Abstract
Effective antibody responses provide crucial immunity against influenza virus infection. The hemagglutinin (HA) protein is the major target of protective antibody responses induced by viral infection and by vaccination with both inactivated and live-attenuated flu vaccines, but knowledge about the optimal designs of protective HA antigens from different flu serotypes is still limited. In this study, we have significantly improved the immunogenicity of HA-expressing DNA vaccines by using codon-optimized HA sequences for either an H1 serotype (A/NewCal/20/99) or an H3 serotype (A/Panama/2007/99) human influenza A virus and then used these constructs as model antigens to identify the optimal HA antigen designs to elicit high-level protective antibody responses. Two forms of HA antigen, a wild-type, full-length HA and a secreted form with transmembrane (TM) domain-truncated HA, were produced. Both forms of HA DNA vaccines, from either H1 or H3 serotypes, were able to elicit high levels of HA-specific immunoglobulin G responses in immunized rabbits as measured by enzyme-linked immunosorbent assay. Interestingly, the abilities of H1 HA and H3 HA antigens to elicit hemagglutination inhibition (HI) and neutralizing antibody (NAb) responses differ. For the H1 HA antigens, the full-length HA induced significantly higher HI and NAb responses than did the TM-truncated HA. For the H3 HA antigen, both the full-length HA and TM-truncated HA induced high levels of HI and NAb responses. These data indicate that H1 and H3 antigens have different expression requirements for the induction of an optimal protective antibody response and that the structure integrity of HA antigens is critical for eliciting type-specific protective antibody responses. Our findings will have an important impact on future subunit-based flu vaccine development.
Influenza virus infection continues to be a major public health threat to human society and animals. Influenza A viruses are the most important because they cause both epidemic and pandemic flu in humans (37). Influenza A viruses have a wide natural host range, including humans and other animals, such as birds, pigs, and horses, and they have a high degree of genetic and antigenic variability (37). Seasonal or epidemic flu infections produce high morbidity and mortality, and the potential for pandemic flu with influenza viruses containing antigenic determinants from an avian source has become a new emerging threat to the worldwide human population. The best option for reducing the impact of influenza virus infection to humans is vaccination (36).
The major form of current human influenza virus vaccines is the traditional trivalent inactivated influenza vaccine (TIV) that incorporates currently circulating viral strains in humans: an H1 subtype and an H3 subtype of influenza A viruses plus an influenza B virus. Recently developed cold-adapted live influenza virus vaccines are more effective than TIV for inducing local immunity and cell-mediated immunity which may be associated with a longer-lasting and more cross-protective immunity than that which is elicited by TIV (5). Because some of the new target influenza viruses (e.g., H5 and H7 avian flu viruses) grow poorly in eggs, a new technology called reverse genetics has been developed to generate high-growth reassortants (8, 12, 14, 31, 32, 43) that combine viral genes from the high-growth-yield laboratory strain of influenza A virus A/PR/8/34 with the genes encoding the antigenic glycoproteins of the target avian viral strains. The hemagglutinin (HA) gene is modified to eliminate high virulence associated with the cleavage site of the HA, allowing the virus to replicate to high titers in eggs (15, 24, 26, 34, 35, 48, 49). However, this technology advancement did not change the fact that large stocks of vaccine viruses still have to be produced for the inactivation and purification of protective HA antigens. This process is cumbersome, lengthy, and costly. Moreover, inactivated pandemic flu vaccines appear to be poorly immunogenic and require high doses to elicit protective antibody responses in humans (33, 51). At the same time, subunit-based flu vaccines, such as those based on recombinant HA antigens, have not been very successful and the reason for such failure is not very clear. In early studies, HA-expressing DNA vaccines, while effective for protecting animals from lethal challenge, were unable to elicit detectable HA-specific, protective antibody responses before challenge (9). This greatly limited the potential for DNA vaccines to serve as a useful tool to study antigen design leading to optimized flu vaccines.
In the past decade, important new discoveries have been made to further improve the efficacy of DNA vaccines. Optimized codon usage is one such improvement, and it has been effective for increasing the overall antigen production and immunogenicity of DNA vaccines (54). In the current study, we tested whether codon-optimized, HA-expressing DNA vaccines will be able to elicit high-level, protective antibody responses prior to viral challenge. Furthermore, we used such improved HA DNA vaccines to initiate studies on the optimal design of HA antigens for the induction of best protective antibody responses, including their use in bivalent formulations against more than one serotype of influenza A viruses. Our data suggested that codon optimization is effective for improving anti-HA antibody responses in both mouse and rabbit models. Interestingly, HA antigens from H1 or H3 serotypes had different preferred antigen designs for generating protective antibodies. Therefore, our results established an important technology platform to identify and optimize the most protective HA antigen designs. Such information not only is important for the selection and modification of HA antigens to be incorporated as part of inactivated or live-attenuated flu vaccines against emerging endemic or pandemic flu viruses but also opens new opportunities for the development of subunit-based flu vaccines.
MATERIALS AND METHODS
Construction of codon-optimized HA DNA vaccine constructs.
The codon usage of HA genes from influenza A human viruses A/NewCal/20/99 (H1N1) and A/Panama/2007/99 (H3N2) was analyzed with MacVector 7.2 software against the codon preference of Homo sapiens. The less optimal codons in HA genes were changed to the preferred codons of mammalian systems to promote higher expressions of the HA proteins. The codon optimization strategy was not limited to the change of codons for mammalian usage. Sequence optimization was also performed to make the mRNA more stable and the gene more favorable for the transcriptional and translational process. During sequence optimization, the following cis-acting sequence motifs were avoided: internal TATA boxes, chi sites, and ribosomal entry sites; AT-rich or GC-rich sequence stretches; ARE, INS, and CRS sequence elements; cryptic splice donor and acceptor sites; and branch points. Despite such DNA-level sequence changes, the final codon-optimized H1 or H3 HA DNA sequences still produced the same HA amino acid (aa) sequences as those in the original flu viruses. These codon-optimized HA genes were chemically synthesized by Geneart (Regensburg, Germany), with added restriction enzyme sites of PstI and BamHI for subcloning purposes immediately upstream of the start codon and downstream of the stop codon, respectively.
For either H1 or H3 HA DNA vaccines, two versions of codon-optimized HA gene inserts were cloned into DNA vaccine vector pSW3891 (53), a derivative of pJW4303 (27), using a cytomegalovirus immediate-early promoter. For the first version, full-length HA gene inserts (H1-HA and H3-HA, 565 aa for both inserts) with their natural HA leader sequences were subcloned individually into the pSW3891 vector at the PstI and BamHI sites. For the second version, a transmembrane (TM)- and cytoplasmic region-truncated form of HA (H1-HA.dTM, aa 23 to 527, or H3-HA-dTM, aa 21 to 527) was PCR amplified from the full-length, codon-optimized H1 or H3 HA genes by using the following primers: H1-HA-opt-1 (5′ GTCGCTCCGCTAGCGGCTACCACGCCAACAACAGC 3′) and H1-HA-opt-2 (5′ AGTCACGGATCCTCACTGGTACACGCCCATGCTCTC 3′) for H1-HA.dTM or H3-HA-opt-1 (5′ GTCGCTCCGCTAGCGGCAACGACAACAGCACCGCCACC 3′) and H3-HA-opt-2 (5′ AGTCACGGATCCTCAATCCTTGTAGCCGCTCTTCAG 3′) for H3-HA.dTM. Both HA.dTM gene inserts were individually cloned into the pSW3891 vector at the NheI and BamHI sites downstream of a human tissue plasminogen activator (tPA) leader sequence substituting for the natural HA leader sequence. Each individual DNA vaccine plasmid was prepared from Escherichia coli (HB101 strain) with a Mega purification kit (QIAGEN, Valencia, CA) for both in vitro transfection and in vivo animal immunization studies.
DNA immunization of New Zealand White (NZW) rabbits and BALB/c mice.
NZW rabbits (∼2 kg of body weight) were purchased from Millbrook Breeding Labs (Amherst, MA), and BALB/c mice (6 to 8 weeks old) were purchased from Taconic Farms (Germantown, NY) for immunogenicity studies. Both rabbits and mice were housed by the Department of Animal Medicine at the University of Massachusetts Medical School in accordance with IACUC-approved protocol. The rabbits were immunized with a Helios gene gun (Bio-Rad) at the shaved abdominal skin as previously reported (55). For each immunization, a total of 36 μg of HA DNA vaccine plasmid or vector control plasmid was delivered. In the bivalent H1 and H3 HA DNA vaccine study, 18 μg of each HA DNA vaccine plasmid was delivered. For the mouse immunogenicity study, 100 μg of HA DNA vaccine plasmid or vector control plasmid was delivered at each immunization by intramuscular injection. Immunizations for both rabbits and mice were given at weeks 0, 2, 4, and 8. For the study of HA-specific antibody responses, serum samples were taken prior to the first immunization and 2 weeks after each immunization.
Western blot analysis of in vitro-expressed HA antigens.
Transient expression of the HA antigens from various HA DNA vaccine constructs was verified by Western blot analysis. HA DNA vaccine constructs were first transfected into the human embryonic kidney 293T cells by using the calcium phosphate precipitation method. Briefly, 2 × 106 293T cells at 50% confluence in a 60-mm dish were transfected with 10 μg of plasmid DNA and harvested 72 h later. Equal amounts of each transiently expressed HA antigen (10 ng of protein) were loaded for sodium dodecyl sulfate-polyacrylamide gel electrophoresis under denatured conditions and then transferred onto polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA). After being blocked overnight at 4°C in blocking buffer (0.2% I-block, 0.1% Tween 20 in 1× phosphate-buffered saline [PBS]), the membranes were incubated with a 1:500 dilution of rabbit sera immunized with HA DNA vaccines for 30 min, followed by washes. Then the membranes were incubated with alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (IgG) at a 1:5,000 dilution for 30 min. Following washes, the signals were detected by using a chemiluminescence-based Western-Light kit (Tropix, Bedford, MA).
ELISA.
An enzyme-linked immunosorbent assay (ELISA) was conducted to measure the HA-specific antibody (IgG) responses in immunized rabbits and mice. The 96-well, flat-bottomed plates were coated with 100 μl of concanavalin A (50 μg/ml) for 1 h at room temperature and washed five times with PBS containing 0.1% Triton X-100. Subsequently, the plates were incubated overnight at 4°C with 100 μl of transiently expressed HA antigen at 1 μg/ml. After being washed five times as above, the plates were blocked with 200 μl/well of blocking buffer (5% nonfat dry milk, 4% whey, 0.5% Tween 20 in PBS at pH 7.2) for 1 h. After five washes, 100 μl of serially diluted rabbit or mouse serum was added in duplicate wells and incubated for 1 h. After another set of washes, the plates were incubated for 1 h at 37°C with 100 μl of biotinylated anti-rabbit or anti-mouse IgG (Vector Laboratories, Burlingame, CA) diluted 1:1,000 in whey dilution buffer (4% whey, 0.5% Tween 20 in PBS). Then, 100 μl of horseradish peroxidase-conjugated streptavidin (Vector Laboratories) diluted 1:2,000 in whey buffer was added to each well and incubated for 1 h. After the final washing, the plates were developed with a 3,3′,5,5′-tetramethylbenzidine solution at 100 μl per well (Sigma, St. Louis, MO) for 3.5 min. The reactions were stopped by adding 25 μl of 2 M H2SO4, and the plates were read at an optical density of 450 nm. The end titer was determined as the highest serum dilution that had an optical density reading more than twice that of the negative control serum.
Preparation of the influenza A virus stocks.
The human influenza A viruses A/NewCaledonia/20/99 (H1N1), A/Panama/2007/99 (H3N2), and A/Moscow/22/99 (H3N2) were obtained from the CDC in Atlanta, Georgia. The A/NewCal/20/99 virus has constituted the H1N1 component of the licensed human flu vaccine since the period from 2000 to 2005. Either of the two H3N2 viruses was the basis of the H3N2 component of the licensed human flu vaccine from 2000 to 2001 and 2003 to 2004. These viruses were cultured in the allantoic cavities of 10-day-old embryonated hen eggs and incubated for 2 days at 37°C. The allantoic fluid was collected and stored at −80°C. The viruses were titrated in Madin-Darby canine kidney (MDCK) cell cultures to determine the PFU per milliliter.
Protective antibody assays.
Both hemagglutination inhibition (HI) and neutralizing antibody (NAb) responses were measured. Three human influenza A viruses were used in these assays: A/NewCal/20/99, A/Panama/2007/99, and A/Moscow/22/99.
Sera were treated with receptor-destroying enzyme (RDE; Sigma-Aldrich, St. Louis, MO) as described elsewhere (19). The lyophilized product was reconstituted with 5 ml sterile distilled water, diluted with 95 ml calcium saline (pH 7.2), aliquoted, and stored at −20°C. RDE was combined with each serum sample in a 4:1 ratio (0.2 ml RDE to 0.05 ml serum) and incubated overnight at 37°C. Following the overnight incubation, 3 volumes (0.15 ml) of 2.5% sodium citrate was added to each sample and incubated for 30 min at 56°C to inactivate the remaining RDE. Finally, we added 2 volumes (0.1 ml) of PBS to raise the starting serum dilution to 1:10.
HI assays.
HI assays were performed by standard methods (19). Briefly, 25 μl of each influenza virus strain with an HA titer of 8 HA units was mixed with 25 μl of twofold dilutions of the specific RDE-treated serum in PBS in V-bottomed 96-well plates. After 30 min of incubation at room temperature, 50 μl of 0.5% chicken erythrocytes was added to the mixtures. The plates were kept at 4°C until a positive hemagglutination was developed in non-serum-containing control wells. The HI titer was defined as the highest dilution of the serum able to inhibit hemagglutination.
Microneutralization assays.
Titers of neutralizing antibodies were determined essentially as described previously (28). In brief, 50 μl of influenza virus containing 100 PFU was incubated with 50 μl of twofold dilutions of the specific RDE-treated serum for 1 h at room temperature in a 96-well plate containing an MDCK cell monolayer. After the incubation, the virus-serum samples were removed from the wells. The cells were incubated at 37°C for 2 days in minimal essential medium-bovine albumin supplemented with 1 μg/ml of tosylsulfonyl phenylalanyl chloromethyl ketone trypsin in the presence of twofold dilutions of the specific RDE-treated serum. The microneutralization titer was defined as the highest dilution of serum that neutralized 100 PFU of virus in MDCK cell cultures (as detected by negative hemagglutination).
Statistical analysis.
The Student t test was used to analyze the differences between animal immunization groups regarding HA-specific binding antibody responses as measured by ELISA, HI antibody titers, and neutralization antibody titers. A P value of less than 0.05 was considered significant.
RESULTS
Codon-optimized HA DNA vaccine was able to achieve better antigen expression and immunogenicity than was the wild-type HA DNA vaccine.
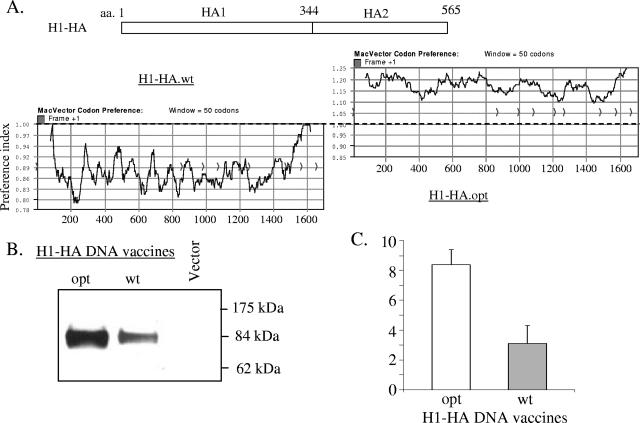
In order to improve the flu HA antigen expression in mammalian cells and potentially the immunogenicity of HA DNA vaccines, a codon-optimized HA gene from influenza A/NewCal/20/99 virus (H1 serotype) was chemically synthesized. During this process, the codon usage of HA amino acids was changed to those preferred for expression in mammalian cells but the original flu HA amino acid sequences were not altered. Figure 1A depicted the codon preference changes between the wild-type HA gene and the codon-optimized HA gene as analyzed by the MacVector software. Table 1 summarized the percentage of codon usage before and after sequence optimization. Both the wild-type and codon-optimized H1 HA genes were then cloned individually into the DNA vaccine vector pSW3891 to form two DNA vaccine plasmids, H1 HA wild type (H1-HA.wt) and H1 HA optimized (H1-HA.opt), respectively. They were transfected into 293T cells, and their expression in mammalian cells was examined. As shown by Western blot analysis (Fig. 1B and 1C), the H1-HA.opt construct was able to achieve a higher level of expression than that of the H1-HA.wt construct.
FIG. 1.
A. Schematic diagram of influenza A H1 HA gene with HA1 and HA2 domains. The coding preferences of wild-type (H1-HA.wt) and codon-optimized (H1-HA.opt) H1 HA genes were analyzed using the computer software MacVector. Plots with values above 1.0 indicate bias toward codons more frequently used in mammalian systems, whereas plots below 1.0 indicate less preferred codons in mammalian systems. (B) Western blot analysis of HA expression from either wild-type (wt) or codon-optimized (opt) H1-HA DNA vaccines in lysates of transiently transfected 293T cells. Lysates from cells transfected with empty DNA vector pSW3891 without HA antigen insert were included as a negative control. (C) Quantification of HA expression from either wild-type (wt) or codon-optimized (opt) H1-HA DNA vaccines by densitometry. The relative amounts of HA antigens were measured by scanning HA-specific bands on Western blots by using an image processing system (Fujix Pictrography 3000, Science Lab 2003, Image Gauge 4.22). The measured AU showed the geometric means of four independent assays. Error bars indicate standard deviations.
TABLE 1.
Codon usage of H1 HA wild-type and codon-optimized genesa
| Amino acid | Codon | HA coding seq.
|
|
Amino acid | Codon | HA coding seq.
|
||
|---|---|---|---|---|---|---|---|---|
| wt | opt | wt | opt | |||||
| Ala | GCT | 19 | 0 | |||||
| GCC | 38 | 100 | Leu | TTA | 14 | 0 | ||
| GCA | 31 | 0 | TTG | 24 | 0 | |||
| GCG | 12 | 0 | CTT | 10 | 2 | |||
| CTC | 2 | 0 | ||||||
| Arg | CGT | 0 | 0 | CTA | 21 | 0 | ||
| CGC | 0 | 0 | CTG | 29 | 98 | |||
| CGA | 0 | 0 | ||||||
| CGG | 0 | 47 | Lys | AAA | 69 | 19 | ||
| AGA | 58 | 53 | AAG | 31 | 81 | |||
| AGG | 42 | 0 | ||||||
| Pro | CCT | 24 | 5 | |||||
| Asn | AAT | 57 | 11 | CCC | 14 | 95 | ||
| AAC | 43 | 89 | CCA | 57 | 0 | |||
| CCG | 5 | 0 | ||||||
| Asp | GAT | 56 | 17 | |||||
| GAC | 44 | 83 | Phe | TTT | 45 | 0 | ||
| TTC | 55 | 100 | ||||||
| Cys | TGT | 69 | 19 | |||||
| TGC | 31 | 81 | Ser | TCT | 15 | 4 | ||
| TCC | 23 | 10 | ||||||
| Gln | CAA | 63 | 0 | TCA | 27 | 0 | ||
| CAG | 37 | 100 | TCG | 0 | 0 | |||
| AGT | 18 | 0 | ||||||
| Glu | GAA | 65 | 12 | AGC | 17 | 86 | ||
| GAG | 35 | 88 | ||||||
| The | ACT | 26 | 0 | |||||
| Gly | GGT | 14 | 0 | ACC | 16 | 90 | ||
| GGC | 9 | 89 | ACA | 55 | 10 | |||
| GGA | 47 | 9 | ACG | 3 | 0 | |||
| GGG | 30 | 2 | ||||||
| Tyr | TAT | 70 | 7 | |||||
| His | CAT | 57 | 0 | TAC | 30 | 93 | ||
| CAC | 43 | 100 | ||||||
| Val | GTT | 20 | 0 | |||||
| Ile | ATT | 34 | 6 | GTC | 22 | 0 | ||
| ATC | 25 | 94 | GTA | 36 | 0 | |||
| ATA | 41 | 0 | GTG | 22 | 100 | |||
The frequencies (%) of the individual codons are shown for each of the degenerately encoded amino acids. The most prevalent codon is shown in bold. seq., sequence.
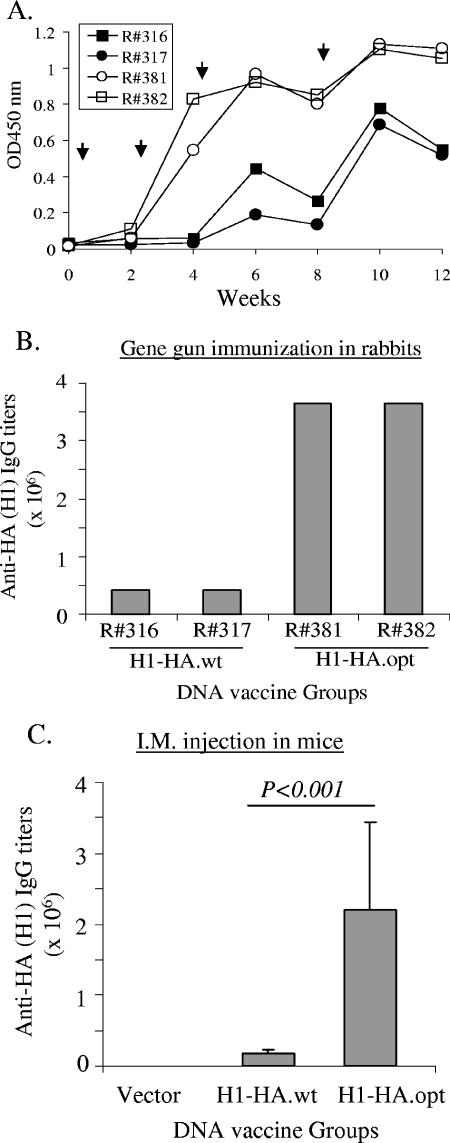
The relative immunogenicity between the wild-type and codon-optimized H1 HA DNA vaccines was then studied in rabbit and mouse models. In a pilot rabbit experiment, the codon-optimized H1 HA DNA vaccine was able to elicit high-level anti-HA antibody responses after only two DNA immunizations, but for the wild-type H1 HA DNA vaccine, three to four immunizations were required to elicit good but still lower levels of anti-HA antibody response (Fig. 2A). Even after multiple DNA-boosting immunizations, the peak-level anti-HA antibody response elicited by the wild-type HA DNA vaccine was still lower than that of the codon-optimized HA DNA vaccine (Fig. 2B). A similar study was conducted in mice with increased group size (10 animals per group). The geometric mean titer for the codon-optimized HA DNA vaccine group was significantly better than that of the wild-type HA DNA vaccine group (P < 0.001) (Fig. 2C). Therefore, our data proved that codon optimization was effective for improving the immunogenicity of flu HA DNA vaccine.
FIG. 2.
Serum anti-HA IgG antibody responses induced with the vaccinations with either H1-HA.wt or H1-HA.opt DNA vaccines in NZW rabbits (A and B) and BALB/c mice (C) as measured by ELISA. (A) Temporal anti-HA IgG responses in rabbits measured at a 1:5,000 serum dilution. The arrows indicate the time of gene gun-mediated DNA immunizations. Animals R#316 and R#317 received H1-HA.wt DNA vaccine, while R#381 and R#382 received H1-HA.opt DNA vaccine. OD450, optical density at 450 nm. (B) End titers of serum anti-HA IgG responses at 2 weeks after the fourth DNA immunization from the same rabbits as shown in panel A. (C) End titers of serum anti-HA IgG responses in BALB/c mice at 2 weeks after the fourth DNA immunization. Animals received either H1-HA.wt or H1- HA.opt DNA vaccines by intramuscular inoculation. Data shown are the geometric mean titers of 10 mice in each group with standard deviations (error bars).
The cleavage of H1 and H3 HA proteins was affected by the forms of HA antigens.
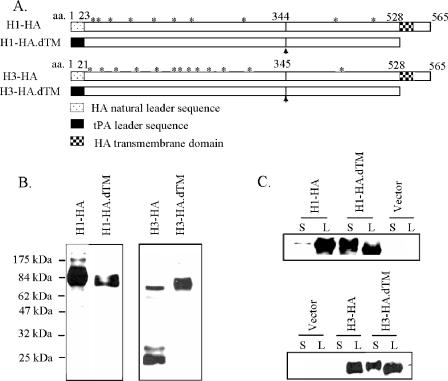
Based on the above results, an additional codon-optimized HA gene from the H3 serotype flu isolate A/Panama/2007/99 was chemically synthesized. Furthermore, both codon-optimized H1 and H3 HA genes were modified to produce two different forms of HA gene inserts (Fig. 3A). One version, the H1-HA and H3-HA insert, coded for the full-length HA proteins, including the natural HA protein leader sequences. The other design, H1-HA.dTM and H3-HA.dTM insert, coded for only the extracellular portion of the HA antigen. The TM and downstream cytoplasmic regions were deleted. The natural leader of HA protein was also replaced by a tPA leader sequence based on the previous finding that a tPA leader was highly effective in expressing secreted proteins in mammalian cells for DNA vaccines (40, 54, 55). The end products of HA.dTM constructs include the entire HA1 domain and the extracellular portion of the HA2 domain (Fig. 3A). The designation of HA1 and HA2 was based on the known cleavage sites (Q/E-X-R) for influenza HA proteins of H1 and H3 serotypes (45).
FIG. 3.
(A) Schematic diagram of various HA gene inserts used in codon-optimized H1 (A/New Caledonia/20/99) and H3 (A/Panama/2007/99) HA DNA vaccines, including the full-length HA antigens (H1-HA or H3-HA) and the transmembrane/cytoplasmic region-truncated HA antigens (H1-HA.dTM or H3-HA.dTM) where a human tPA leader also replaced the natural HA leader sequence. Potential N-linked glycosylation sites (*) on HA proteins and the cleavage sites between HA1 and HA2 subunits are marked. (B) Western blot analysis on the difference of susceptibility to cleavage between H1 and H3 HA proteins expressed by different H1 and H3 HA DNA vaccine constructs in transiently transfected 293T cells. (C) Western blot analysis on the secretion of H1 and H3 HA proteins by different H1 and H3 HA DNA vaccines in the lysate (L) or supernatant (S) of transiently transfected 293T cells.
The expression of HA antigens by the above-mentioned different forms of HA DNA vaccines was assessed by their transient transfection in 293T cells and Western blot analysis. Regardless of H1 or H3 serotypes, all four HA DNA vaccine constructs had good expressions of HA proteins (Fig. 3B). An interesting finding was that the HA proteins expressed from the H1-HA, H1-HA.dTM, and H3-HA.dTM inserts were in mainly the noncleaved form, while the HA protein expressed from the H3-HA insert was in the form of cleaved HA products of HA1 and HA2 subunits (Fig. 3B). Both H1 and H3 HA proteins expressed from our DNA vaccines were N-glycosylated as confirmed by the treatment of PNGase-F, which removed the glycosylation and led to lower-molecular-weight HA proteins (data not shown).
The full-length HA antigens, encoded by the H1-HA and H3-HA DNA vaccines, were expressed mainly in cell-associated form, while truncation of the C-terminal segment, including the removal of the TM domain (the H1-HA.dTM and H3-HA.dTM DNA vaccines), was highly effective for producing more secreted HA antigen than did the full-length HA design for both H1 and H3 serotypes (Fig. 3C). However, the abilities of these DNA vaccines to elicit anti-HA antibody responses were very similar (see below).
The immunogenicity of various forms of H1 and H3 HA DNA vaccines.
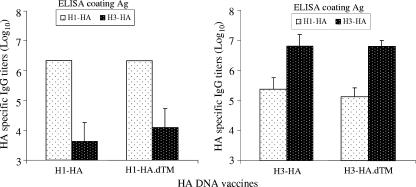
NZW rabbits were immunized with various forms of codon-optimized H1 and H3 HA DNA vaccines with a gene gun. Sera were collected 2 weeks after the fourth immunization to measure the peak-level antibody responses. High-level anti-HA IgG responses were elicited with all four HA DNA vaccines, and they were generally H1 or H3 type specific (Fig. 4). Rabbit sera immunized with H1-HA or H1-HA.dTM DNA vaccines recognized the H1 serotype HA antigen almost exclusively. On the other hand, rabbit sera immunized with H3-HA or H3-HA.dTM DNA vaccines mainly recognized H3 serotype HA antigen but also had low-level cross-reactivity against the H1 serotype HA antigen (Fig. 4). There was no difference in the peak-level anti-HA antibody responses between the full-length and TM-truncated HA antigens (Fig. 4), although the HA.dTM DNA vaccines induced a quicker rise of antibody responses than did the full-length HA designs (data not shown).
FIG. 4.
Peak-level serum anti-HA IgG antibody responses in NZW rabbits at 2 weeks after the fourth DNA immunization induced by different H1 and H3 HA DNA vaccines as measured by ELISA. Data shown are the geometric mean titers of each group, with standard deviations (error bars) if applicable (three rabbits in each group), against either autologous or heterologous HA antigens.
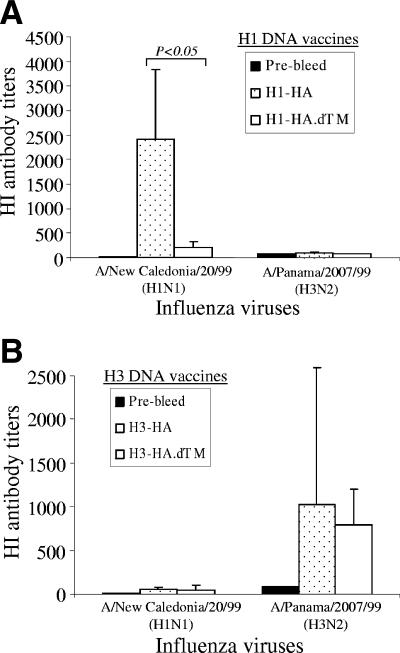
High-level HI antibody responses induced by HA DNA vaccines.
The functional activities of the anti-HA rabbit immune sera were investigated by determining the HI antibody titers against H1 or H3 influenza viruses. It was interesting to find that, for H1 serotype HA DNA vaccines, the full-length HA antigen elicited higher HI antibody titers against the autologous H1N1 virus A/NewCal/20/99 than did the TM region-truncated HA.dTM antigen (Fig. 5A) (P < 0.05). In contrast, for H3 DNA vaccines, both the full-length and the TM-truncated HA antigens were equally effective in inducing high HI antibody titers against the autologous H3N2 virus A/Panama/2007/99 (Fig. 5B) and a similar H3N2 virus A/Moscow/10/99 (data not shown). The HI activities in immunized rabbit sera were highly specific for expected flu serotypes, i.e., rabbit sera immunized with the H1-HA antigens did not show significant HI titers against the H3 virus and the H3-HA antigen-immunized rabbit sera did not show high HI activities against the H1 virus (Fig. 5). The HI titers for control rabbits immunized twice with the licensed inactivated human flu vaccine (Fluzone) were at levels in the 1:200 to 1:300 range (data not shown).
FIG. 5.
The HI antibody responses in NZW rabbit sera immunized with different designs of H1 and H3 HA DNA vaccines. The HI antibody titers are shown as the geometric means for each group (three rabbits per group), with standard deviations (error bars), against H1N1 (A/New Caledonia/20/99) and H3N2 (A/Panama/2007/99) viruses. The statistical difference between each testing group was determined, and the P value that was less than 0.05 is indicated.
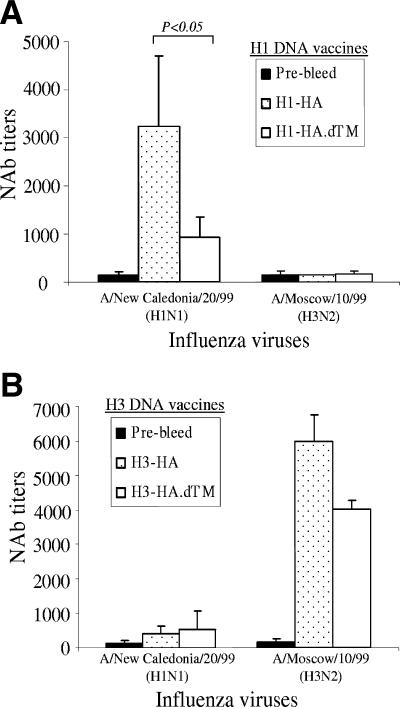
Virus-neutralizing antibody responses induced by H1 and H3 HA DNA vaccines.
The protective antibody responses induced by different forms of HA DNA vaccines were further confirmed by a microneutralization assay. The patterns of neutralizing antibody activities for different forms of HA DNA vaccines were similar to that shown by the HI assay. For H1 HA DNA vaccines, the full-length form of HA antigen elicited much higher neutralizing antibody titers against the autologous H1N1 virus A/NewCal/20/99 than did the membrane-truncated form of HA antigen (Fig. 6A). The neutralizing antibody titers induced between these two forms of H1 HA antigens were also statistically different (P < 0.05). However, for H3 HA DNA vaccines, the difference in neutralizing antibody titers between the full-length and TM-truncated forms was much less when tested against the H3N2 virus A/Moscow/10/99 (Fig. 6B). The neutralizing activities were also highly serotype specific. Rabbit sera immunized with the H1 HA DNA vaccine neutralized only the H1 virus, and rabbit sera immunized with the H3 HA DNA vaccines neutralized mainly the H3 virus (Fig. 6).
FIG. 6.
The NAb responses in NZW rabbit sera immunized with different designs of H1 and H3 HA DNA vaccines. The NAb titers against H1N1 (A/New Caledonia/20/99) or H3N2 (A/Moscow/10/99) virus infection for MDCK cells are shown as the geometric means from each group (three rabbits per group) with standard deviations (error bars). The statistical difference between each testing group was determined, and the P value that was less than 0.05 is indicated.
Broad protective antibody responses induced by the bivalent H1 plus H3 HA DNA vaccines.
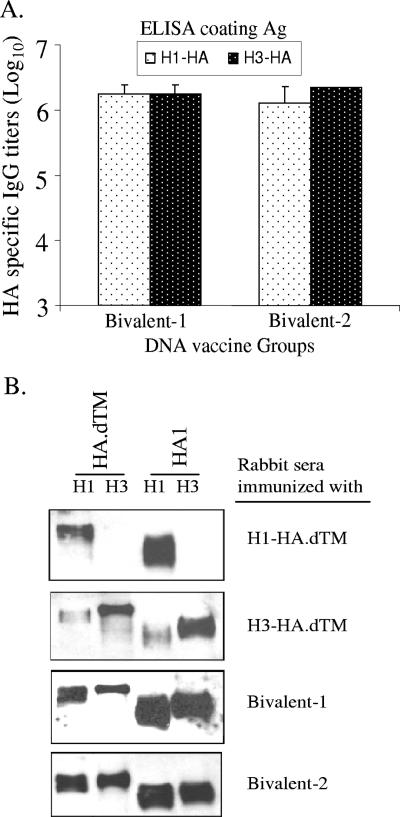
One advantage of DNA vaccination is its ability to deliver multiple HA antigens in one formulation. We next immunized rabbits with two bivalent HA formulations. The bivalent 1 formulation included DNA vaccines expressing two TM-truncated HA inserts, H1-HA.dTM and H3-HA.dTM. The bivalent 2 formulation included the full-length H1-HA and TM-truncated H3-HA.dTM DNA vaccines. Both bivalent formulations were able to elicit comparable anti-HA antibody responses recognizing various forms of HA antigens of both H1 and H3 serotypes as confirmed by ELISA and Western blot analyses (Fig. 7).
FIG. 7.
Levels and specificity of anti-HA IgG responses elicited by bivalent HA DNA vaccines. (A) Peak serum-level anti-HA IgG responses in rabbits immunized with bivalent HA DNA vaccines (bivalent 1, H1-HA.dTM plus H3-HA.dTM, and bivalent 2, H1-HA plus H3-HA.dTM) at 2 weeks after the fourth DNA immunization as measured by ELISA against the full-length H1-HA or H3-HA antigens. Data shown are the geometric mean titers of each group (three rabbits per group) with standard deviations (error bars). (B) Western blot analysis of NZW rabbit sera immunized with either monovalent (H1-HA.dTM or H3-HA.dTM) or bivalent formulations. The transmembrane-truncated HA proteins (H1-HA.dTM and H3-HA.dTM) or the HA1 subunits of HA proteins (H1-HA1 and H3-HA1) expressed from 293T cells were used in the study.
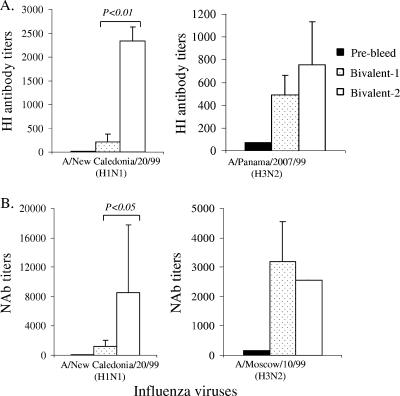
Although the HI titers of the two bivalent formulations were similar against the H3 viruses (A/Panama/2007/99 and A/Moscow/10/99), bivalent 2 formulation was more effective than bivalent 1 formulation for generating HI antibodies against the H1 virus (A/NewCal/20/99) (Fig. 8A). Similarly, the neutralizing antibody results also confirmed that bivalent 2 formulation was able to elicit better neutralizing antibody responses than those of bivalent 1 formulation against the H1 virus (Fig. 8B). Because bivalent 2 formulation included the full-length H1-HA DNA vaccine, these findings further confirmed that the full-length H1-HA antigen was able to elicit better HI and neutralizing antibody responses than those of the H1-HA.dTM antigen (Fig. 5 and 6). Therefore, in order to provide the best protection against targeted viruses, the selection of an optimal design for HA antigen is important for the development of polyvalent influenza vaccines.
FIG. 8.
The HI and NAb responses induced by bivalent H1 plus H3 HA DNA vaccines. The statistical difference between each testing group was determined, and the P values that are less than 0.05 are indicated. (A) HI antibody responses are shown as the geometric mean titers of each group (three rabbits per group), with standard deviations, against either H1N1 (A/New Caledonia/20/99) or H3N2 (A/Panama/2007/99) viruses. (B) NAb responses are shown as the geometric mean titers of each group (three rabbits per group), with standard deviations (error bars), against either H1N1 (A/New Caledonia/20/99) or H3N2 (A/Moscow/10/99) virus infection of MDCK cells.
DISCUSSION
Data presented in this report proved that codon optimization was effective for improving the antigen expression and immunogenicity of flu HA DNA vaccines. In early flu DNA vaccine studies, there were only low or undetectable HA-specific antibody responses after DNA immunizations. Positive antibody responses were detected only after a flu virus challenge (9). In the last several years, codon optimization has been shown effective for improving the immunogenicity of DNA vaccines expressing human immunodeficiency virus antigens (1, 6, 10, 11, 41, 46, 54). In addition, DNA vaccines with codon-optimized antigen gene inserts have been used successfully against viral (such as severe acute respiratory syndrome) (53, 56), papillomaviral (3, 4, 20, 25), bacterial (21, 47), and parasitic pathogens (17, 29). By using an H1 serotype HA antigen as the model system in the current study, we compared the relative levels of antigen expression and immunogenicity between wild-type and codon-optimized flu HA DNA vaccines in two animal models (mouse and rabbit) with two DNA vaccine delivery methods (gene gun and intramuscular needle injection). The data clearly demonstrated that the codon-optimized HA DNA vaccines were able to induce better HA-specific antibody responses than were the wild-type gene sequences because codon-optimized HA DNA vaccine was able to induce a quicker rise and higher peak-level anti-HA antibody responses.
The ability of eliciting high-titer, anti-HA antibodies by DNA immunization prior to the viral challenge allowed us to expand our study to further measure the levels of protective antibody components in animal sera immunized with HA antigens from either H1 or H3 serotypes. Both high-level HI and neutralizing activities were easily elicited by codon-optimized DNA vaccines. The titers of protective antibodies achieved in this report were not only significantly improved over that from early versions of flu DNA vaccines (2, 9, 18, 22, 50) but also comparable to or higher than those of positive anti-HA sera reported in previous animal studies with various flu vaccines, including recombinant viral vector-based flu HA vaccines (16, 30, 39) and inactivated and live flu vaccines (23, 38, 42).
It is very interesting to discover that the protective antibody responses were dependent on the forms of HA antigen inserts. For the H1 HA antigen used in the current study, only the full-length form was able to induce high levels of protective antibodies, while both the full-length and the TM-truncated H1 HA antigens elicited very similar levels of HA-specific binding antibody responses. Therefore the full-length H1 HA antigen was more effective for preserving certain protective antigen conformation than the TM-truncated form of H1 HA antigen was. On the other hand, both the full-length and the TM-truncated forms of H3 HA antigens were able to elicit similar levels of protective antibodies. Singh et al. reported that the truncated anchor-free HA could form monomers or trimers depending on the strain and construct used (44). It is likely that the truncated H1 HA DNA vaccine produces the monomeric forms, and truncated H3 HA DNA vaccine produces the trimeric forms, which would contribute to the differences in conformation-sensitive protective antibody responses.
Consistent with the above finding that H1 serotype HA and H3 serotype HA antigens are conformationally different, we found that the H1 HA antigen was resistant to natural cleavage into its two subunits, while the H3 HA antigen was easily detected in the forms of HA1 and HA2 subunits in transiently transfected 293T cells. Treanor et al. conducted a recent vaccine study in humans using a trivalent, baculovirus-expressed, recombinant HA vaccine based on the HAs of influenza A/Panama/2007/99 (H3N2), A/New Caledonia/20/99 (H1N1), and B/Hong Kong/330/2001 (52) and concluded that the H3 component, but not the H1 and influenza virus type B components, elicited a robust neutralizing antibody response in humans. Although the authors could not explain the reasons for these differences, their results are in agreement with our study, in which we found that H1 HA-based vaccines are less immunogenic when lacking the appropriate transmembrane and cytoplasmic domains.
Our results will have an important impact on the selection and design of HA antigens for developing the most protective flu vaccines. Traditionally, in order to study the structure requirement for immunogenicity and protection by individual HA antigens, the flu viral stocks expressing different HA antigens in their native or modified forms would have to be produced, inactivated, concentrated, and purified. This process was shown in a recent effort studying the role of specific HA amino acids in the immunogenicity and protection of H5N1 influenza virus vaccines (13). Such a process is complicated and may introduce multiple variables to the final immunization studies. There has been a limited effort to produce HA antigens by a recombinant protein approach, presumably due to the challenges of cost and time when facing the large number of HA antigens existing in the world, as the results of antigen drift, even within one serotype of flu viruses. The discovery of DNA immunization technology in the early 1990s initially led to high enthusiasm that this novel approach may be a simple alternative to the traditional flu vaccines. However, the low immunogenicity of early DNA vaccine designs, especially in higher animal species or humans when delivered without the help of physical delivery methods (gene gun or electroporation), quickly dampened such hope. One important early observation was that flu DNA vaccines were unable to elicit high-level, HA-specific antibody responses prior to viral challenge (9). In the last decade, significant improvement has been made in DNA vaccination approaches. However, such improvement has not been well incorporated into flu vaccine applications. Data presented in this report confirmed that codon optimization was effective for improving the immunogenicity of HA DNA vaccines. With a codon-optimized DNA vaccination approach, including the use of the gene gun delivery method, we report here that it is feasible to reliably elicit high-level protective antibody responses against flu HA antigens. This will provide a powerful tool to greatly accelerate the identification, selection, and rational design of the most protective HA antigens, which can then be incorporated into either conventional or novel flu vaccine strategies in either wild-type or codon-optimized gene sequences.
The development of high-immunogenicity HA DNA vaccines also provides an alternative approach to the development of vaccines against pandemic influenza caused by avian flu viruses since these emerging flu viruses do not grow well enough to produce high-yield viral stocks for the manufacturing of inactivated vaccines. Recent HA DNA vaccine studies in humans have demonstrated that significant levels of HA-specific antibody responses are induced when such DNA vaccines are delivered by a gene gun (7). With DNA vaccination, it is easy to mix several protective antigens in one delivery. Our data with HA antigens from both H1 and H3 serotypes confirmed that, relative to monovalent antigen formulation, such polyvalent formulations can be equally effective in eliciting protective antibody responses.
Acknowledgments
This work was partially supported by NIH/NIAID U01 AI 056536 (S.L.) and CIVIA, a human immunology center supported by NIH/NIAID U19 grant AI62623 (A.G.-S.).
We thank Richard Cadagan for expert technical assistance and Te-hui Chou for critical reading of the manuscript.
Footnotes
Published ahead of print on 20 September 2006.
REFERENCES
- 1.André, S., B. Seed, J. Eberle, W. Schraut, A. Bultmann, and J. Haas. 1998. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J. Virol. 72:1497-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bot, A., S. Antohi, S. Bot, A. Garcia-Sastre, and C. Bona. 1997. Induction of humoral and cellular immunity against influenza virus by immunization of newborn mice with a plasmid bearing a hemagglutinin gene. Int. Immunol. 9:1641-1650. [DOI] [PubMed] [Google Scholar]
- 3.Cheung, Y. K., S. C. Cheng, F. W. Sin, and Y. Xie. 2004. Plasmid encoding papillomavirus type 16 (HPV16) DNA constructed with codon optimization improved the immunogenicity against HPV infection. Vaccine 23:629-638. [DOI] [PubMed] [Google Scholar]
- 4.Cid-Arregui, A., V. Juarez, and H. zur Hausen. 2003. A synthetic E7 gene of human papillomavirus type 16 that yields enhanced expression of the protein in mammalian cells and is useful for DNA immunization studies. J. Virol. 77:4928-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox, R. J., K. A. Brokstad, and P. Ogra. 2004. Influenza virus: immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scand. J. Immunol. 59:1-15. [DOI] [PubMed] [Google Scholar]
- 6.Deml, L., A. Bojak, S. Steck, M. Graf, J. Wild, R. Schirmbeck, H. Wolf, and R. Wagner. 2001. Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 Gag protein. J. Virol. 75:10991-11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drape, R. J., M. D. Macklin, L. J. Barr, S. Jones, J. R. Haynes, and H. J. Dean. 2006. Epidermal DNA vaccine for influenza is immunogenic in humans. Vaccine 24:4475-4481. [DOI] [PubMed] [Google Scholar]
- 8.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. Garcia-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fynan, E. F., R. G. Webster, D. H. Fuller, J. R. Haynes, J. C. Santoro, and H. L. Robinson. 1993. DNA vaccines: protective immunizations by parenteral, mucosal, and gene-gun inoculations. Proc. Natl. Acad. Sci. USA 90:11478-11482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, F., Y. Li, J. M. Decker, F. W. Peyerl, F. Bibollet-Ruche, C. M. Rodenburg, Y. Chen, D. R. Shaw, S. Allen, R. Musonda, G. M. Shaw, A. J. Zajac, N. Letvin, and B. H. Hahn. 2003. Codon usage optimization of HIV type 1 subtype C gag, pol, env, and nef genes: in vitro expression and immune responses in DNA-vaccinated mice. AIDS Res. Hum. Retrovir. 19:817-823. [DOI] [PubMed] [Google Scholar]
- 11.Haas, J., E. C. Park, and B. Seed. 1996. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr. Biol. 6:315-324. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann, E., S. Krauss, D. Perez, R. Webby, and R. G. Webster. 2002. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 20:3165-3170. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann, E., A. S. Lipatov, R. J. Webby, E. A. Govorkova, and R. G. Webster. 2005. Role of specific hemagglutinin amino acids in the immunogenicity and protection of H5N1 influenza virus vaccines. Proc. Natl. Acad. Sci. USA 102:12915-12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann, E., G. Neumann, Y. Kawaoka, G. Hobom, and R. G. Webster. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 97:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horimoto, T., A. Takada, K. Fujii, H. Goto, M. Hatta, S. Watanabe, K. Iwatsuki-Horimoto, M. Ito, Y. Tagawa-Sakai, S. Yamada, H. Ito, T. Ito, M. Imai, S. Itamura, T. Odagiri, M. Tashiro, W. Lim, Y. Guan, M. Peiris, and Y. Kawaoka. 2006. The development and characterization of H5 influenza virus vaccines derived from a 2003 human isolate. Vaccine 24:3669-3676. [DOI] [PubMed] [Google Scholar]
- 16.Hunt, L. A., D. W. Brown, H. L. Robinson, C. W. Naeve, and R. G. Webster. 1988. Retrovirus-expressed hemagglutinin protects against lethal influenza virus infections. J. Virol. 62:3014-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivory, C., and K. Chadee. 2004. DNA vaccines: designing strategies against parasitic infections. Genet. Vaccines Ther. 2:17-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Justewicz, D. M., M. J. Morin, H. L. Robinson, and R. G. Webster. 1995. Antibody-forming cell response to virus challenge in mice immunized with DNA encoding the influenza virus hemagglutinin. J. Virol. 69:7712-7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kendal, A. P., J. J. Skehel, and M. S. Pereira. 1982. World Health Organization Collaborating Centers for Reference and Research on Influenza: concepts and procedures for laboratory-based influenza surveillance, p. B17-B35. World Health Organization Collaborating Centers for Reference and Research in Influenza, Centers for Disease Control, Atlanta, Ga.
- 20.Kim, M. S., and J. I. Sin. 2005. Both antigen optimization and lysosomal targeting are required for enhanced anti-tumour protective immunity in a human papillomavirus E7-expressing animal tumour model. Immunology 116:255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ko, H. J., S. Y. Ko, Y. J. Kim, E. G. Lee, S. N. Cho, and C. Y. Kang. 2005. Optimization of codon usage enhances the immunogenicity of a DNA vaccine encoding mycobacterial antigen Ag85B. Infect. Immun. 73:5666-5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kodihalli, S., H. Goto, D. L. Kobasa, S. Krauss, Y. Kawaoka, and R. G. Webster. 1999. DNA vaccine encoding hemagglutinin provides protective immunity against H5N1 influenza virus infection in mice. J. Virol. 73:2094-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, C. W., D. A. Senne, and D. L. Suarez. 2004. Generation of reassortant influenza vaccines by reverse genetics that allows utilization of a DIVA (Differentiating Infected from Vaccinated Animals) strategy for the control of avian influenza. Vaccine 22:3175-3181. [DOI] [PubMed] [Google Scholar]
- 24.Li, S., C. Liu, A. Klimov, K. Subbarao, M. L. Perdue, D. Mo, Y. Ji, L. Woods, S. Hietala, and M. Bryant. 1999. Recombinant influenza A virus vaccines for the pathogenic human A/Hong Kong/97 (H5N1) viruses. J. Infect. Dis. 179:1132-1138. [DOI] [PubMed] [Google Scholar]
- 25.Lin, C. T., T. Y., L. He, R. Calizo, H. H. Chou, T. C. Chang, Y. K. Soong, C. F. Hung, and C. H. Lai. 2006. A DNA vaccine encoding a codon-optimized human papillomavirus type 16 E6 gene enhances CTL response and antitumor activity. J. Biomed. Sci. 13:481-488. [DOI] [PubMed]
- 26.Liu, M., J. M. Wood, T. Ellis, S. Krauss, P. Seiler, C. Johnson, E. Hoffmann, J. Humberd, D. Hulse, Y. Zhang, R. G. Webster, and D. R. Perez. 2003. Preparation of a standardized, efficacious agricultural H5N3 vaccine by reverse genetics. Virology 314:580-590. [DOI] [PubMed] [Google Scholar]
- 27.Lu, S., S. Manning, and J. Athos. 1999. Antigen engineering in DNA immunization, p. 355-374. In D. B. Lowrie and R. G. Whalen (ed.), DNA vaccines: methods and protocols. Humana, Totowa, New Jersey.
- 28.Mozdzanowska, K., M. Furchner, G. Washko, J. Mozdzanowski, and W. Gerhard. 1997. A pulmonary influenza virus infection in SCID mice can be cured by treatment with hemagglutinin-specific antibodies that display very low virus-neutralizing activity in vitro. J. Virol. 71:4347-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagata, T., M. Uchijima, A. Yoshida, M. Kawashima, and Y. Koide. 1999. Codon optimization effect on translational efficiency of DNA vaccine in mammalian cells: analysis of plasmid DNA encoding a CTL epitope derived from microorganisms. Biochem. Biophys. Res. Commun. 261:445-451. [DOI] [PubMed] [Google Scholar]
- 30.Nakaya, T., J. Cros, M. S. Park, Y. Nakaya, H. Zheng, A. Sagrera, E. Villar, A. Garcia-Sastre, and P. Palese. 2001. Recombinant Newcastle disease virus as a vaccine vector. J. Virol. 75:11868-11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neumann, G., K. Fujii, Y. Kino, and Y. Kawaoka. 2005. An improved reverse genetics system for influenza A virus generation and its implications for vaccine production. Proc. Natl. Acad. Sci. USA 102:16825-16829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicholson, K. G., A. E. Colegate, A. Podda, I. Stephenson, J. Wood, E. Ypma, and M. C. Zambon. 2001. Safety and antigenicity of non-adjuvanted and MF59-adjuvanted influenza A/Duck/Singapore/97 (H5N3) vaccine: a randomised trial of two potential vaccines against H5N1 influenza. Lancet 357:1937-1943. [DOI] [PubMed] [Google Scholar]
- 34.Nicolson, C., D. Major, J. M. Wood, and J. S. Robertson. 2005. Generation of influenza vaccine viruses on Vero cells by reverse genetics: an H5N1 candidate vaccine strain produced under a quality system. Vaccine 23:2943-2952. [DOI] [PubMed] [Google Scholar]
- 35.Palese, P. 2006. Making better influenza virus vaccines? Emerg. Infect. Dis. 12:61-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palese, P., and A. Garcia-Sastre. 2002. Influenza vaccines: present and future. J. Clin. Investig. 110:9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palese, P., and J. F. Young. 1982. Variation of influenza A, B, and C viruses. Science 215:1468-1474. [DOI] [PubMed] [Google Scholar]
- 38.Parkin, N. T., P. Chiu, and K. Coelingh. 1997. Genetically engineered live attenuated influenza A virus vaccine candidates. J. Virol. 71:2772-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiao, C. L., K. Z. Yu, Y. P. Jiang, Y. Q. Jia, G. B. Tian, M. Liu, G. H. Deng, X. R. Wang, Q. W. Meng, and X. Y. Tang. 2003. Protection of chickens against highly lethal H5N1 and H7N1 avian influenza viruses with a recombinant fowlpox virus co-expressing H5 haemagglutinin and N1 neuraminidase genes. Avian Pathol. 32:25-32. [DOI] [PubMed] [Google Scholar]
- 40.Qiu, J. T., B. Liu, C. Tian, G. N. Pavlakis, and X. F. Yu. 2000. Enhancement of primary and secondary cellular immune responses against human immunodeficiency virus type 1 Gag by using DNA expression vectors that target Gag antigen to the secretory pathway. J. Virol. 74:5997-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramakrishna, L., K. K. Anand, K. M. Mohankumar, and U. Ranga. 2004. Codon optimization of the Tat antigen of human immunodeficiency virus type 1 generates strong immune responses in mice following genetic immunization. J. Virol. 78:9174-9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rota, P. A., B. K. De, M. W. Shaw, R. A. Black, W. C. Gamble, and A. P. Kendal. 1990. Comparison of inactivated, live and recombinant DNA vaccines against influenza virus in a mouse model. Virus Res. 16:83-93. [DOI] [PubMed] [Google Scholar]
- 43.Schickli, J. H., A. Flandorfer, T. Nakaya, L. Martinez-Sobrido, A. García-Sastre, and P. Palese. 2001. Plasmid-only rescue of influenza A virus vaccine candidates. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:1965-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh, I., R. W. Doms, K. R. Wagner, and A. Helenius. 1990. Intracellular transport of soluble and membrane-bound glycoproteins: folding, assembly and secretion of anchor-free influenza hemagglutinin. EMBO J. 9:631-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skehel, J. J., and D. C. Wiley. 2002. Influenza haemagglutinin. Vaccine 20(Suppl. 2):S51-S54. [DOI] [PubMed] [Google Scholar]
- 46.Smith, J. M., R. R. Amara, D. Campbell, Y. Xu, M. Patel, S. Sharma, S. T. Butera, D. L. Ellenberger, H. Yi, L. Chennareddi, J. G. Herndon, L. S. Wyatt, D. Montefiori, B. Moss, H. M. McClure, and H. L. Robinson. 2004. DNA/MVA vaccine for HIV type 1: effects of codon-optimization and the expression of aggregates or virus-like particles on the immunogenicity of the DNA prime. AIDS Res. Hum. Retrovir. 20:1335-1347. [DOI] [PubMed] [Google Scholar]
- 47.Stratford, R., G. Douce, L. Zhang-Barber, N. Fairweather, J. Eskola, and G. Dougan. 2000. Influence of codon usage on the immunogenicity of a DNA vaccine against tetanus. Vaccine 19:810-815. [DOI] [PubMed] [Google Scholar]
- 48.Subbarao, K., H. Chen, D. Swayne, L. Mingay, E. Fodor, G. Brownlee, X. Xu, X. Lu, J. Katz, N. Cox, and Y. Matsuoka. 2003. Evaluation of a genetically modified reassortant H5N1 influenza A virus vaccine candidate generated by plasmid-based reverse genetics. Virology 305:192-200. [DOI] [PubMed] [Google Scholar]
- 49.Tian, G., S. Zhang, Y. Li, Z. Bu, P. Liu, J. Zhou, C. Li, J. Shi, K. Yu, and H. Chen. 2005. Protective efficacy in chickens, geese and ducks of an H5N1-inactivated vaccine developed by reverse genetics. Virology 341:153-162. [DOI] [PubMed] [Google Scholar]
- 50.Tonegawa, K., E. Nobusawa, K. Nakajima, T. Kato, T. Kutsuna, K. Kuroda, T. Shibata, Y. Harada, A. Nakamura, and M. Itoh. 2003. Analysis of epitope recognition of antibodies induced by DNA immunization against hemagglutinin protein of influenza A virus. Vaccine 21:3118-3125. [DOI] [PubMed] [Google Scholar]
- 51.Treanor, J. J., J. D. Campbell, K. M. Zangwill, T. Rowe, and M. R. Wolff. 2006. Safety and immunogenicity of an inactivated subvirion influenza A (H5N1) vaccine. N. Engl. J. Med. 354:1343-1351. [DOI] [PubMed] [Google Scholar]
- 52.Treanor, J. J., G. M. Schiff, R. B. Couch, T. R. Cate, R. C. Brady, C. M. Hay, M. Wolff, D. She, and M. M. Cox. 2006. Dose-related safety and immunogenicity of a trivalent baculovirus-expressed influenza-virus hemagglutinin vaccine in elderly adults. J. Infect. Dis. 193:1223-1228. [DOI] [PubMed] [Google Scholar]
- 53.Wang, S., T. H. Chou, P. V. Sakhatskyy, S. Huang, J. M. Lawrence, H. Cao, X. Huang, and S. Lu. 2005. Identification of two neutralizing regions on the severe acute respiratory syndrome coronavirus spike glycoprotein produced from the mammalian expression system. J. Virol. 79:1906-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, S., D. J. Farfan-Arribas, S. Shen, T. H. Chou, A. Hirsch, F. He, and, S. Lu. 2006. Relative contributions of codon usage, promoter efficiency and leader sequence to the antigen expression and immunogenicity of HIV-1 Env DNA vaccine. Vaccine 24:4531-4540. [DOI] [PubMed] [Google Scholar]
- 55.Wang, S., D. Heilman, F. Liu, T. Giehl, S. Joshi, X. Huang, T. H. Chou, J. Goguen, and S. Lu. 2004. A DNA vaccine producing LcrV antigen in oligomers is effective in protecting mice from lethal mucosal challenge of plague. Vaccine 22:3348-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang, Z. Y., W. P. Kong, Y. Huang, A. Roberts, B. R. Murphy, K. Subbarao, and G. J. Nabel. 2004. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature 428:561-564. [DOI] [PMC free article] [PubMed] [Google Scholar]