long and short inverted repeats, respectively) identifying the regions of the genome that are “repeats”. Solid boxes under the linear map show the genomic locations of terminal restriction endonuclease BamHI P and S fragments and an internal joint PS fragment, which hybridizes to radioactive probes that are generated from the BamHI PS fragment. Probes generated from BamHI B fragment hybridize to both BamHI B and E fragments, because they both contain repeat sequences. Note that BamHI P and S fragments exist only as linked PS fragments (two copies) in the circular form. (B) The size and location of the BamHI-digested terminal fragments and joint fragments of ICP4-deleted mutant d120 and the wild-type strain KOS. Please note that the boundaries of the 4.1-kb deletion in both copies of the ICP4 locus were not determined precisely in previous studies (8). After the mapping study using Southern blot hybridization, the p fragment of the d120 genome is no longer detected when the membrane was probed with the BamHI PS fragment. (C) Southern blot analysis of HSV-1 DNA structures of wild-type strain KOS and mutant d120 following infection. HSV-1 wild-type strain KOS or ICP4 mutant d120 was used to infect Vero cells or ICP4-expressing E5 cells at an MOI of 5 in the presence of PAA. Cells were harvested 3 hpi, and infected nuclear DNA was isolated. One-microgram aliquots of uninfected Vero DNA, Vero DNA with 20 ng of KOS virion, KOS-infected Vero DNA, Vero DNA with 20 ng of d120 virion, d120-infected E5 DNA, and d120-infected Vero DNA were analyzed with the 32P-labeled probes generated from BamHI PS and B fragments as described in Materials and Methods. The intensities of the bands were determined by PhosphorImager analysis and are listed in Table 1. The mobility of terminal P and S, joint PS, internal B, and E fragments identified by the probes are indicated. The joint (J) and terminal (T) fragments for each virus are also indicated. (D) Statistical analysis comparing the percent of circularization of the genome of wild-type strain KOS and mutant strain d120 following infection in Vero and E5 cells as described for panel C and as analyzed in Table 1. Note the percent of circular DNA was calculated using a previously published equation (29), as noted in Table 1. The data from 9 (n = 9) or 10 (n = 10) experiments were analyzed and are displayed as the range of data with the median, indicated as a thick line in the middle of the box, which indicates the range from the 25% quartile to the 75% quartile of the data. The P value displayed was obtained by analyzing the data using the Mann-Whitney two-tailed test.
The physical state of the viral genomes following infection of Vero cells was determined by comparing the relative abundance of the terminal (BamHI S and P) and penultimate (BamHI E and B) restriction fragments of the viral genome present in the virion and after infection, generally as described previously (26). Infection was performed in the presence of the DNA replication inhibitor PAA (400 μg/ml), which prevents all detectable viral DNA replication (Su et al., unpublished; see also Materials and Methods). DNA was isolated from virions and from infected cells 3 hpi and was digested to completion with BamHI, resolved through agarose gels, transferred to nylon membranes, and hybridized with radioactive probes specific for BamHI SP and B/E.
The results presented in Fig. 1C show that terminal fragments (S and P) derived from KOS-infected Vero cells are almost undetectable, whereas they exist in amounts approximately equimolar to those of the internal joint SP fragments present in virion DNA (Fig. 1C; Table 1). This result is not surprising, since circularization, or “endless” DNA formation, is expected to occur shortly after infection, even in the absence of DNA synthesis. Circularization would result in joining of the terminal S and P fragments and a loss of their detection (as illustrated in Fig. 1A and B).
TABLE 1.
Quantitation of DNA fragmentsa
| Corresponding lane in Fig. 1C | Raw value for BamHI fragment:
|
Molar ratiob of BE/PS + P + S | Ratiob of PS/P + S | % Circularizationc | |||
|---|---|---|---|---|---|---|---|
| BE | PS | P | S | ||||
| KOS virion | 17,857 | 15,055 | 10,227 | 7,228 | 1.0 | 1.0 | 0 |
| KOS/Vero (3 h/PAA) | 15,830 | 14,225 | 2,497 | 604 | 1.5 | 5.2 | 67 |
| d120 virion | 37,584 | 19,306 | 15,870 | 1.0 | 1.0 | 0 | |
| d120/E5 (3 h/PAA) | 18,131 | 19,007 | 2,311 | 0.9 | 8.2 | 78 | |
| d120/Vero (3 h/PAA) | 30,536 | 21,100 | 15,541 | 0.9 | 1.0 | 0 | |
The specific bands (BamHI B, E, PS, P, and S) that correspond with those of the image in Fig. 1C were quantified by the PhosphorImager, and the raw values are listed.
The molar ratio of BE/PS + P + S and the ratio of PS/P + S are normalized based on virion DNA, which is set to a value of 1 for both ratios.
The percent of circularization in the absence of viral DNA replication was calculated based on a previously described formula (29): % circularization = [0.5 (R − 1)]/1 + 0.5(R − 1) × 100, where R is PS/(P + S).
The situation with d120 is dramatically different. There is no detectable reduction of d120 genomic terminal S and P fragments following infection of Vero cells. The restriction pattern (Fig. 1C) and the molar ratio of PS/P+S (Table 1) of the digests of genomes derived from infected Vero cells and virions are almost indistinguishable. This is consistent with an absence of genomic “state change” or “endless formation” following infection.
Note that due to the 4.1-kb deletion in both ICP4 loci of the d120 genome, the BamHI P fragment no longer existed when the d120 genome was digested with restriction endonuclease BamHI. The 500-nucleotide ladder of d120 genome-derived BamHI S and PS fragments is presumably due to multiple insertions of the “a” sequence with a BamHI restriction site and not the presence of multiple isolates within the virus stock, since plaque-purified viral clones (n = 3) bred true with respect to this restriction pattern (data not shown). In any event, d120 genomes did not “circularize” or assume the “endless” state following infection of Vero cells.
On the other hand, terminal BamHI restriction fragments of the d120 genome are greatly reduced in relative abundance following infection of E5 cells (Fig. 1). In this particular experiment, when the band intensity was quantified by the PhosphorImager, the molar ratio of PS/P+S was 8.2 and there was approximately 78% d120 DNA circularized, as shown in Table 1. Since the Vero-derived E5 cell line produces functional ICP4 polypeptide (25), these data suggest that d120 genomes can form endless, presumably circular, genomes following infection of cells that provide ICP4 function in trans.
Loss of the terminal fragments following infection as shown in Fig. 1C could be due to exonuclease degradation from genomic termini and not circularization or concatenation. This possibility was explored by examining the abundance of BamHI B and E fragments, which are adjacent to the terminal fragments. That is, since BamHI B and E are adjacent to the ends, if exonuclease degradation from the ends was responsible for the loss of the terminal fragments (P and S), then some commensurate loss of B and E fragments would be expected. Since there was no reduction in the abundance of BamHI B and E fragments which are immediately adjacent to the termini despite a significant reduction in the termini (Fig. 1C; Table 1), it is concluded that it is unlikely that exonuclease degradation is responsible for the loss of the terminal S and P fragments. In the absence of viral DNA replication, the concatameric replication intermediate should be rare, if present at all, thus, the endless structure should not be the long concatemer, although more work is needed to definitively determine the genomic structures.
Repeated analysis of the behavior of the d120 genome following infection in Vero and E5 cells.
The experiments shown in Fig. 1C were repeated numerous times, and the results are summarized in Fig. 1D, in which the relative amount of “endless” DNA formation (presumably circularization) of the viral genome following either wild-type KOS or mutant d120 infection of Vero and E5 cells was quantified and the percent of circularization of the HSV-1 DNA was calculated, as described by examples shown in Table 1. The data were analyzed by the Mann-Whitney two-tailed test and suggest statistically significant differences between the percentages of circularized DNA of the d120 genome in E5 cells and Vero cells (P < 0.0001). There was no evidence of the formation of the circular d120 genome in Vero cells in any of the experiments (0% for all the experiments performed), while 33% to 78%, with a median of 51%, of the infected d120 genomes in E5 cells were shown to be in a circular form shortly after infection. In d120-infected E5 cells, the percentage of d120 DNA that circularized was not statistically different from that of KOS in Vero cells (P = 0.156). Thus, the ICP4 protein provided from d120-infected E5 cells was able to compensate for the defect of d120 in the formation of the endless genome structure to the level of the wild type.
Circularization of the d120 genome in Vero cells preinfected with an ICP4-producing DNA replication-incompetent HSV-1 mutant.
Since d120 did not form a genomic endless state in Vero cells but did so in ICP4-producing E5 cells, it was reasoned that ICP4, provided by the E5 cells, was mediating the genomic state change. However, it was possible that factors other than ICP4, present in E5 cells (but not in Vero cells), were responsible for the complementation. Given that the ICP4 protein is a pleiotropic phosphoprotein with multiple phosphorylation sites that possess both DNA and protein binding domains, it was possible that the physiology of E5 cells was altered after being stably transfected with the ICP4 gene, and these secondary effects were responsible for causing the d120 genome to assume an endless form in the E5 cells.
To determine whether or not the endless formation of the d120 genome is E5 specific, Vero cells were first infected with another DNA replication-incompetent mutant to provide ICP4 protein and then to provide the degree to which the d120 genome circularized was determined. To prevent viral DNA replication due to the complementation between these two mutants, the infections were performed in the presence of PAA, as before.
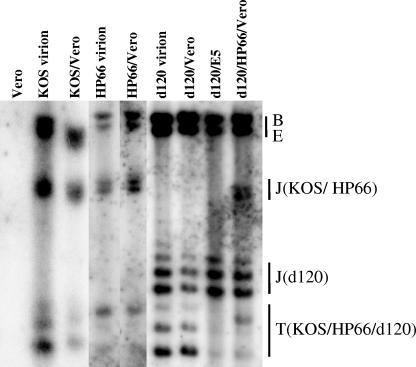
Vero cells were infected first with the replication-defective mutant HP66 at an MOI of 5. HP66 is defective in the polymerase gene but produces a fully functional ICP4 protein. Three hours after HP66 infection, cultures were superinfected with mutant d120 at an MOI of 5. The infected cells were harvested 3 h after d120 infection. Nuclear DNA was isolated and analyzed for the formation of the endless viral genome, measured by the reduction of terminal fragments as described in Fig. 1C.
Based upon the reduction in the relative amount of terminal to internal BamHI restriction fragments (Fig. 2), approximately 40% of the input HP66 genomes formed the endless structure following infection. This is in contrast to the situation with monoinfection of Vero cells with d120, where there was little, if any, evidence of circularization of the viral genome. As expected for the controls, an increased amount of the joint fragment accompanying a decreased amount of terminal fragments was observed in the BamHI-digested KOS-infected Vero DNA, suggesting the circularization of the wild-type KOS genome was evident in Vero cells after wild-type KOS infection.
FIG. 2.
Circularization of the genome of ICP4-deleted mutant d120 in Vero cells preinfected with DNA polymerase mutant HP66. Vero cells were infected first with DNA polymerase mutant HP66 at an MOI of 5. Three hours later, HP66-infected cultures were either harvested for DNA analysis or superinfected with d120 at an MOI of 5 in the presence of PAA. The superinfected cells (d120/HP66/Vero) were harvested 3 h following d120 infection and analyzed for viral genome circularization as described in the legend to Fig. 1C. As controls, uninfected Vero cell DNA, wild-type strain KOS virion, KOS-infected Vero DNA (KOS/Vero), HP66 virion DNA, d120 virion DNA, d120-infected Vero DNA (d120/Vero), and d120-infected E5 DNA (d120/E5) were also analyzed. Note that all the infections were performed in the presence of PAA and harvested 3 hpi, except for superinfected samples, as noted. The BamHI digests of Vero, KOS virion, and KOS/Vero were loaded in a separate gel. The terminal fragments (T) and the joint fragment (J) are indicated for each virus. The results shown are representative of three independent experiments.
As mentioned above, the joint and terminal fragments of a complete BamHI-digested viral genome as revealed by the BamHI PS fragment hybridization are different in size for d120 and HP66, since HP66 has no deletion in those regions of the genome. Thus, we can determine the percent of circularization of each virus in the HP66 and d120 mix-infected nuclear DNA.
There was a clear loss of the detectable d120 terminal fragment, suggesting that the circularization had occurred in mixed infection. Thus, the capability of the d120 genome to form the endless structure in the E5 cells was not due to the effect of the permanently transfected ICP4 gene on the physiology of the E5 cells. The ICP4 protein provided by mutant HP66 in trans was capable of mediating the endless formation event of the d120 genome in Vero cells.
ICP4 protein expression in d120-infected Vero and E5 cells.
Previous studies reported that the viral DNA “state change” or circularization can occur in the absence of de novo protein synthesis (12, 14, 20). Since the ICP4 protein is present in the virion (35), d120 progeny propagated in the ICP4-complementary E5 cells might have been expected to acquire sufficient ICP4 protein to mediate the circularization event.
However, our results suggest that, to the extent that ICP4 protein was associated with the d120 virion, it failed to mediate the circularization event following infection of Vero cells. It seemed possible, therefore, that the ICP4 associated with d120 derived from E5 cells was of a different quality or quantity than that associated with wild-type KOS.
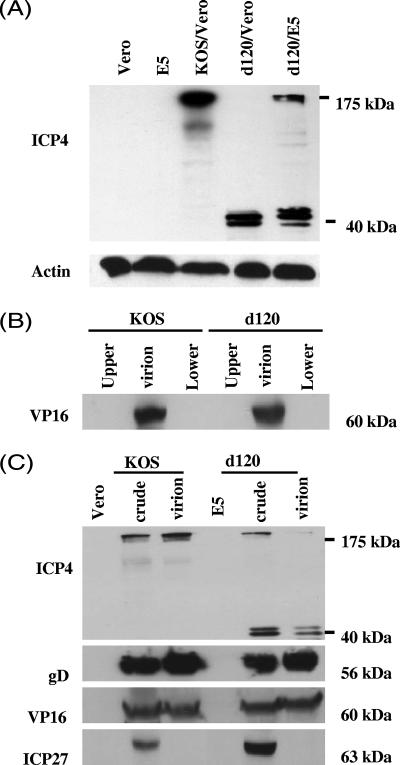
To help resolve this issue, we compared the virion-associated ICP4 protein in d120 and wild-type KOS by Western blot analysis. Western blot analysis was first performed on infected cell lysates from the d120-infected Vero cells and the d120-infected E5 cells that produce ICP4 polypeptide upon infection of HSV-1 (25), as detailed in Materials and Methods. Briefly, Vero and E5 cells were infected with mutant d120 at an MOI of 5. Total cell lysate was harvested 6 hpi and analyzed, as shown in Fig. 3A. The antibody to the ICP4 protein recognizes the full-length 175-kDa ICP4 protein in both KOS-infected Vero cells and d120-infected E5 cell lysates. No band was detected in mock-infected Vero cell lysate, showing the specificity of the antibody, although a similar amount of Vero cell lysate was loaded, as indicated by reprobing the blot with antibody against actin.
FIG. 3.
Western blot analysis of virion-associated ICP4 protein in wild-type KOS and mutant d120 virions. (A) ICP4 expression in d120-infected Vero and E5 cells. Vero and E5 cells were infected with ICP4-deleted mutant d120 or wild-type KOS as indicated at an MOI of 5. At 6 hpi, protein lysates of Vero, E5, or infected cells were prepared, run on a 9% SDS-polyacrylamide gel, transferred to a nylon membrane, and analyzed with antibodies to ICP4 and actin, respectively, as described in Materials and Methods. The positions of a full-length ICP4 protein (175 kDa) and a truncated form (40 kDa) on the gel are indicated. (B) Purity of virion preparation. Virions were purified by two sequential sucrose gradients as described in Materials and Methods. The fractions above (upper) and below (lower) the virion band were collected at the end of the second sucrose purification, concentrated, and analyzed with 1/10th of the virion lysate (30 μg) harvested from the same gradient by Western blot analysis with antibody to virion-associated protein VP16. (C) Comparison of the virion-associated ICP4 protein between wild-type KOS and mutant d120 virions. Seventy-microgram aliquots of protein of uninfected Vero and E5 cell lysates, KOS-infected Vero cell lysate (crude), d120-infected E5 cell lysate (crude), purified KOS virion lysate, and d120 virion lysate were analyzed by Western blot using antibodies to HSV-1 ICP4, glycoprotein D (gD), VP16, and ICP27 proteins as described in Materials and Methods. These results represent three independent experiments.
The amount of ICP4 protein made in uninfected E5 cells was below the level of detection by Western blot analysis. Note that the expression of ICP4 protein in E5 cells is under the control of the ICP4 promoter, thus, its expression is significantly increased only following HSV infection, presumably as a consequence of activation of the promoter by the trans-activator virion-associated VP16 protein.
Consistent with a previous report (8), the 40-kDa amino-terminal portion of the ICP4 is detected in d120-infected cells. Interestingly, we detected the truncated 40-kDa ICP4 protein in three different size forms on an SDS-polyacrylamide gel in d120-infected Vero cells and d120-infected E5 cells. No full-length ICP4 protein was detected in Vero cells infected with d120 virus. The different-sized forms of the truncated ICP4 protein are likely due to heterogeneity of phosphorylation (34). Further studies are needed to confirm whether these different forms were due to the difference in phosphorylation.
The truncated form is the major ICP4 polypeptide in d120 virions grown in E5 cells.
To compare the virion-associated ICP4 protein in d120 and wild-type KOS virions, virions were purified by centrifuging through a 20% sucrose gradient and then followed by a 20% to 60% sucrose gradient, as detailed in Materials and Methods. Purified virions were lysed and analyzed for ICP4 protein by Western blot analysis.
To examine the purity of the virion preparation, two approaches were used. First, the fractions above (Fig. 3A, upper) and below (Fig. 3A, lower) the virion band, between 45% to 50% sucrose concentration, were collected at the end of a 20% to 60% sucrose gradient purification and concentrated through a microconcentrator-10 column.
The quantity of protein in the concentrates of the upper and lower fractions was below the level of detection (absorption at 280 nm) by the spectrophotometer. This was further confirmed by the Western blot analysis. While the viral VP16 protein was clearly detected in 1/10th of the virion preparation (30 ng of virion lysate), no detectable VP16 protein was found in the upper and lower fractions (Fig. 3B).
Second, since ICP27 protein is not associated with virions (36), the ICP27 protein can be used to control for cellular contamination during virion purification. Seventy micrograms of protein lysates from Vero cells, E5 cells, KOS-infected Vero cells, d120-infected E5 cells, KOS virions, and d120 virions were resolved in a 9% SDS-polyacrylamide gel, transferred, and Western blotted with the antibodies to viral ICP4, VP16, glycoprotein D (gD), and ICP27 proteins (Fig. 3C). In agreement with previous studies (36) that the ICP27 protein is not a virion-associated protein, no detectable ICP27 protein was found in both KOS and d120 virion lysates by Western blot analysis. This further confirmed the purity of virion preparations.
As shown in Fig. 3C, all the viral proteins tested were readily detected in both the KOS-infected Vero cell lysate and the d120-infected E5 cell lysate but not in uninfected Vero or E5 cell lysates. When the membrane was probed with the anti-ICP4 antibody, a full-length 175-kDa ICP4 protein was readily detected in wild-type KOS virion lysate. Interestingly, the ICP4 protein detected in d120 virion lysate was predominately the truncated 40-kDa form, with only a small fraction of the ICP4 protein being full length (Fig. 3C). In contrast, the amounts of other known virion-associated proteins, VP16 and gD, were similar in both KOS and d120 virion preparations, suggesting that similar quantities of virion lysates were loaded in each lane. Since an expected amount of full-length ICP4 protein was readily detected in KOS virion, the lesser amount of full-length ICP4 detected in d120 virion was not due to the inefficiency of transferring the protein to the membrane. It is thus concluded that the truncated ICP4 protein is the major species of the ICP4 proteins found in the d120 virion.
The d120 virion-associated ICP4 protein is insufficient to mediate the circularization of the infecting viral genome in the E5 cells.
The results of the Western blot study indicate that the major species of ICP4 protein present in assembled d120 virions is a truncated, defective protein. As shown in Fig. 3C, only a minority of the virion-associated ICP4 protein is full length. It seemed likely that this small amount of full-length ICP4 protein is not sufficient to mediate the genomic structure change. Thus, the ICP4 protein that mediates endless viral genome formation (as seen in d120-infected E5 cells) should be the result of de novo-synthesized ICP4 protein from E5 cells following d120 infection.
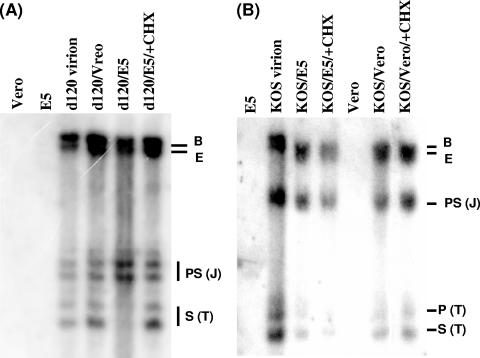
To test this hypothesis, we infected E5 cells with d120 mutant viruses in the presence or absence of a protein synthesis inhibitor, cycloheximide (CHX; 50 μg/ml), and analyzed the structure of the viral genome 3 hpi. As controls, infections of wild-type KOS in the presence or absence of CHX were performed in both Vero and E5 cells in the presence of PAA. The infected DNA was harvested 3 hpi and subjected to DNA structure analysis.
As shown in Fig. 4A, terminal fragments of the d120 genome were reduced significantly in the E5 cells by 3 hpi in the absence of CHX, suggesting that the structure of the genome changed from linear to endless, as expected. In contrast, when de novo protein synthesis was inhibited (by the presence of CHX), no detectable difference were seen in the relative molarity of terminal S, joint PS, and B and E fragments between linear virion DNA and the DNA harvested 3 hpi in the E5-infected cultures. As a reference, the d120 genomes present in infected Vero cells are also shown.
FIG. 4.
Southern blot analysis of the d120 genome structure in E5 cells following infection in the absence of de novo protein synthesis. Vero or E5 cells were infected with d120 mutant (A) or wild-type KOS (B) at an MOI of 5 in the presence of PAA and in the presence or absence of 50 μg/ml CHX. Infected cell nuclei were harvested 3 hpi and subjected to DNA isolation. The structures of the HSV-1 genomes were analyzed by Southern blot hybridization as described in the legend to Fig. 1C. DNA isolated from Vero, E5, and d120 virions was used as the control for the specificity of the probe and the reference of the hybridization intensity of BamHI B, E, PS, and S fragments. As controls for d120-infected E5 cells in the absence (d120/E5) or presence (d120/E5/+CHX) of CHX, DNA isolated from d120-infected Vero (d120/Vero), KOS-infected Vero in the absence (KOS/Vero) or presence (KOS/Vero/+CHX) of CHX, and KOS-infected E5 in the absence (KOS/E5) or presence (KOS/Vero/+CHX) of CHX were also analyzed. This figure is representative of two independent experiments.
In agreement with previous studies (12, 14, 20) reporting that the HSV-1 wild-type genome circularization could occur in the absence of de novo protein synthesis, a noticeable amount of input KOS genomes circularized in the presence of CHX in both Vero and E5 cells (Fig. 4B). Thus, the de novo protein synthesis is not needed for the wild-type HSV-1 genome to circularize following infection. In contrast, the structural change of the d120 genome from linear to endless (or circular) seen in infected E5 cells was mediated by de novo-synthesized ICP4 protein from the E5 cells after d120 infection. The d120 virion-associated ICP4 protein alone was not sufficient to mediate the circularization of the d120 genome.
DISCUSSION
This study provides evidence that one of the HSV-1 immediate-early gene products, the ICP4 protein, is essential for endless formation, presumably circularization, of the mutant d120 viral genome following infection. To our knowledge, this is the first report of a role for ICP4 in viral genome circularization.
ICP4 protein provided by the ICP4-expressing E5 cell line, or by another replication-incompetent HSV-1 polymerase mutant (HP66), was sufficient to complement the defect of the ICP4-deleted mutant virus, d120. However, the ICP4 present within the d120 virion was apparently insufficient to mediate circularization. These data suggest, for the first time, an important role for the virion-associated ICP4 protein.
The inadequacy of the ICP4 present in the d120 virion to mediate circularization of the infecting genome was probably due to the fact that the majority of d120 virion-associated ICP4 was present as a truncated form (40 kDa). Only a small amount of the ICP4 present in the virions derived from the ICP4-producing E5 cells was full-length ICP4 protein. This raises questions regarding the mechanism whereby ICP4 polypeptide associates with progeny virions.
It has been shown previously that viral genome circularization does not require de novo protein synthesis following infection, since it occurs in the cells treated with inhibitors of protein synthesis at the time of infection, as demonstrated by previous studies (12, 14, 20). This suggests that the ICP4 protein associated with wild-type virions is sufficient to mediate circularization. However, the ICP4 proteins associated with the infectious d120 virion were unable to mediate the circularization of the d120 genome. Western blot analysis using the antibody to the ICP4 protein showed that the d120 virion-associated ICP4 protein was predominately in a truncated form with only a small fraction of the ICP4 protein being full length. Interestingly, although an amount of the full-length ICP4 protein sufficient to support the productive viral life cycle was made in E5 cells after d120 infection, it seems that during the virion assembly the truncated form was preferentially packed into the virion. The circularization of the linear d120 genome observed in d120-infected E5 cells thus required de novo synthesis of the ICP4 protein from E5 cells after d120 infection, as demonstrated in Fig. 4. Therefore, we reason that the amount of full-length ICP4 protein present in d120 virion is insufficient to mediate the circularization.
ICP4 is a multifunctional protein characterized by its role as a trans-activator as well as a repressor. In addition to its role in transcription regulation, ICP4 has been previously associated with the formation of viral DNA replication compartments (2, 30). After infection, ICP4 protein was colocalized with the viral genome at a site next to ND10 structures. The DNA replication compartment was formed next to ND10 early in the infection, and ND10 structures were destroyed soon after by ICP0 protein (10, 11). Interestingly, the formation of the ICP4 foci early in the infection was dependent on the correct folding of its DNA binding domain, and this required the input of viral genomes. In this study, our data indicate an important role for virion-associated ICP4 protein in the event of viral DNA circularization following infection, and it is possible that this event occurs at the site where DNA replication compartments later develop.
The failure of d120 to circularize in Vero cells is in contrast to the behavior of dl09, another ICP4-defective mutant, which apparently remains circularization competent (14). However, d120 and d109 differ in several important ways. For example, d109 contains deletions in other immediate-early genes, including ICP27, ICP0, ICP22, and ICP47. This study also reports that ICP0 inhibits viral genomic circularization (14). Perhaps d109 virions, derived from their ICP4-complementing cells, contain a sufficient amount of virion-associated ICP4 to mediate the circularization. This may or may not be due to the lack of the circularization inhibitor (ICP0) in d109-infected Vero cells. The genotype of dl09 is complicated, and work is needed to resolve the differences between the phenotypes of d120 and dl09 with respect to virion-associated proteins and circularization.
The inability of d120 to circularize in Vero cells and the ability of ICP4 provided in trans to complement this defect suggest that ICP4 has a role in the circularization process. To our knowledge, this is the first report of a role for the virion-associated ICP4 protein in mediating the circularization of the viral genome. The management of the physical state of the viral genome is likely to be of central importance to the virus life cycle, given how important genome structure is to infection, replication, latency, or encapsidation. Thus, it is important to study the mechanism whereby ICP4 protein achieves this function.
Acknowledgments
We thank Pamela Norton for critical reading of the manuscript and Mengjun Wang for assistance in statistical analysis.
This work was supported by National Institutes of Health grant NS 33768 and an appropriation from The Commonwealth of Pennsylvania (through the Institute of Hepatitis and Virus Research).
Footnotes
Published ahead of print on 20 September 2006.
REFERENCES
- 1.Advani, S. J., R. Hagglund, R. R. Weichselbaum, and B. Roizman. 2001. Posttranslational processing of infected cell proteins 0 and 4 of herpes simplex virus 1 is sequential and reflects the subcellular compartment in which the proteins localize. J. Virol. 75:7904-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aslani, A., S. Simonsson, and P. Elias. 2000. A novel conformation of the herpes simplex virus origin of DNA replication recognized by the origin binding protein. J. Biol. Chem. 275:5880-5887. [DOI] [PubMed] [Google Scholar]
- 3.Blaho, J. A., N. Michael, V. Kang, N. Aboul-Ela, M. Smulson, M. Jacobson, and B. Roizman. 1992. Differences in the poly(ADP-ribosyl)ation patterns of ICP4, the herpes simplex virus major regulatory protein, in infected cells and in isolated nuclei. J. Virol. 66:6398-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaho, J. A., and B. Roizman. 1991. ICP4, the major regulatory protein of herpes simplex virus, shares features common to GTP-binding proteins and is adenylated and guanylated. J. Virol. 65:3759-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Block, T., S. Barney, J. Masonis, J. Maggioncalda, T. Valyi-Nagy, and N. W. Fraser. 1994. Long term herpes simplex virus type 1 infection of nerve growth factor-treated PC12 cells. J. Gen. Virol. 75:2481-2487. [DOI] [PubMed] [Google Scholar]
- 6.Clements, J. B., R. J. Watson, and N. Wilkie. 1977. Temporal regulation of herpes simplex virus type 1 transcription: location of transcripts on the viral genome. Cell 12:275-285. [DOI] [PubMed] [Google Scholar]
- 7.DeLuca, N. A., and P. A. Schaffer. 1987. Activities of herpes simplex virus type 1 (HSV-1) ICP4 genes specifying nonsense peptides. Nucleic Acids Res. 15:4491-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLuca, N. A., A. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Efstathiou, S., A. C. Minson, H. J. field, J. R. Anderson, and P. Wildy. 1986. Detection of herpes simplex virus-specific DNA sequences in latently infected mice and in humans. J. Virol. 57:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett, R. D., G. Sourvinos, and A. Orr. 2003. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J. Virol. 77:3680-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett, R. D., G. Sourvinos, C. Leiper, J. B. Clements, and A. Orr. 2004. Formation of nuclear foci of the herpes simplex virus type 1 regulatory protein ICP4 at early times of infection: localization, dynamics, recruitment of ICP27, and evidence for the de novo induction of ND10-like complexes. J. Virol. 78:1903-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garber, D. A., S. M. Beverley, and D. M. Coen. 1993. Demonstration of circularization of herpes simplex virus DNA following infection using pulsed field gel electrophoresis. Virology 197:459-462. [DOI] [PubMed] [Google Scholar]
- 13.Hwang, Y. T., B.-Y. Liu, D. M. Coen, and C. B. C. Hwang. 1997. Effects of mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene on enzyme activities, viral replication, and replication fidelity. J. Virol. 71:7791-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson, S. A., and N. A. DeLuca. 2003. Relationship of herpes simplex virus genome configuration to productive and persistent infections. Proc. Natl. Acad. Sci. USA 100:7873-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knipe, D. M., W. T. Ruyechan, B. Roizman, and I. W. Halliburton. 1978. Molecular genetics of herpes simplex virus: demonstration of regions of obligatory and nonobligatory identity within diploid regions of the genome by sequence replacement and insertion. Proc. Natl. Acad. Sci. USA 75:3896-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcy, A. I., D. R. Yager, and D. M. Coen. 1990. Isolation and characterization of herpes simplex virus mutants containing engineered mutations at the DNA polymerase locus. J. Virol. 64:2208-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maul, G. G. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660-667. [DOI] [PubMed] [Google Scholar]
- 18.Mellerick, D. M., and N. W. Fraser. 1987. Physical state of the latent herpes simplex virus genome in a mouse model system: evidence suggesting an episomal state. Virology 158:265-275. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell, C., J. A. Blaho, A. L. McCormick, and B. Roizman. 1997. The nucleotidylylation of herpes simplex virus 1 regulatory protein alpha 22 by human casein kinase II. J. Biol. Chem. 272:25394-25400. [DOI] [PubMed] [Google Scholar]
- 20.Poffenberger, K. L., and B. Roizman. 1985. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J. Virol. 53:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Post, L. E., S. Mackem, and B. Roizman. 1981. Regulation of alpha genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with alpha gene promoters. Cell 24:555-565. [DOI] [PubMed] [Google Scholar]
- 22.Preston, C. M. 2000. Repression of viral transcription during herpes simplex virus latency. J. Gen. Virol. 81:1-19. [DOI] [PubMed] [Google Scholar]
- 23.Preston, C. M., and E. L. Natariamni. 1983. Poly(ADP-ribosyl)ation of a herpes simplex virus immediate early polypeptide. Virology 131:492-501. [DOI] [PubMed] [Google Scholar]
- 24.Preston, C. 1979. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J. Virol. 29:275-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivera-Gonzalez, R., A. N. Imbalzano, B. Gu, and N. A. DeLuca. 1994. The role of ICP4 repressor activity in temporal expression of the IE-3 and latency-associated transcript promoters during HSV-1 infection. Virology 202:550-564. [DOI] [PubMed] [Google Scholar]
- 26.Rock, D. L., and N. W. Fraser. 1985. Latent herpes simplex type 1 DNA contains two copies of the virion DNA joint region. J. Virol. 55:849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2443. In D. M. Knipe and P. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 28.Strang, B. L., and N. D. Stow. 2005. Circularization of the herpes simplex virus type 1 genome upon lytic infection. J. Virol. 79:12487-12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su, Y.-H., M. Moxley, A. K. Ng, J. Lin, R. Jordan, N. W. Fraser, and T. M. Block. 2002. Stability and circularization of herpes simplex virus type 1 genomes in quiescently infected PC12 cultures. J. Gen. Virol. 83:2943-2950. [DOI] [PubMed] [Google Scholar]
- 30.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umene, K., and T. Nishimoto. 1996. Replication of herpes simplex virus type 1 DNA is inhibited in a temperature-sensitive mutant of BHK-21 cells lacking RCC1 (regulator of chromosome condensation) and virus DNA remains linear. J. Gen. Virol. 77:2261-2270. [DOI] [PubMed] [Google Scholar]
- 32.Uprichard, S. L., and D. M. Knipe. 2003. Conformational changes in the herpes simplex virus ICP8 DNA-binding protein coincident with assembly in viral replication structures. J. Virol. 77:7467-7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watson, R. J., and J. Clements. 1980. A herpes simplex virus type 1 function continuously required for early and late virus RNA synthesis. Nature 285:329-330. [DOI] [PubMed] [Google Scholar]
- 34.Xia, K., D. M. Knipe, and N. A. DeLuca. 1996. Role of protein kinase A and the serine-rich region of herpes simplex virus type 1 ICP4 in viral replication. J. Virol. 70:1050-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao, F., and R. Courtney. 1989. A major transcriptional regulatory protein (ICP4) of herpes simplex virus type 1 is associated with purified virions. J. Virol. 63:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao, F., and R. Courtney. 1992. Association of ICP0 but not ICP27 with purified virions of herpes simplex virus type 1. J. Virol. 66:2709-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]