Abstract
38K (ac98) of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) is a highly conserved baculovirus gene whose function is unknown. To determine the role of 38K in the baculovirus life cycle, a 38K knockout bacmid containing the AcMNPV genome was generated through homologous recombination in Escherichia coli. Furthermore, a 38K repair bacmid was constructed by transposing the 38K open reading frame with its native promoter region into the polyhedrin locus of the 38K knockout bacmid. After transfection of these viruses into Spodoptera frugiperda cells, the 38K knockout bacmid led to a defect in production of infectious budded virus, while the 38K repair bacmid rescued this defect, allowing budded-virus titers to reach wild-type levels. Slot blot analysis indicated that 38K deletion did not affect the levels of viral DNA replication. Subsequent immunoelectron-microscopic analysis revealed that masses of electron-lucent tubular structures containing the capsid protein VP39 were present in cells transfected with 38K knockout bacmids, suggesting that nucleocapsid assembly was interrupted. In contrast, the production of normal nucleocapsids was restored when the 38K knockout bacmid was rescued with a copy of 38K. Recombinant virus that expresses 38K fused to green fluorescent protein as a visual marker was constructed to monitor protein transport and localization within the nucleus during infection. Fluorescence was first detected along the cytoplasmic periphery of the nucleus and subsequently localized to the center of the nucleus. These results demonstrate that 38K plays a role in nucleocapsid assembly and is essential for viral replication in the AcMNPV life cycle.
The family Baculoviridae comprises a diverse group of arthropod-specific DNA viruses. They have been reported worldwide from over 600 host species, mostly from insects of the order Lepidoptera but also from the orders Diptera and Hymenoptera (5). Baculoviruses are characterized by a circular double-stranded DNA genome (ranging from 80 to 180 kbp) packaged within a rod-shaped capsid and enclosed by a lipid envelope (16, 17). Their life cycle typically involves the production of two virus forms, the budded virus (BV) and the occlusion-derived virus (ODV). Although the nucleocapsids of BVs and ODVs are similar in structure, the composition of their envelopes is different to accommodate their respective functions in an infection cycle (7). The ODVs, which are embedded within a paracrystalline matrix consisting mainly of virus-encoded polyhedrin protein, initiate a primary infection of the animal by infecting epithelial cells of the midgut when virus is released by the alkaline pH of the gut. The BVs are produced from the infected midgut epithelial cells and can initiate a secondary infection. Thus, these two virus forms play significantly different roles in the virus life cycle (10).
The baculovirus infection cycle in cultured cells can be subdivided into three major phases of viral gene transcription: early, late, and very late. The switchover from early to late viral gene expression coincides with the start of viral DNA replication and intranuclear development of the viral replication center, called the virogenic stroma. During the late phase of infection, newly replicated viral DNA is condensed and packaged into tubular capsid structures to form nucleocapsids within the virogenic stroma. These nucleocapsids egress from the cell nucleus and bud through a modified plasma membrane to acquire a loosely fitting envelope important for BV infectivity. Later in infection, nucleocapsids remain within the nucleus, become bundled together, and are enveloped by a membrane elaborated within the nucleus. The resulting ODVs are embedded within large proteinaceous paracrystalline occlusion bodies, also known as polyhedra (35).
The first baculovirus to be completely sequenced was Autographa californica nucleopolyhedrovirus (AcMNPV), which is the type species and the most widely studied of the Baculoviridae (2). AcMNPV contains a covalently closed circular genome of 134 kbp, about 154 putative open reading frames (ORFs) based on the criterion that the ORF be a single, contiguous, nonoverlapping coding region (2). Comparative analysis of the 29 currently completely sequenced baculovirus genomes shows that there are 29 identified genes in common, which are grouped as the baculovirus core set genes (12, 16). The poor conservation of the gene order between baculovirus genomes indicates that extensive genome rearrangements have occurred during the course of baculovirus evolution. It is thus particularly notable that only one core gene cluster of four genes has remained in same relative position in all sequenced baculovirus genomes (16), including one dipteran baculovirus (1) and two hymenopteran baculoviruses (12, 20). This core gene cluster is composed of helicase, lef5, ac96, and 38K.
Helicase, also referred to as P143, is a DNA binding protein with DNA-unwinding activity (19, 26); LEF-5 functions as a transcription initiation factor (13) and has a zinc ribbon that is homologous to a domain in the eukaryotic transcription elongation factor SII (14). Although both 38K and ac96 are members of the 29-gene baculovirus core set, which is indicative of their having a potentially vital biological role in the baculovirus life cycle, the functions of the two genes are unknown.
The AcMNPV 38K ORF was first identified by nucleotide sequence analysis of the 60.1 to 65.5 map units of AcMNPV (23), and its 1.1-kbp transcript was expressed at late phase during infection (24). 38K was found to be able to increase early, late, and very late gene expression by the late expression factor assay (15). Tagged versions of 38K were used to show that 38K accumulates in the nuclei of transfected cells and interacts with itself to form homo-oligomers (15). The failure to construct a 38K-null mutant virus suggested that 38K may be crucial to the AcMNPV life cycle (15).
In this study, we used an AcMNPV bacmid to generate a 38K knockout mutant by homologous recombination in Escherichia coli to determine the role of 38K in AcMNPV viral replication. Our data indicated that the 38K gene is essential for budded virus production, although deletion of 38K does not affect viral DNA replication. Immunoelectron microscopic analysis demonstrated that the 38K gene is required for normal nucleocapsid assembly.
MATERIALS AND METHODS
Viruses and cells.
Bacmid bMON14272, containing an AcMNPV genome, can propagate in E. coli DH10B as a large plasmid that confers resistance to kanamycin and can complement a lacZ deletion present on the chromosome to form blue colonies (Lac+) in the presence of the chromogenic substrate 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and the inducer isopropyl-β-D-thiogalactopyranoside (IPTG) (25). It contains a mini-attTn7 attachment site to allow generation of a recombinant bacmid following transposition of a pFastBac expression construct by helper plasmid pMON7124, which encodes the transposase (25). The Sf-9 insect cell line, the clonal isolate 9 from IPLB-Sf21-AE cells derived from the fall armyworm (Spodoptera frugiperda) (33), was cultured at 27°C in Grace's medium (Invitrogen Life Technologies) supplemented with 10% fetal bovine serum, penicillin (100 μg/ml), and streptomycin (30 μg/ml). In all experiments, the virus inoculum was allowed to adsorb for 1 h upon infection or 5 h upon transfection at 27°C and then was replaced with fresh medium. Time zero was defined as the time when the virus was replaced with fresh medium.
Generation of the 38K knockout AcMNPV bacmid.
Using the bacmid bMON14272, a 38K knockout AcMNPV was generated through homologous recombination in E. coli as previously described (4, 22). First, a transfer vector in which the 38K locus region was replaced with a chloramphenicol resistance gene (Cm) for antibiotic selection in E. coli was constructed as follows. A 540-bp fragment homologous to the 5′ region of the 38K ORF (AcMNPV nucleotides 85490 to 86017) was PCR amplified from the AcMNPV bacmid using the primers 38K-US-1 (5′-GAGCTCTAAATAAGGCGTAACTGGTC-3′) and 38K-US-2 (5′-GGATCCTCCTGCAAGCTGTCATACAC-3′) (SacI and BamHI sites, respectively, are underlined). The PCR product was digested with SacI and BamHI and then ligated into vector pUC18 to generate the recombinant plasmid pUC18-US. With the primers CmU (5′-GGATCCCTTCGAATAAATACCTGTGA-3′) and CmD (5′-CTGCAGAACCAGCAATAGACATAAGC-3′) (BamHI and PstI sites, respectively, are underlined), a 1,039-bp fragment containing the Cm gene cassette was PCR amplified from plasmid pKOV-KanF (18). The resulting product was digested with BamHI and PstI and ligated into plasmid pUC18-US, which was also digested with the same BamHI-PstI mixture to generate the recombinant plasmid pUC18-US-Cm. With primers 38K-DS-1 (5′-CTGCAGGTGCAAGATTGGGAACATTATCAC-3′) and 38K-DS-2 (5′-AAGCTTCGCTCTATTATTACTTGGACTCGGC-3′) (PstI and HindIII sites, respectively, are underlined), a 527-bp fragment homologous to the 3′ region of the 38K ORF (AcMNPV nucleotides 84581 to 85095) was PCR amplified from the AcMNPV bacmid. The PCR product was digested with PstI and HindIII and cloned into plasmid pUC18-US-Cm, which was digested with PstI/HindIII to generate a final 38K knockout transfer vector named pUC18-US-Cm-DS. This transfer vector was digested with SacI and HindIII, and the resulting linear 2.1-kbp fragment containing the Cm gene cassette and 38K flanking region was gel purified and suspended in distilled water to a final concentration of 200 ng/μl.
To facilitate homologous recombination between the Cm gene and the bacmid target sequence, DH10Bac cells (DH10B contains AcMNPV bacmid bMON14272) were transformed with pBAD-gbaA (27). pBAD-gbaA contains the λ red recombinase genes gamma, beta, and alpha, which encode a RecBC inhibitor, a single-strand-DNA-annealing protein, and a 5′→3′ double-strand-DNA exonuclease, respectively (27). The resulting clone cells were induced by addition of l-arabinose to allow expression of the λ red system, made competent, and electrotransformed with 1 μg of the purified linear 2.1-kbp fragment as previously described (29). The electroporated cells, containing an altered genotype of the recombinant bacmid, were incubated at 37°C for 1 h in 1 ml SOC medium (25) and subsequently spread onto agar medium containing 20 μg of chloramphenicol per ml, 50 μg of kanamycin per ml, and 7 μg of tetracycline per ml. Plates were incubated at 37°C for 2 days, and colonies resistant to chloramphenicol and kanamycin were selected and verified by PCR analysis and Southern blot analysis. The resulting 38K knockout bacmid was named vAc38K-KO.
PCR confirmation of recombinant bacmids.
Two pairs of specific PCR primers were used to confirm the replacement of 38K by the chloramphenicol-resistant gene in the 38K locus of bMON14272. Primers CmU and CmD were used to detect the correct insertion of the Cm gene cassette. Primers Ac9852 (5′-GGATCCATGGCCTCCTCGCTTCAAA-3′) and Ac9834 (5′-TCTAGATTTAATAAAATATTGTTCGTA-3′), which are just outside the flanking sequence for recombination, were used to confirm the deletion region. Primer pair Ac9852/CmD and primer pair CmU/Ac9834 were used to examine the recombination junctions of the upstream and downstream flanking regions.
Southern blot analysis.
Southern blot hybridization analysis was used to confirm the absence of 38K in the AcMNPV bacmid and the replacement of it by the Cm gene. A deletion fragment of 38K was PCR amplified from the AcMNPV bacmid with primers ET-38K-P1 (5′-GTTGCACGAGATGGGTTGCGTTTTA-3′) and ET-38K-P2 (5′-GGGGGTGGGGCATCGTTTGACTTTG-3′). The PCR product was purified and digoxigenin (DIG High Prime labeling and detection starter kit I; Roche Biochemicals) labeled overnight for use as a probe for Southern blot hybridization to detect the deletion of 38K. Meanwhile, the fragment containing the Cm gene was PCR amplified with primers CmU and CmD, and the PCR product was also labeled with digoxigenin-dUTP and was used as a probe to detect the replacement of Cm gene. AcMNPV bacmid DNA (wild-type [wt] AcMNPV, 38K-positive control) and vAc38K-KO were isolated from E. coli cells according to the Bac-to-Bac protocol (Invitrogen Life Technologies). HindIII-digested bacmid DNA was run overnight in ethidium bromide-stained 0.8% agarose gels, and DNA was transferred to a Nytran N nylon transfer membrane (Schleicher & Schuell). Hybridization and colorimetric detection with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate were performed according to the manufacturer's recommendations.
Construction of knockout, repair, and wt AcMNPV bacmids containing polyhedrin and gfp.
Bacmid bMON14272 has an occlusion-minus phenotype because the polyhedrin gene has been inserted so as to be inactive (25). In order to determine if 38K deletion has any effect on occlusion morphogenesis and to facilitate examination of virus infection, polyhedrin and an enhanced green fluorescence protein gene (egfp) were inserted into the polyhedrin locus by site-specific transposition as previously described (9). egfp was from pUC19egfp (donated by J. M. Vlak), and we refer to egfp as gfp in this study. Briefly, polyhedrin and gfp genes were inserted into the pFastBac1 (Invitrogen Life Technologies) plasmid under the control of the polyhedrin promoter and the AcMNPV ie1 promoter, respectively, to generate the donor plasmid pFB1-PH-GFP according to an identical method described by Dai et al. (9). A 1,522-bp fragment containing the wt 38K gene with its own promoter and poly(A) tail was PCR amplified using primers Ac9855 (5′-CTGCAGGGGCGGCTTTATGTCAAACAGT-3′) and Ac98310 (5′-TCTAGATCATCGTAGTAATCATTTTAATACATTT-3′) (PstI and XbaI sites, respectively, are underlined). The PCR product was digested with PstI/XbaI and ligated with pFB1-PH-GFP, which was digested with the enzymes used previously to give pFB1-38K-PH-GFP. Electrocompetent DH10B cells containing helper plasmid pMON7124 and the bacmid bMON14272 were transformed with pFB1-PH-GFP to generate a wt control virus named vAcPH-GFP. Electrocompetent DH10B cells containing pMON7124 helper plasmid and vAc38K-KO were transformed with donor plasmids pFB1-PH-GFP and pFB1-38K-PH-GFP to generate the 38K-null bacmid vAc38K-KO-PH-GFP and the 38K repair bacmid vAc38K-REP-PH-GFP, respectively. After 4 h of incubation at 37°C in 1 ml SOC, cells were spread onto agar medium containing 50 μg of kanamycin per ml, 7 μg of gentamicin per ml, 10 μg of tetracycline per ml, 100 μg of X-Gal per ml, and 40 μg of IPTG per ml. Plates were incubated at 37°C for 48 h; white colonies were selected and restreaked onto fresh plates to verify the phenotype, which was then confirmed by PCR.
Bacmid purification.
The correct recombinant bacmids vAcPH-GFP, vAc38K-KO-PH-GFP, and vAc38K-REP-PH-GFP were electroporated into E. coli DH10B cells, and the resulting colonies were screened for tetracycline sensitivity to ensure that the isolated bacmids were free of helper plasmids. Bacmid DNA was extracted and purified with a QIAGEN large-construct kit and quantified by optical density.
Analysis of viral growth curve.
To assess whether 38K is required for virus production, a viral growth curve analysis was performed. Sf-9 cells (2 × 106) were transfected in triplicate with 2.0 μg of the appropriate bacmid DNA using Cellfectin liposome reagent (Invitrogen Life Technologies) or infected with BV at a multiplicity of infection (MOI) of 5. The cell monolayers were incubated for 5 h after transfection or 1 h after infection, washed twice with Grace's medium, and replenished with 2 ml of fresh Grace's medium supplemented with 10% fetal bovine serum, penicillin (100 μg/ml), and streptomycin (30 μg/ml). Virus supernatants were collected at selected time points. The titers of BV were determined by a 50% tissue culture infective dose (TCID50) end point dilution assay on Sf-9 cells (28).
Plaque assay.
Sf-9 cells (2 × 106 per 35-mm-diameter dish) were transfected with 0.01 μg of each bacmid (vAcPH-GFP, vAc38K-KO-PH-GFP, or vAc38K-REP-PH-GFP). After a 5-h incubation period, the medium was removed and the cells were washed with 1 ml of Grace's medium. Cells were overlaid with 2 ml of Grace's medium containing 1% low-melting-point agarose (SeaPlaque) preequilibrated to 37°C, followed by incubation at 27°C in a moist box for 4 days before observation by fluorescence microscopy.
Slot blot analysis.
To examine viral DNA replication in infected cells, 2 × 106 Sf-9 cells were transfected in triplicate with 2.0 μg of each bacmid DNA (vAcPH-GFP, vAc38K-KO-PH-GFP, or vAc38K-REP-PH-GFP). At selected time points, cells were harvested, and each cell pellet was washed twice with PBS; 1 × 104 cells were resuspended in 250 μl of a 0.4 M NaOH-10 mM EDTA solution and heat treated at 100°C for 10 min. The samples were chilled on ice for 5 min, neutralized by adding 250 μl of cold 2 M ammonium acetate (pH 7.0), and then blotted under a vacuum onto a Nytran N nylon transfer membrane (Schleicher & Schuell) by using a slot blot apparatus (Bio-Dot SF microfiltration apparatus; Bio-Rad, Inc.) according to the instruction manual. The blotted membrane was prehybridized for 4 h at 68°C and then hybridized with a [32P]dCTP-labeled AcMNPV DNA probe, which was labeled by random priming (Random Primer DNA labeling kit, version 2.0; TAKARA). The blot was visualized by exposure to Amersham Phosphorscreen, developed with a Typhoon 8600 variable-mode imager, and quantified by Quantity One 1-D analysis software (Bio-Rad, Inc.).
Electron microscopy.
Sf-9 cells (5 × 106) were transfected with 5 μg vAcPH-GFP, vAc38K-KO-PH-GFP, or vAc38K-REP-PH-GFP, respectively. Mock-transfected cells were treated similarly but without the addition of virus DNA. By 72 h posttransfection (hpt), cells were dislodged and pelleted at 1,000 × g for 5 min. Cells were fixed, dehydrated, embedded, sectioned, and stained as described previously (21). For immunoelectron microscopy, the polyclonal antibody against AcMNPV capsid antigen VP39 was produced in this lab (unpublished data); rabbit polyclonal antibody against glutathione S-transferase was used as a negative control. Colloidal gold particles (10 nm) coated with protein A were prepared and donated by Dongwei Hu. vAc38K-KO-PH-GFP-transfected Sf-9 cells at 72 hpt were fixed in 2.5% (vol/vol) glutaraldehyde in 100 mM phosphate-buffered saline, pH 7.2, for 4 h at 4°C. Dehydration and embedding were performed according to a previously described procedure (21). Ultrathin sections were carried out using a diamond knife and collected on Formvar-coated nickel grids. The ultrathin sections were first floated on a drop of double-distilled water for 5 min and then transferred to blocking solution (1% bovine serum albumin, 0.02% PEG 20000, 100 mM NaCl, 1% NaN3) for 60 min, incubated for 60 min on VP39 antiserum at a dilution of 1:100 with blocking solution, floated on blocking solution again for 60 min and on protein A-gold at a dilution of 1:50 with blocking solution for 60 min, washed thoroughly with double-distilled water for several changes, and air-dried. Grids were then stained for 20 s on drops of aqueous 1% uranyl acetate and for 20 s with 0.2% lead citrate. Samples were viewed with a JEM-1230 transmission electron microscope at an accelerating voltage of 80 kV.
Generation of recombinant GFP fusion-containing bacmids and confocal microscopy.
To monitor the localization of 38K in AcMNPV-infected insect cells, 38K was expressed in-frame with GFP to create a 38K-GFP chimera. The 38K gene without the stop codon (TAA) was amplified from AcMNPV bacmid bMON14272 with the PCR primers Ac9852 and Ac9833 (5′-TCTAGATTTAATAAAATATTGTTCGTA-3′) (XbaI site is underlined). The 38K PCR product was digested with BamHI and XbaI and then inserted into BamHI/XbaI-digested pUC18 to create pUC18-38K. The ORF of gfp was digested with XbaI and PstI from pUC19egfp, and the resulting fragment was cloned into the XbaI/PstI site of pUC18-38K to generate pUC18-38KGFP. The fragment that the 38K ORF was upstream of and that was in-frame with the GFP ORF was cut from pUC18-38KGFP with BamHI/PstI and then cloned into pFastBac1 to generate donor plasmid pFB1-38KGFP. According to the Bac-to-Bac instruction manual (Invitrogen Life Technologies), DH10Bac cells which contain AcMNPV bacmid bMON14272 and helper plasmid pMON7124 were transformed with pFB1-38KGFP, and the 38K-GFP chimera was site-specifically transposed into the AcMNPV bacmid polyhedrin locus. Thus, 38K is expressed with a GFP tag under the control of the polyhedrin promoter in the resulting bacmid, which is referred to as vAcph38KGFP. The control bacmid vAcphGFP, in which only GFP is expressed under the control of the polyhedrin promoter, was generated in a procedure similar to that for vAcph38KGFP. In order to determine if there is any difference between 38K-GFP chimera expression from the polyhedrin promoter and that from the 38K native promoter in virus-infected insect cells, a recombinant bacmid in which 38K can be expressed in frame with GFP under the control of its own promoter was also constructed as follows. The polyhedrin promoter was moved from pFastBac1 to create pFB1-ph− as previously described (9). The 38K ORF (without the stop codon, TAA) and its own promoter was amplified from the AcMNPV bacmid using primers Ac9856 (5′-GAATTCGGGCGGCTTTATGTCAAACAGT-3′) (EcoRI site is underlined) and Ac9833. The EcoRI/XbaI-digested PCR product was inserted into pFB1-ph− to generate pFB1-ph−38K. The gfp fragment described above was cloned into pFB1-ph−38K to create donor plasmid pFB1-ph− 38KGFP. This donor plasmid was transposed into the AcMNPV bacmid polyhedrin locus to generate the recombinant bacmid vAc38KGFP. All three recombinant AcMNPV bacmids mentioned above retained the wt 38K gene locus.
Sf-9 cells (2 × 106) were transfected with 2 μg vAcphGFP, vAcph38KGFP, or vAc38KGFP bacmid DNA. At 96 hpt, supernatants were collected, and the titers of BV were determined by TCID50 end point dilution assay on Sf-9 cells. For confocal microscopy, Sf-9 cells (1 × 105) were seeded onto glass coverslips and left to stand for several hours to allow cell attachment. Cells were infected with vAcphGFP, vAcph38KGFP, or vAc38KGFP virus at an MOI of 10. At 16, 24, 48, and 72 h postinfection (hpi), cells were visualized with a Leica TCS SP2 confocal laser scanning microscope for fluorescence using a laser line wavelength of 488 nm for GFP. All images were digitally recorded and merged by the use of Leica software.
RESULTS
Construction of 38K knockout AcMNPV bacmid.
To determine whether 38K is essential for viral replication, we generated an infectious bacmid containing a knockout in the 38K gene via the λ red recombination system in E. coli as described previously (4, 22). For this procedure, we constructed a transfer vector (pUC18-US-Cm-DS) in which the 38K locus region was replaced with a Cm gene for antibiotic selection in E. coli. Four hundred ninety-two base pairs of the 5′ end and 75 bp of the 3′ end of the 38K coding region were retained so that the deletion of 38K in AcMNPV bacmid would not affect transcription of ac97 and lef-5 (Fig. 1A). DH10Bac cells containing AcMNPV bacmid were transformed with pBAD-gbaA and were induced by l-arabinose to allow expression of the λ red system, and then the cultures were transformed with the linear DNA fragment containing the Cm gene cassette and 38K flanking region. The remaining 394 bp of the 38K coding region in the AcMNPV bacmid was replaced by the Cm gene cassette via homologous recombination. The resulting bacmid, vAc38K-KO, was selected by growth on medium containing kanamycin and chloramphenicol (Fig. 1A) and was confirmed by PCR analysis and Southern blot analysis.
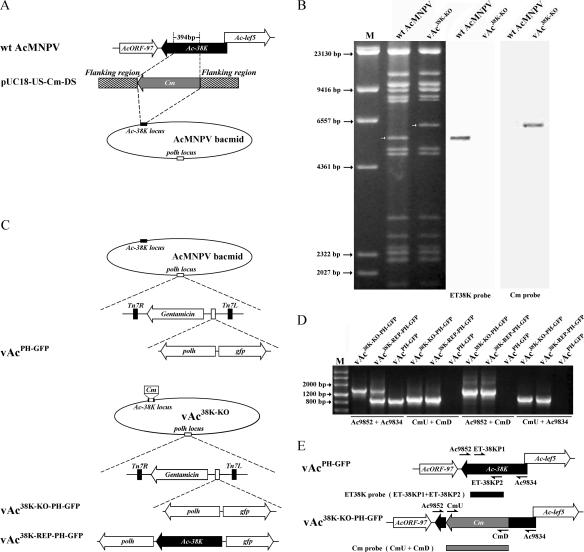
FIG. 1.
Construction of 38K knockout, repair, and wt AcMNPV bacmids. (A) Strategy for construction of a 38K knockout bacmid containing a deletion of the AcMNPV 38K gene by recombination in E. coli. A 394-bp portion of the 38K ORF was deleted and replaced with the chloramphenicol resistance gene (Cm). (B) Southern blot analysis of wt AcMNPV and vAc38K-KO using the ET38K and Cm probes to confirm the deletion of 38K and its replacement with Cm. (C) Schematic diagram of three viruses, vAc38K-KO-PH-GFP, vAc38K-REP-PH-GFP, and vAcPH-GFP, showing the polyhedrin (polh) and gfp genes inserted into the polyhedrin locus by Tn7-mediated transposition. (D) PCR analysis of the presence or absence of sequence modifications in vAc38K-KO-PH-GFP, vAc38K-REP-PH-GFP, and vAcPH-GFP. The virus templates are shown above each lane, and the primer pairs used are shown below. (E) Positions of primer pairs and probes used to confirm disruption of the 38K ORF and the correct insertion of the Cm cassette.
Four primer pairs were used to confirm that 38K had been deleted from the 38K locus of the AcMNPV bacmid genome. Primers CmU and CmD were used to detect the correct insertion of the Cm gene cassette. Primers Ac9852 and Ac9834 were used to confirm the loss of the 394 bp within the 38K coding region. Primers Ac9852 and CmD and primers CmU and Ac9834 were used to examine the recombination junctions of the upstream and downstream flanking regions (data not shown).
The absence of 38K and its replacement with Cm in the AcMNPV bacmid were also confirmed by Southern blot hybridization analysis. The PCR-amplified 394-bp fragment from the AcMNPV bacmid, which is the deleted part of the 38K gene, was purified and digoxigenin labeled and was used as a probe (ET38K probe) to detect the 38K gene (Fig. 1E). The PCR-amplified 1,039-bp fragment containing the Cm gene was also labeled with digoxigenin-dUTP and was used as a probe (Cm probe) to detect the Cm gene (Fig. 1E). wt AcMNPV bacmid (bMON14272, 38K-positive control) and vAc38K-KO were digested with HindIII and hybridized with the ET38K probe and Cm probe, respectively. A 5.5-kbp HindIII-J fragment, which contains the 38K gene located in the AcMNPV bacmid genome, hybridized strongly to the ET38K probe; however, there was no signal with vAc38K-KO (Fig. 1B). Since a 394-bp fragment of 38K was replaced by a 1,039-bp Cm gene in vAc38K-KO, the AcMNPV bacmid 5.5-kbp HindIII-J fragment was lengthened to 6.1 kbp. As expected, in vAc38K-KO a 6.1-kbp HindIII-J fragment of vAc38K-KO hybridized strongly to the Cm probe, but there was no signal with wt AcMNPV (Fig. 1B). These results demonstrated that 38K was successfully replaced by the Cm gene and no intact 38K gene cassette existed in the vAc38K-KO genome.
Construction of knockout, repair, and wt AcMNPV bacmids containing polyhedrin and gfp.
To determine if 38K deletion has any effect on occlusion morphogenesis and to facilitate examination of virus infection, a 38K knockout mutant, vAc38K-KO-PH-GFP, containing polyhedrin and gfp genes was constructed by transposition of polyhedrin and gfp genes into the polyhedrin locus of vAc38K-KO, using the Bac-to-Bac system (Fig. 1C). For rescue and confirmation of the phenotype resulting from the 38K knockout, a repair bacmid, vAc38K-REP-PH-GFP, was generated by inserting 38K under the control of its own promoter as well as polyhedrin and gfp into the polyhedrin locus by transposition (Fig. 1C). polyhedrin and gfp were also inserted into the polyhedrin locus of AcMNPV bacmid bMON14272 to generate a wt bacmid named vAcPH-GFP, which was used as a positive control (Fig. 1C).
All constructs were confirmed by PCR analysis as described above. Positions of primer pairs used to confirm these recombinant bacmids are shown in Fig. 1E, and the results are shown in Fig. 1D. Primers Ac9852 and Ac9834 produced single fragments of 975 bp and 1,620 bp for vAcPH-GFP and vAc38K-KO-PH-GFP, respectively, but both fragments for vAc38K-REP-PH-GFP (Fig. 1D). Primers CmU and CmD produced no PCR product in vAcPH-GFP but produced a single 1,039-bp fragment in both vAc38K-KO-PH-GFP and vAc38K-REP-PH-GFP (Fig. 1D). Primers Ac9852 and CmD produced no PCR product in vAcPH-GFP but produced a single 1,575-bp fragment in vAc38K-KO-PH-GFP and vAc38K-REP-PH-GFP (Fig. 1D). Similarly, primers CmU and Ac9834 produced no PCR product in vAcPH-GFP but produced a single 1,128-bp fragment in vAc38K-KO-PH-GFP and vAc38K-REP-PH-GFP (Fig. 1D). Transposition events were confirmed later by GFP expression and occlusion body formation in bacmid DNA-transfected Sf-9 cells as described below.
Analysis of knockout, repair, and wt AcMNPV bacmids replication in transfected Sf-9 cells.
To determine the effect of disruption of 38K on virus replication, Sf-9 cells were transfected with vAcPH-GFP, vAc38K-KO-PH-GFP, and vAc38K-REP-PH-GFP, respectively. Infected cells were monitored by fluorescence due to expression of gfp, which was under the control of the AcMNPV ie1 promoter. No difference was observed among the three viruses at 24 hpt, indicating comparable transfection efficiencies of approximately 30% (Fig. 2A). Fluorescence was observed in almost all vAcPH-GFP- or vAc38K-REP-PH-GFP-transfected cells by 72 hpt (Fig. 2A), indicating that the wt and 38K repair bacmids could generate infectious budded virions from the initial transfection. In contrast, however, vAc38K-KO-PH-GFP-transfected cells showed almost no increase in the number of infected cells (Fig. 2A), indicating that there was no spread of the virus beyond the cells initially transfected with the 38K knockout bacmid DNA. To further compare the spread of virus between vAcPH-GFP, vAc38K-KO-PH-GFP, and vAc38K-REP-PH-GFP, plaque assays were performed to observe the spreading of virus to adjoining cells. By 4 days posttransfection, vAcPH-GFP and vAc38K-REP-PH-GFP both produced large normal plaques (Fig. 2A). In contrast, the vAc38K-KO-PH-GFP plaques consisted of only a single cell (Fig. 2A), indicating that this virus is unable to spread.
FIG. 2.
Analysis of viral replication in Sf-9 cells. (A) The top and middle panels show Sf-9 cells transfected with vAcPH-GFP, vAc38K-KO-PH-GFP, or vAc38K-REP-PH-GFP at 24 and 72 hpt, respectively. The bottom panels show plaque assay results at 96 hpt. The arrows show single infected cells, indicating the inability of vAc38K-KO-PH-GFP to spread the infection. (B) Light microscopy of vAcPH-GFP-, vAc38K-KO-PH-GFP-, and vAc38K-REP-PH-GFP-transfected cells at 48 and 96 hpt. (C) Virus growth curves generated from a transfection or infection of virus in Sf-9 cells. For the transfection curves, Sf-9 cells were transfected with 2 μg of bacmid DNA from each virus. Cells culture supernatants were harvested at the selected time points and assayed for the production of infectious virus by TCID50 assay. For the infection curves, cells were infected with each virus at an MOI of 5, and cells culture supernatants were harvested and assayed for the production of infectious virus by TCID50 assay. Each datum point represents the average from three independent transfections or infections. Error bars represent standard errors.
Microscopic analysis showed that occlusion bodies formed in vAcPH-GFP- and vAc38K-REP-PH-GFP-transfected cells as well as in vAc38K-KO-PH-GFP-transfected cells. At 48 hpt, only a small proportion of the cells contained occlusion bodies, corresponding closely to the number of cells initially transfected (Fig. 2B). However, by 96 hpt, significant differences were observed between vAcPH-GFP- or vAc38K-REP-PH-GFP-transfected cells and vAc38K-KO-PH-GFP-transfected cells. Almost every single cell transfected with vAcPH-GFP- or vAc38K-REP-PH-GFP contained occlusion bodies, whereas the number of the vAc38K-KO-PH-GFP-transfected cells containing occlusion bodies showed no increase (Fig. 2B).
These results showed that the deletion of 38K leads to a defect in infectious BV production. To further assess the effect of deleting 38K on virus replication and determine the replication kinetics of virus constructs, a virus growth curve analysis was performed. For these experiments, Sf-9 cells were either transfected with bacmid DNA or infected with BVs; at selected time points, the BV titers were determined by a TCID50 end point dilution assay. Sf-9 cells transfected with vAcPH-GFP and vAc38K-REP-PH-GFP revealed a steady increase in virus production (Fig. 2C). In contrast, no titer was detectable at any time point up to 120 hpt for vAc38K-KO-PH-GFP-transfected cells (Fig. 2C). To determine whether 38K repair bacmid could rescue the defect of infectious BV production, a second growth curve was obtained with the BV produced from vAcPH-GFP- and vAc38K-REP-PH-GFP-transfected cells at an MOI of 5. These growth curves revealed that the 38K repair virus was as proficient in virus production as the wt virus (Fig. 2C). Hence, these data showed that the defect in 38K knockout virus was due to the deletion of 38K, because the defect can be rescued by the insertion of 38K back into the polyhedrin locus of the 38K knockout bacmid.
38K deletion does not affect viral DNA replication in Sf-9 cells.
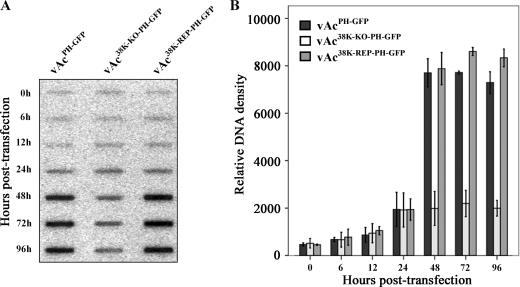
To determine whether 38K is essential for viral DNA replication, a slot blot analysis was performed to compare the initiation and levels of viral DNA replication in vAcPH-GFP-, vAc38K-KO-PH-GFP-, and vAc38K-REP-PH-GFP-transfected cells. Equal amounts of virus-transfected Sf-9 cells were collected at designated time points, and cell lysates were prepared and subjected to slot blot analysis. Replication of viral DNA was detected by hybridization with [32P]dCTP-labeled AcMNPV genomic DNA and quantified by measuring relative intensity. At 0 hpt, similar signals for viral DNA were detected from cells transfected with each bacmid construct, indicating equal amounts of input DNA (Fig. 3). Three viruses had the same time of onset of viral replication at 24 hpt (Fig. 3), indicating that 38K is not required for viral DNA synthesis. Analysis of viral DNA replication from 48 to 96 hpt revealed a substantial and steady increase in viral DNA generated by the wt and 38K repair bacmids; however, the level of viral DNA generated by the 38K knockout bacmid did not increase (but plateaued to about 25% of the wt control and 38K repair bacmids) (Fig. 3B). Because the same number of cells was analyzed at each time point, these data suggest that the wt and 38K repair bacmids can initiate a secondary infection leading to an increase in the number of cells containing viral DNA, whereas the 38K knockout bacmid can replicate only in cells that were initially transfected. Thus, the slot blot analysis indicated that deletion of 38K does not affect viral DNA replication in Sf9 cells.
FIG. 3.
Slot blot analysis of viral DNA replication. (A) Slot blot analysis of intracellular viral DNA replication in transfected Sf-9 cells. Cells (2 × 106) were transfected with 2.0 μg of each bacmid (vAcPH-GFP, vAc38K-KO-PH-GFP, or vAc38K-REP-PH-GFP). At the indicated times, 1 × 104 cells were harvested, processed, and applied to a nylon membrane using a slot blot apparatus. Hybridization was performed with AcMNPV DNA labeled with [32P]dCTP. (B) Quantitative analysis of viral DNA replication detected in the slot blot. The blots were visualized by exposure to Amersham Phosphorscreen, developed with a Typhoon 8600 variable-mode imager, and quantified with Quantity One 1-D analysis software (Bio-Rad, Inc.). Means ± standard deviations from three independent experiments are shown.
Electron microscopic analysis of 38K wt, knockout, and repair bacmid-transfected cells.
The results described above indicated that there could be a total defect in BV production or that noninfectious particles could be produced in insect cells transfected with 38K knockout virus. To further analyze whether 38K affects nucleocapsid structure, electron microscopy was performed with thin sections generated from bacmid-transfected cells (Fig. 4). Virus-transfected cells could be easily distinguished from mock-transfected cells by the presence of the virogenic stroma, which is an electron-dense, baculovirus-induced structure seen within the nucleus (data not shown). The virogenic stroma is thought to be the active site for viral DNA replication, condensation, and packaging into capsids (11, 37). Nucleocapsids are usually observed at the electron-dense edges of the stroma and occasionally seem to be in the process of acquiring a condensed core of DNA (11). Maturation of the virogenic stroma yielded a significant and morphologically distinct peristromal compartment of the nucleoplasm, called the ring zone. Nucleocapsids align with vesicle-like structures of de novo envelopes, and bundles of nucleocapsids prior to occlusion in the protein-crystalline matrix of the developing occlusion bodies are often observed within this region (35).
FIG. 4.
Electron microscopic analysis of transfected Sf-9 cells to characterize 38K mutants. (A) Portion of a cell transfected with vAc38K-REP-PH-GFP. Normal nucleocapsids are dispersed at the electron-dense edges of the stroma. The inset shows several nucleocapsids enveloped in one virion. (B) Image of polyhedra within the nucleus of the cell transfected with vAc38K-REP-PH-GFP, showing normal virions embedded in the polyhedra. (C) Image of the nucleus of a cell transfected with vAc38K-KO-PH-GFP showing masses of electron-lucent tubular structures at the electron-dense edges of the stroma. (D) Polyhedra within a nucleus of a cell transfected with vAc38K-KO-PH-GFP, showing no normal virions embedded, although in some cases a few lucent tubular structures could be discerned. Bar, 500 nm.
As expected, events such as rod-shaped nucleocapsids associating with electron-dense edges of the virogenic stroma, nucleocapsids aligning with vesicle-like structures of de novo envelopes, and bundles of nucleocapsid embedding into the developing polyhedra within the ring zone were frequently observed in vAcPH-GFP-transfected cells (data not shown). Observations of cells transfected with the 38K repair bacmid (vAc38K-REP-PH-GFP) revealed the presence of electron-dense rod-shaped nucleocapsids that were morphologically indistinguishable from those observed in cells transfected with the wt bacmid (Fig. 4A). However, in contrast to cells transfected with the wt virus, all vAc38K-KO-PH-GFP-transfected cells completely lacked nucleocapsids and showed masses of electron-lucent tubular structures at the electron-dense edges of the stroma (Fig. 4C). A cross-section of these tubular structures indicated that they lack an electron-dense core, which would be indicative of nucleoprotein (Fig. 4C). These electron-lucent tubular structures seem to be the empty capsid sheaths which are devoid of a nucleoprotein core, indicating that the viral DNA genomes failed to be condensed or packaged into the tubular structures. Clusters of elongated tubular structures that appear to lack an electron-dense core were observed to localize to the inner nuclear membrane in vAc38K-KO-PH-GFP-transfected cells (data not shown). Interestingly, such structures have been observed in cells transfected with a vlf-1 knockout bacmid (21, 32). VLF-1 of AcMNPV is a putative tyrosine recombinase and is required for both very late gene expression and budded-virus production (31).
Polyhedra were observed in nuclei of 38K wt, knockout, and repair bacmid-transfected cells. The sizes and shapes of the polyhedra in vAc38K-KO-PH-GFP-transfected cells (Fig. 4D) are similar to those in wt-transfected (data not shown) and repair-virus-transfected (Fig. 4B) cells. Enveloped virions had been occluded or were being occluded into polyhedra within the ring zone of wt-transfected (data not shown) and repair-virus-transfected (Fig. 4B) cells. However, in 38K knockout virus-transfected cells, no virions were observed to be embedded in polyhedra (Fig. 4D). Examination of the vAc38K-KO-PH-GFP-transfected cells at 24 hpt revealed formation of profiles of membrane material juxtaposed to the nuclear membrane. These membrane profiles likely represent the source of virion envelopes (data not shown). However, nucleocapsids aligning with de novo membranous profiles end-on to acquire their envelope were not observed up to 72 hpt. In contrast, in wt- or repair virus-transfected cells, multiple nucleocapsids with nucleoprotein cores were enveloped in one virion (Fig. 4A, inset). These observations indicated that deletion of 38K does not abrogate occlusion body morphogenesis.
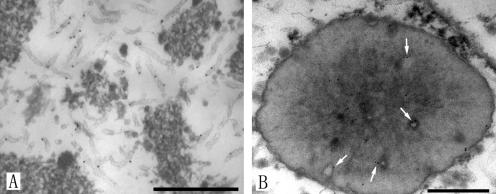
To determine whether the electron-lucent tubular structures observed in cells transfected with the 38K knockout bacmid (vAc38K-KO-PH-GFP) are related to capsids, immunoelectron microscopy was performed. Figure 5A is an electron micrograph of a portion of virogenic stroma in a cell transfected with vAc38K-KO-PH-GFP at 72 hpt. The lucent tubular structures within the virogenic stroma appeared to be reactive with the VP39 polyclonal antibody, as indicated by the position of the gold label. Similar positioning of label was observed in the abnormal polyhedra (Fig. 5B), indicating that the tubular structures can be embedded within the polyhedra. No immunogold localization was seen in the control section (data not shown). Together, these data indicated that the aberrant capsid structures observed in cells transfected with the 38K knockout bacmid were the result of a structural or enzymatic deficiency associated with 38K and suggested that 38K is required for proper capsid assembly.
FIG. 5.
Immunogold staining of the VP39 protein in thin sections of 38K knockout bacmid-transfected Sf-9 cells. (A) Portion of the virogenic stroma in the nucleus. Electron-lucent tubular structures appeared to be reactive with the VP39 polyclonal antibody, as indicated by the position of the gold label. (B) Cross-section of polyhedron. Arrows show the tubular structures in the abnormal polyhedra labeled with immunogold. Bar, 500 nm.
Localization of 38K in AcMNPV-infected insect cells.
To detect the subcellular localization of 38K, three recombinant baculoviruses, vAcphGFP, vAcph38KGFP, and vAc38KGFP, were constructed (Fig. 6A). The 38K-GFP chimera was inserted into the polyhedrin locus of the AcMNPV bacmid by site-specific transposition, and the native 38K locus was left intact. GFP was fused to the C terminus of 38K and was expressed under the control of the polyhedrin promoter and 38K native promoter in vAcph38KGFP and vAc38KGFP, respectively. As a control, GFP alone was expressed under the control of the polyhedrin promoter (Fig. 6A).
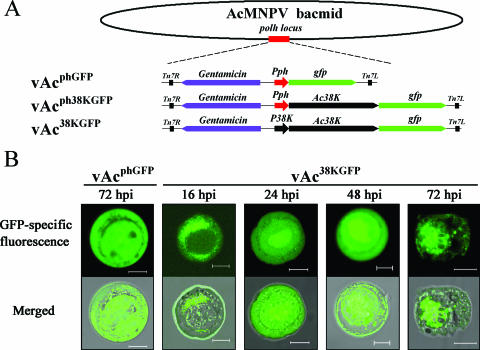
FIG. 6.
Subcellular localization of 38K-GFP fusion protein in Sf-9 cells transfected with GFP-tagged bacmids. (A) Schematic map of construction of GFP-tagged recombinant bacmids. Nonfused gfp was transposed into the polyhedrin locus of the AcMNPV bacmid to generate vAcphGFP. 38K fused with the GFP tag under the control of polyhedrin promoter or its own promoter was transposed into the polyhedrin locus of the AcMNPV bacmid, and the resulting recombinant bacmids were named vAcph38KGFP or vAc38KGFP, respectively. (B) Confocal images of Sf-9 cells. Cells were infected with vAc38KGFP at an MOI of 10. At 16, 24, 48, and 72 hpi, cells were examined for fluorescence by confocal laser scanning microscopy. Cells infected with vAcphGFP were used as a control. For each time point, GFP-specific fluorescence micrographs are shown above the merged micrographs. Bar, 10 μm.
Sf-9 cells infected with vAcphGFP, vAcph38KGFP, or vAc38KGFP at an MOI of 10 were examined for GFP-specific fluorescence by confocal laser scanning (Fig. 6B). The observed intranuclear localization of 38K-GFP changed over time in a similar pattern in both vAcph38KGFP-infected cells and vAc38KGFP-infected cells. Fluorescence localized along the outer periphery of the nucleus at 16 hpi, moved into the nucleus at 24 hpi, accumulated in the center of the nucleus at 48 hpi, and at 72 hpi became highly condensed within the center of the nucleus and had the appearance of a net, which may be the virogenic stroma, the viral DNA replication and nucleocapsid assembly site (Fig. 6B). Sporadic small foci of 38K-GFP fluorescence were detected in the ring zone of the nucleus and within the cytoplasm of the infected cells (Fig. 6B). Fluorescence was observed throughout the cytosol and the nucleus of vAcphGFP-infected cells at any time point selected from 16 hpi to 72 hpi (Fig. 6B). These results are consistent with our previous observation that 38K is involved in nucleocapsid assembly.
DISCUSSION
The main finding of the present study is that the 38K gene is essential for AcMNPV replication and is required for nucleocapsid assembly. First, the role of 38K in the context of an AcMNPV infection in Sf-9 cells was examined with an AcMNPV 38K knockout virus which can propagate as a bacmid in E. coli. The 38K knockout virus was unable to propagate after transfection into Sf-9 cells, and the infection was restricted to the cells that were initially transfected, which was confirmed by plaque assay. This defect in replication could be rescued by the reinsertion of the 38K gene into the polyhedrin locus of the same bacmid. The results confirmed that the observed phenotype resulted from the 38K knockout, not from a second site mutation or from disruption of regulatory elements located at the 38K locus. Therefore, our studies indicated that the 38K gene is essential for AcMNPV replication in Sf-9 cells.
Second, slot blot analysis using [32P]dCTP-labeled viral DNA as a probe showed that the onset of viral DNA replication appeared to be similar between the 38K knockout and wt virus or repaired virus, which indicated that 38K is not required for viral DNA replication. The reasons that the 38K knockout virus could replicate only to about 25% of the levels of the wt control and 38K repair bacmids may be as follows. (i) 38K is essential for AcMNPV replication and can therefore replicate only in initially vAc38K-KO-PH-GFP-transfected cells. (ii) Comparable transfection efficiencies were approximately 30% among vAcPH-GFP-, vAc38K-KO-PH-GFP-, and vAc38K-REP-PH-GFP-transfected cells at 24 hpt (Fig. 2A), so the cell cycle of infected cells would be arrested by expression of some AcMNPV genes, such as ie2 (30); meanwhile, the remaining uninfected cells would propagate normally. Due to the cell population doubling time and the maximum cell density, only a portion of the remaining uninfected cells could propagate once, so the total amount of viral DNA in vAc38K-KO-PH-GFP-transfected cells is about 25% of that in vAcPH-GFP- or vAc38K-REP-PH-GFP-transfected cells from 48 hpt onwards. This result is similar to that in some previous studies (9, 22).
Third, electron microscopy and immunoelectron microscopy performed with a 38K knockout virus demonstrated that the failure of production of budded viruses was due to a defect in nucleocapsid assembly. Immunoelectron microscopy revealed masses of electron-lucent tubular structures, which reacted specifically with a polyclonal antibody to the major capsid protein VP39, present in cells transfected with the 38K knockout virus. These structures represent incomplete capsid particles which were waiting for nucleic acids. Baculovirus nucleocapsids are composed of a tubular capsid shell with distinct cap structures on each end, enclosing an inner nucleoprotein core in which the genome is condensed about 100-fold (8). One previously proposed model suggests that viral DNA is packaged into a preassembled capsid sheath (11). Another previous study on normal AcMNPV nucleocapsid assembly suggested that nucleocapsid components, such as the capsid and the DNA core, assemble in successive stages rather than simultaneously (3). Our results support the hypothesis that the defect in nucleocapsid assembly is due to the failure of viral DNA packaging.
Excessively long tubular structures containing capsid protein were located primarily in the nucleus and were associated with the inner nuclear membrane of the 38K knockout virus-transfected cells. Another investigation also found that long tubular structures largely devoid of viral DNA were located primarily in the nucleus and were associated with the inner nuclear membrane of AcMNPV-infected cells in the presence of cytochalasin D (34). Although capsid antigen is synthesized and transported to the nucleus and viral DNA is synthesized, normal-sized nucleocapsids are only rarely seen. Cytochalasin D is a specific inhibitor of actin microfilament elongation. The observation that cytochalasin D interfered with the assembly of normal capsids indicated that actin microfilaments were somehow involved in this intranuclear event (34).
Recently, a similar observation, i.e., the appearance of aberrant long capsid structures in the nuclei of cells transfected with a vlf-1 knockout bacmid, was reported (21, 32). VLF-1 has been shown to play a role in baculovirus capsid structure (32). VLF-1 of AcMNPV is a putative tyrosine recombinase and is an essential capsid component (31, 36). Interestingly, it was found that these aberrant capsid structures remained apart from the virogenic stroma, localizing instead to the inner nuclear membrane. The virogenic stroma is thought to be the center for DNA replication and packaging, so it was proposed that the long capsid structures need further processing to generate legitimate capsid precursors and the aberrant capsid structures remain incomplete due to the lack of VLF-1 (31). However, in our study, masses of electron-lucent tubular structures of normal length appeared at the electron-dense edges of the stroma in cells transfected with the 38K knockout virus, indicating that the processing of long capsid precursors had occurred. 38K does not interfere with viral DNA replication. Therefore, it may be involved in DNA transactions during the late stages of the replication cycle; namely, it may facilitate packaging into viral capsids.
Using a recombinant virus that expresses 38K fused to GFP as a visual marker to monitor protein transport and localization within the nucleus during infection, we found that in the late phase of virus infection, 38K mainly localized to the center of nucleus, where it may be the virogenic stroma. In previous studies, computer analysis has indicated that 38K possesses a zinc finger motif (2) and is a nuclear protein (15), suggesting a regulatory role in baculovirus infection. The local accumulation of 38K at the center of the nucleus in the present study implies that this protein may interact with viral DNA or another protein to play a role in nucleocapsid assembly in the virogenic stroma. The sporadic small foci of 38K-GFP fluorescence detected in the ring zone of the nucleus and within the cytoplasm of the infected cells suggest that 38K may be incorporated into ODVs as a consequence of protein abundance or colocalization, or these foci may be a component of BVs or ODVs which can progress out of the nucleus and then pass through the cytoplasm or can be embedded within the polyhedra in the ring zone, respectively. However, we are not aware of any previous reports, including an investigation using multiple approaches to identify the proteins associated with AcMNPV (6), which showed that 38K is a component of AcMNPV. Thus, there may be other explanations for the sporadic cytosolic foci. Antiserum generated to 38K protein and used to probe purified virions would determine whether 38K is present in BVs and/or ODVs.
Additionally, we demonstrated that the 38K knockout virus could progress through the very late phase in Sf-9 cells, as evidenced by the abnormal development of occlusion bodies in the nuclei of transfected cells; although no embedded virions could be observed within the polyhedra. 38K does not appear to associate with polyhedra, even though its deletion results in aberrant capsid structures and defects in ODV formation. In two previous studies, however, when cells were treated with cytochalasin D (34) or transfected with a vlf-1 knockout bacmid (32), the aberrant capsid structures appeared and no polyhedron formation was observed.
Nucleoprotein core encapsidation generally involves a set of highly ordered protein-DNA and protein-protein interactions that lead to the assembly of a nucleocapsid. 38K is one of the 29 baculovirus core set genes and one of four genes which make up the only core gene cluster in the 29 currently completely sequenced baculovirus genomes. 38K is a highly conserved gene, but no significant sequence similarities between 38K and nonbaculovirus proteins with known functions have been reported. Herein we demonstrate that 38K is required for assembly of the viral nucleocapsid, although its exact role in the assembly process is unclear. The present study will lead to a better understanding of the molecular aspects of this protein and the molecular basis for factors governing the assembly of the nucleocapsid.
Acknowledgments
We thank Just M. Vlak (Wageningen University and Research Center, Wageningen, The Netherlands) for the generous gift of pUC19-EGFP, Xiaojiang Dai for critical review of the manuscript, and Dongwei Hu and Qingfen Zhang for assistance with the electron microscopy work.
This research was supported by the National Major Basic Research Project (“973”) of China (no. 2003CB114202) and in part by the National Nature Science Foundation of China (no. 30530540 and 30470069).
Footnotes
Published ahead of print on 20 September 2006.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, C. A. Balinsky, B. A. Moser, J. J. Becnel, D. L. Rock, and G. F. Kutish. 2001. Genome sequence of a baculovirus pathogenic for Culex nigripalpus. J. Virol. 75:11157-11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 3.Bassemir, U., H. G. Miltenburger, and P. David. 1983. Morphogenesis of nuclear polyhedrosis virus from Autographa californica in a cell line from Mamestra brassicae (cabbage moth). Further aspects on baculovirus assembly. Cell Tissue Res. 228:587-595. [DOI] [PubMed] [Google Scholar]
- 4.Bideshi, D. K., and B. A. Federici. 2000. The Trichoplusia ni granulovirus helicase is unable to support replication of Autographa californica multicapsid nucleopolyhedrovirus in cells and larvae of T. ni. J. Gen. Virol. 81:1593-1599. [DOI] [PubMed] [Google Scholar]
- 5.Blissard, G. W., B. Black, N. Crook, B. A. Keddie, R. Possee, G. F. Rohrmann, D. A. Theilmann, and L. Volkman. 2000. Family Baculoviridae, p. 195-202. In M. H. V. van Regenmortel, C. M. Fauquet, and D. H. L. Bishop (ed.), Virus taxonomy. Academic Press, San Diego, Calif.
- 6.Braunagel, S. C., W. K. Russell, G. Rosas-Acosta, D. H. Russell, and M. D. Summers. 2003. Determination of the protein composition of the occlusion-derived virus of Autographa californica nucleopolyhedrovirus. Proc. Natl. Acad. Sci. USA 100:9797-9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braunagel, S. C., and M. D. Summers. 1994. Autographa californica nuclear polyhedrosis virus, PDV, and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology 202:315-328. [DOI] [PubMed] [Google Scholar]
- 8.Bud, H. M., and D. C. Kelly. 1980. An electron microscope study of partially lysed baculovirus nucleocapsids: the intranucleocapsid packaging of viral DNA. J. Ultrastruct. Res. 73:361-368. [DOI] [PubMed] [Google Scholar]
- 9.Dai, X., T. M. Stewart, J. A. Pathakamuri, Q. Li, and D. A. Theilmann. 2004. Autographa californica multiple nucleopolyhedrovirus exon0 (orf141), which encodes a RING finger protein, is required for efficient production of budded virus. J. Virol. 78:9633-9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Federici, B. A. 1997. Baculovirus pathogenesis, p. 33-56. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 11.Fraser, M. J. 1986. Ultrastructural observations of virion maturation in Autographa californica nuclear polyhedrosis virus infected Spodoptera frugiperda cell cultures. J. Ultrastruct. Mol. Struct. Res. 95:189-195. [Google Scholar]
- 12.Garcia-Maruniak, A., J. E. Maruniak, P. M. A. Zanotto, A. E. Doumbouya, J. C. Liu, T. A. Merritt, and J. S. Lanoie. 2004. Sequence analysis of the genome of the Neodiprion sertifer nucleopolyhedrovirus. J. Virol. 78:7036-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guarino, L. A., W. Dong, and J. Jin. 2002. In vitro activity of the baculovirus late expression factor LEF-5. J. Virol. 76:12663-12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harwood, S. H., L. Li, P. S. Ho, A. K. Preston, and G. F. Rohrmann. 1998. AcMNPV late expression factor-5 interacts with itself and contains a zinc ribbon domain that is required for maximal late transcription activity and is homologous to elongation factor TFIIS. Virology 250:118-134. [DOI] [PubMed] [Google Scholar]
- 15.Hefferon, K. L. 2003. ORF98 of Autographa californica nucleopolyhedrosis virus is an auxiliary factor in late gene expression. Can. J. Microbiol. 49:157-163. [DOI] [PubMed] [Google Scholar]
- 16.Herniou, E. A., J. A. Olszewski, J. S. Cory, and D. R. O'Reilly. 2003. The genome sequence and evolution of baculoviruses. Annu. Rev. Entomol. 48:211-234. [DOI] [PubMed] [Google Scholar]
- 17.Jakubowska, A. K., S. A. Peters, J. Ziemnicka, J. M. Vlak, and M. M. van Oers. 2006. Genome sequence of an enhancin gene-rich nucleopolyhedrovirus (NPV) from Agrotis segetum: collinearity with Spodoptera exigua multiple NPV. J. Gen. Virol. 87:537-551. [DOI] [PubMed] [Google Scholar]
- 18.Lalioti, M., and J. Heath. 2001. A new method for generating point mutations in bacterial artificial chromosomes by homologous recombination in Escherichia coli. Nucleic Acids Res. 29:E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laufs, S., A. Lu, K. Arrell, and E. B. Carstens. 1997. Autographa californica nuclear polyhedrosis virus p143 gene product is a DNA-binding protein. Virology 228:98-106. [DOI] [PubMed] [Google Scholar]
- 20.Lauzon, H. A. M., C. J. Lucarotti, P. J. Krell, Q. L. Feng, A. Retnakaran, and B. M. Arif. 2004. Sequence and organization of the Neodiprion lecontei nucleopolyhedrovirus genome. J. Virol. 78:7023-7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Y., J. Wang, R. Deng, Q. Zhang, K. Yang, and X. Wang. 2005. vlf-1 deletion brought AcMNPV to defect in nucleocapsid formation. Virus Genes 31:275-284. [DOI] [PubMed] [Google Scholar]
- 22.Lin, G., and G. W. Blissard. 2002. Analysis of an Autographa californica nucleopolyhedrovirus lef-11 knockout: LEF-11 is essential for viral DNA replication. J. Virol. 76:2770-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, A., and E. B. Carstens. 1991. Nucleotide sequence of a gene essential for viral DNA replication in the baculovirus Autographa californica nuclear polyhedrosis virus. Virology 181:336-347. [DOI] [PubMed] [Google Scholar]
- 24.Lu, A., and E. B. Carstens. 1992. Transcription analysis of the EcoRI D region of the baculovirus Autographa californica nuclear polyhedrosis virus identifies an early 4-kilobase RNA encoding the essential p143 gene. J. Virol. 66:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDougal, V. V., and L. A. Guarino. 2000. The Autographa californica nuclear polyhedrosis virus p143 gene encodes a DNA helicase. J. Virol. 74:5273-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muyrers, J. P., Y. Zhang, G. Testa, and A. F. Stewart. 1999. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 27:1555-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Reilly, D. R., L. K. Miller, and V. A. Luckow. 1992. Baculovirus expression vector: a laboratory manual. W. H. Freeman and Co., New York, N.Y.
- 29.Pijlman, G. P., J. C. Dortmans, A. M. Vermeesch, K. Yang, D. E. Martens, R. W. Goldbach, and J. M. Vlak. 2002. Pivotal role of the non-hr origin of DNA replication in the genesis of defective interfering baculoviruses. J. Virol. 76:5605-5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prikhod'ko, E. A., and L. K. Miller. 1998. Role of baculovirus IE2 and its RING finger in cell cycle arrest. J. Virol. 72:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanarsdall, A. L., K. Okano, and G. F. Rohrmann. 2004. Characterization of a baculovirus with a deletion of vlf-1. Virology 326:191-201. [DOI] [PubMed] [Google Scholar]
- 32.Vanarsdall, A. L., K. Okano, and G. F. Rohrmann. 2006. Characterization of the role of very late expression factor 1 in baculovirus capsid structure and DNA processing. J. Virol. 80:1724-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaughn, J. L., R. H. Goodwin, G. J. Tompkins, and P. McCawley. 1977. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae). In Vitro 13:213-217. [DOI] [PubMed] [Google Scholar]
- 34.Volkman, L. E. 1988. Autographa californica MNPV nucleocapsid assembly: inhibition by cytochalasin D. Virology 163:547-553. [DOI] [PubMed] [Google Scholar]
- 35.Willians, G. V., and P. Faulkner. 1997. Cytological changes and viral morphogenesis during baculovirus infection, p. 61-107. In L. K. Miller (ed.), The baculoviruses. Plenum Press, New York, N.Y.
- 36.Yang, S., and L. K. Miller. 1998. Expression and mutational analysis of the baculovirus very late factor 1 (vlf-1) gene. Virology 245:99-109. [DOI] [PubMed] [Google Scholar]
- 37.Young, J. C., E. A. MacKinnon, and P. Faulkner. 1993. The architecture of the virogenic stroma in isolated nuclei of Spodoptera frugiperda cells in vitro infected by Autographa californica nuclear polyhedrosis virus. J. Struct. Biol. 110:141-153. [Google Scholar]