Abstract
The mechanisms involved in the construction of the icosahedral capsid of the African swine fever virus (ASFV) particle are not well understood at present. Capsid formation requires protein p72, the major capsid component, but other viral proteins are likely to play also a role in this process. We have examined the function of the ASFV structural protein pB438L, encoded by gene B438L, in virus morphogenesis. We show that protein pB438L associates with membranes during the infection, behaving as an integral membrane protein. Using a recombinant ASFV that inducibly expresses protein pB438L, we have determined that this structural protein is essential for the formation of infectious virus particles. In the absence of the protein, the virus assembly sites contain, instead of icosahedral particles, large aberrant tubular structures of viral origin as well as bilobulate forms that present morphological similarities with the tubules. The filamentous particles, which possess an aberrant core shell domain and an inner envelope, are covered by a capsid-like layer that, although containing the major capsid protein p72, does not acquire icosahedral morphology. This capsid, however, is to some extent functional, as the filamentous particles can move from the virus assembly sites to the plasma membrane and exit the cell by budding. The finding that, in the absence of protein pB438L, the viral particles formed have a tubular structure in which the icosahedral symmetry is lost supports a role for this protein in the construction or stabilization of the icosahedral vertices of the virus particle.
African swine fever virus (ASFV), the causative agent of a severe hemorrhagic disease of domestic pigs, is a complex enveloped deoxyvirus with icosahedral morphology (40). The ASFV genome, a double-stranded DNA molecule of 170 to 190 kbp with covalently closed ends and terminal inverted repeats, contains more than 150 open reading frames (49). The virus particle, with about 50 structural proteins (15, 21), consists of a central DNA-containing nucleoid surrounded by a protein layer designated the core shell that contains the proteolytic products of the virus polyproteins pp220 and pp62 (3, 8, 14). This layer is enwrapped by an inner envelope, which is thought to derive from a collapsed endoplasmic reticulum (ER) cisterna (6, 37), and by the icosahedral capsid constituted by protein p72, the major capsid component, and by protein pE120R, required for the microtubule-mediated virus transport from the cytoplasmic factory to the plasma membrane (7, 16, 24). The extracellular virus is enveloped by an additional membrane, which is acquired by budding through the plasma membrane (13).
The complex ASFV morphogenetic process is initiated by the recruitment to the virus assembly sites of ER cisternae, which are modified by the insertion of viral proteins, such as protein p54 (35), followed by the collapse of the modified cisternae to form the virus inner envelope. The outer capsid layer is then progressively assembled on one side of this envelope, while, underneath the other side, the core shell domain is simultaneously formed. Subsequently, the viral DNA together with the nucleoproteins is packaged and condensed to form the central nucleoid, thus completing the assembly of the infectious intracellular viral particle (8).
Certain steps in the construction of the viral particle can occur independently of other morphogenetic events. Thus, the assembly of the viral capsid on one side of the inner envelope, although probably requiring a previous modification of the ER membranes, is independent of the events that lead to the formation of the inner domains of the viral particle. This was shown in experiments using a recombinant virus where repression of polyprotein pp220 led to the generation of icosahedral empty capsids (5). In contrast, the assembly of the core shell and the processing of the two viral polyproteins require the simultaneous construction of the capsid (3). As mentioned above, two proteins, p72 and pE120R, have been identified so far as components of the icosahedral viral capsid. However, the capsid can be correctly assembled in the absence of pE120R protein, leading to the generation of infectious virus (7). It is therefore likely that in addition to protein p72, which is the major capsid component and probably forms the hexagonal capsomers (8, 24), other viral proteins might play a role in the construction of the capsid. These may be additional components of the capsid, such as those composing the pentameric structures of the vertices of the icosahedron, as has been described for other icosahedral viruses (28, 31, 39, 48), or nonstructural viral proteins, such as protein pB602L, which has been described as a molecular chaperone involved in the correct folding of protein p72 (17).
Although the function of several structural ASFV proteins has been established, the localization in the particle and the role in the morphogenetic process of many other structural components are unknown. One of these structural proteins of unknown function is protein pB438L, encoded by gene B438L, which is expressed at late times postinfection (23). Protein pB438L is localized at the viral factory, but its precise localization in the virus particle has not been determined. In order to investigate the function of protein pB438L in ASFV morphogenesis, we have examined the consequences of repressing gene B438L for the virus morphogenetic process.
In this report, we show that protein pB438L is associated with membranes during the infection, behaving as an integral membrane protein. Using a recombinant ASFV that inducibly expresses protein pB438L, we have found that repression of the protein leads to the generation of aberrant tubular structures containing a capsid that, although composed of proteins p72 and pE120R, does not posses icosahedral morphology.
MATERIALS AND METHODS
Cells and viruses.
Vero cells were obtained from the American Type Culture Collection and grown in Dulbecco's modified Eagle's medium containing 10% fetal calf serum (FCS). The ASFV strain BA71V adapted to grow in Vero cells and vGUSREP, a BA71V-derived recombinant virus that expresses the Escherichia coli lac repressor, have been described previously (20, 24).
Antibodies.
The monospecific rabbit polyclonal sera against the structural proteins pp220/p150, p37, pp62/p35, pE120R, and p72, as well as the mouse monoclonal antibodies 17LD3 against protein p72 and 17KG12 against protein p17, have been described previously (22, 29, 43, 45, 46). To prepare an antibody against protein pB438L, plasmid pRSETA-B438L (23) was expressed in E. coli BL21(DE3)/pLys-S for 3 h at 37aC. The bacterial pellet was resuspended in 1% Triton X-100 in phosphate-buffered saline (PBS), sonicated, and incubated for 15 min at 4°C with shaking. Then DNase I (20 μg/ml) was added and incubation was continued for 30 min at room temperature. After centrifugation for 15 min at 14,000 × g, the sediment containing the insoluble pB438L protein was resuspended in PBS and used to prepare an antibody in rabbits.
Construction of plasmid pIND3.B438L.
For the construction of plasmid pIND3.B438L, the intermediate transfer vector pIND3, designed to allow the inducible expression of a target gene after homologous recombination with vGUSREP (24), was used. pIND3 is essentially identical to pIND1 (7), except that in the viral inducible promoter p72.I* the distance between the E. coli lac operator and the ASFV late promoter p72.4 is 2 bp, for a stronger repression of gene expression (24). Plasmid pIND3 also contains the lacZ gene under the control of the strong late promoter p72 and two multiple cloning sites for the cloning of the target gene and the corresponding upstream and downstream flanking sequences.
To generate the intermediate plasmid, a DNA fragment of 738 bp, which contains the nucleotide sequence from position −12 to +722 relative to the translation initiation codon of the B438L gene, was obtained by PCR using the EcoRI B fragment as a template and the oligonucleotides 5′-CTTTTACTGCAGATGTATCATGATTTATGC and 5′-GATTAAGCTTAGGAATACCTCTACCTGC, which contain PstI and HindIII restriction sites (underlined), respectively. Plasmid pIND3.B438L.FI was generated by inserting the PstI- and HindIII-digested PCR fragment into the PstI- and HindIII-linearized plasmid pIND3. A fragment of 734 bp, which contains the nucleotide sequence from the adjacent B169L open reading frame, was obtained by PCR using the EcoRI B fragment as a template and the oligonucleotides 5′-GTATAGGTACCTTAATTATTATATTGAGAAGGC and 5′-AAAGAAGCTGTGCGGCCGCCCAAAGAAAATATC, which contain KpnI and NotI restriction sites (underlined), respectively. The PCR fragment was inserted into the KpnI and NotI sites of the plasmid pIND3.B438L.FI to obtain the final transfer vector pIND3.B438L.
Generation of recombinant virus vB438Li.
Recombinant virus was generated essentially as previously described (34) with minor modifications. Briefly, Vero cells were transfected with linearized plasmid pIND3.B438L and infected with virus vGUSREP (24) in the presence of different concentrations of isopropyl-β-d-thiogalactopyranoside (IPTG). At 72 h postinfection (hpi), the cells were harvested and the recombinant virus vB438Li was isolated by sequential rounds of plaque purification in the presence of IPTG. The growth and purification of the recombinant virus were carried out with 250 μM IPTG, a concentration that was found to be optimal for the production of the recombinant virus.
Plaque assay.
Vero cell monolayers, in six-well plates, were infected with recombinant vB438Li or parental BA71V. After 2 h, the inoculum was removed and the cells were overlaid with Dulbecco's modified Eagle's medium containing 0.55% Noble agar and 3% FCS in the presence or absence of 250 μM IPTG. Five days later, the medium was removed and the monolayers were stained with 1% crystal violet.
One-step virus growth curves.
Vero cell monolayers, in 24-well plates, were infected with 5 PFU of recombinant vB438Li or parental BA71V per cell. After 2 h of adsorption, the cells were incubated in medium supplemented with 2% FCS. IPTG (250 μM) was added, when indicated, immediately after the adsorption period. Infected cells with their culture supernatants were harvested at different times postinfection, and titers were determined by plaque assay in the presence of 250 μM.
Subcellular fractionation.
Subcellular fractionation was performed as previously described (4). Briefly, Vero cells were mock infected or infected with 10 PFU/cell of the BA71V strain. At 20 hpi, the cells were resuspended at 107 cells/ml in homogenization buffer (20 mM Tris-HCl, pH 7.5, 0.25 M sucrose, 1 mM EDTA, 25 mM N-ethylmaleimide) supplemented with protease inhibitors (Complete EDTA-free cocktail; Roche) and passaged through a 25-gauge syringe 20 times. The homogenate was centrifuged at 750 × g for 5 min to sediment nuclei and unbroken cells, and the postnuclear supernatant was subsequently centrifuged at 100,000 × g for 15 min to separate the cytosolic and membrane/particulate fractions. Membrane/particulate pellets were gently rinsed in homogenization buffer prior to resuspension in the appropriate buffer.
Triton X-114 and sodium carbonate extraction of membranes.
Membrane/particulate cell fractions were treated with 2% Triton X-114 or sodium carbonate buffer (0.1 M sodium carbonate, pH 11.3) as previously described (4). Equivalent amounts of the resulting fractions were analyzed by Western immunoblotting.
Construction of plasmid pcDNA.B438L.
To generate the pcDNA.B438L plasmid, the complete coding sequence of the gene B438L was excised from pRSET.B438L using EcoRI and BamHI restriction enzymes and cloned into EcoRI- and BamHI-digested pcDNA 3.1/myc His (Invitrogen).
In vitro transcription and translation.
In vitro transcription reactions with reaction mixtures containing 1 μg of linearized pcDNA.B438L template DNA were carried out as described in the mMessage mMachine T7 kit (Ambion). Translation of the mRNAs was performed in the presence of [35S]methionine-cysteine (Redivue Promix; Amersham Pharmacia Biotech) for 120 min at 30°C using the Flexi Rabbit Reticulocyte Lysate System (Promega) in the presence or absence of canine pancreatic microsomal membranes (Promega). As a control, β-lactamase mRNA was translated in parallel.
For membrane sedimentation analysis, an aliquot of the translation mixture was diluted with physiological salt buffer, adjusted to 3 mM tetracaine, and incubated for 5 min on ice. Samples were layered on a 0.5 M sucrose cushion and centrifuged at 100,000 × g for 15 min at room temperature. Membrane sediments were gently rinsed in physiological salt buffer before dissociation. Analysis of equivalent amounts of supernatant and pellet fractions was performed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by autoradiography.
Western blotting.
Preconfluent Vero cell monolayers were either mock infected or infected with BA71V or recombinant vB438Li virus in the presence or absence of 250 μM IPTG at a multiplicity of infection of 10 PFU per cell. The cells were lysed at 12 hpi in Laemmli sample buffer, and equivalent amounts were electrophoresed in SDS-polyacrylamide gels and transferred to nitrocellulose as described previously (8). Protein detection was carried out with peroxidase-conjugated antibodies and the ECL system (Amersham Pharmacia Biotech) according to the manufacturer's indications.
Indirect immunofluorescence.
For the immunofluorescence assay of cells infected with BA71V or recombinant vB438Li virus, preconfluent Vero cells grown on coverslips were infected at 1 PFU per cell in the presence or absence of 250 μM IPTG. At 14 hpi, infected cells were fixed with 3% paraformaldehyde at room temperature for 15 min. Fixed cells were incubated with 1% Triton X-100 in PBS at room temperature. After 15 min, coverslips were blocked for 30 min with blocking buffer (2% FCS, 1% bovine serum albumin, 40 mM glycine in PBS). The cells were then sequentially incubated for 30 min with the primary and the corresponding secondary antibodies diluted with blocking buffer. Finally, the coverslips were mounted on glass slides with Mowiol/Dabco and 4′,6′-diamidino-2-phenylindole (DAPI) to stain DNA in nuclei and virus factories. Preparations were examined with a Bio-Rad Radiance 2000 confocal laser scanning microscope. Images were processed using Adobe Photoshop software.
Electron microscopy.
For conventional Epon section analysis, Vero cells were infected with 10 PFU per cell and fixed at 18 hpi with 2% glutaraldehyde in PBS for 1 h at room temperature. Postfixation was carried out with 1% OsO4 and 1.5% K3Fe(CN)6 in H2O at 4°C for 30 min. Samples were dehydrated with acetone and embedded in Epon according to standard procedures. For immunoelectron microscopy, the cells were fixed at the indicated times with 8% paraformaldehyde and 0.2% glutaraldehyde in 120 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 50 mM HEPES, 4 mM MgCl2, 20 mM EGTA, pH 6.9, for 2 h at room temperature. The fixative was then removed, and a solution of 8% paraformaldehyde in 60 mM PIPES, 25 mM HEPES, 2 mM MgCl2, 10 mM EGTA, pH 6.9, was added. Finally, the fixed cells were prepared for cryosectioning and immunolabeling as previously described (25).
RESULTS
Protein pB438L is a membrane-associated polypeptide that forms disulfide-linked oligomers.
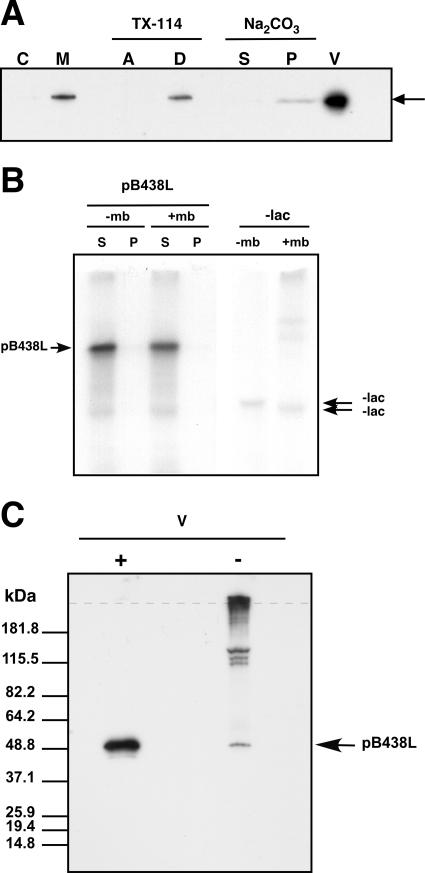
As mentioned in the introduction, protein pB438L, a structural component of the ASFV particle, is localized at the virus assembly sites. To determine more precisely its localization within the viral factory, ASFV-infected cells were collected at 20 hpi and subjected to subcellular fractionation as indicated under Materials and Methods, and the presence of protein pB438L in the cytosolic and membrane fractions was analyzed by Western blotting using a specific antibody against the protein. As can be seen in Fig. 1A, protein pB438L is detected only in the membrane fraction. To examine the type of association of protein pB438L with membranes, the membrane fraction was incubated with 100 mM sodium carbonate, pH 11.3, a treatment that extracts peripherally associated proteins from membranes. As shown in Fig. 1A, pB438L remained associated with the membrane sediment after such treatment. When the membrane fraction was treated with Triton X-114 and subjected to a temperature-induced phase separation, protein pB438L partitioned entirely into the detergent phase (Fig. 1A). The results obtained with the sodium carbonate and Triton X-114 treatments indicate that protein pB438L behaves as an integral membrane protein. This finding was rather unexpected, since the protein lacks any putative transmembrane region (data not shown). Therefore, to investigate the intrinsic capacity of protein pB438L to associate with membranes, the protein was translated in vitro in the presence or in the absence of rough microsomal membranes and its presence in the membrane and soluble fractions, separated by centrifugation on a sucrose cushion, was determined after SDS-PAGE and autoradiography. As shown in Fig. 1B, the pB438L protein was exclusively detected in the soluble fraction under both conditions, indicating that the protein does not possess the capacity to associate in vitro with membranes. This finding suggests that the interaction of pB438L protein with membranes during the infection may be mediated by a cellular or viral factor not present in the membrane samples used in the in vitro assay or may be due to a posttranslational modification of the protein that does not occur under in vitro conditions.
FIG. 1.
Membrane association and disulfide bond formation of protein pB438L. (A) Subcellular distribution of protein pB438L in ASFV-infected cells. Infected Vero cells were disrupted at 20 hpi and fractionated into cytosolic (C) and membrane/particulate (M) fractions, as described in Materials and Methods. The membrane fraction was treated with sodium carbonate, and after centrifugation, equivalent amounts of the supernatant (S) and sediment (P), along with 2 μg of purified ASFV particles (V), were analyzed by immunoblotting using an anti-pB438L antibody. Alternatively, the membrane fraction was extracted with Triton X-114 and subjected to temperature-induced phase separation. After centrifugation, the aqueous (A) and detergent-rich (D) phases were analyzed as before. The band corresponding to protein pB438L is indicated by an arrow. (B) Analysis of membrane association of in vitro-synthesized protein pB438L. Protein pB438L was translated in vitro in the presence of [35S]methionine-cysteine using a rabbit reticulocyte system in the presence (+mb) or absence (−mb) of microsomal membranes. Soluble (S) and membrane-associated (P) fractions were separated by centrifugation and analyzed by SDS-PAGE followed by autoradiography. As a control, the processing of in vitro-translated β-lactamase (-lac) in the presence or absence of membranes was analyzed in parallel. Proteins pB438L and β-lactamase are indicated by arrows. (C) Analysis of disulfide bond formation of protein pB438L in ASFV particles. Twenty micrograms of purified ASFV particles (V) was resolved by reducing (+) and nonreducing (−) SDS-PAGE and analyzed by immunoblotting using an anti-pB438L antibody. The arrow indicates the position of the monomeric pB438L protein and the dotted line the origin of the gel. The position of molecular weight markers is indicated on the left.
Since certain ASFV membrane proteins, such as the structural components p54 and p12, form disulfide-bonded homodimers (9, 35), we examined the possibility that protein pB438L might also form oligomers in the virus particle. For this, highly purified virus samples, treated or not with dithiothreitol, were analyzed by Western blotting using the anti-pB438L antibody. As can be seen in Fig. 1C, a single band of 49 kDa corresponding to the monomeric pB438L protein was detected in the presence of dithiothreitol, while, in its absence, the intensity of the 49-kDa band was considerably decreased and three additional bands with molecular masses of 115 to 150 kDa were detected. In addition, a material of higher molecular weight was retained at the origin of the gel. These results indicate that protein pB438L is able to form disulfide-linked homo- or heterooligomers in the virus particle.
Inducible expression of protein pB438L by recombinant virus vB438Li.
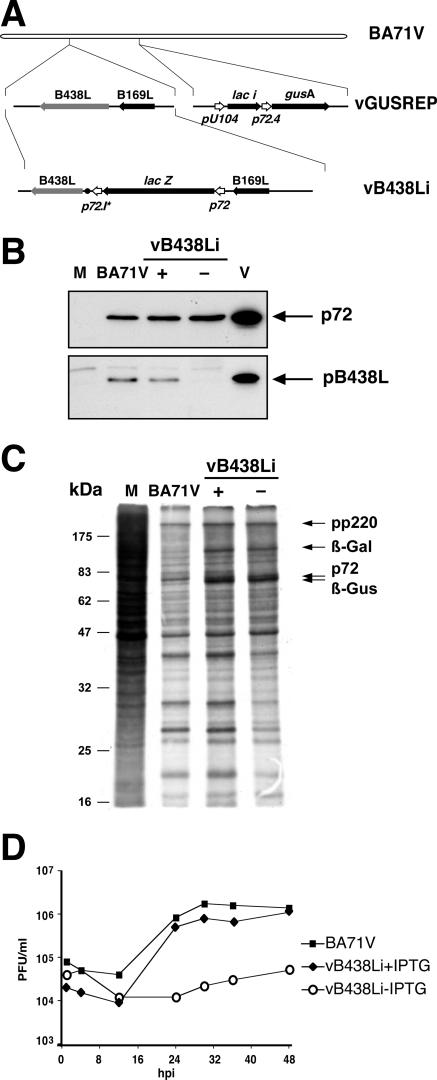
To investigate the possible role of protein pB438L in the virus morphogenetic process, we generated a recombinant ASFV, vB438Li, in which the expression of gene B438L is under the control of the E. coli lac operator/repressor system (Fig. 2A). To this end, the BA71V-derived vGUSREP recombinant virus, constitutively expressing the E. coli lac repressor, was modified by replacing the original promoter of gene B438L with the inducible promoter p72.I*, composed of the late viral promoter p72.4 and the operator sequence O1 (24).
FIG. 2.
(A) Genomic structure of the ASFV recombinant virus vB438Li. The recombinant virus vB438Li was obtained from vGUSREP, a BA71V-derived recombinant virus, which contains the lac repressor-encoding gene lacI inserted into the nonessential thymidine kinase locus (24). In the vB438Li virus, the promoter of gene B438L was replaced by an inducible promoter, p72.I*, which is composed of a strong late promoter (p72.4) and the operator sequence O1 from the E. coli lac operon. (B) Inducible expression of protein pB438L. Vero cells were either mock infected (M) or infected with parental BA71V or recombinant vB438Li virus in the presence (+) or absence (−) of 250 μM IPTG. At 12 hpi, the cells were lysed and analyzed, along with 2 μg of purified ASFV particles (V), by immunoblotting with antibodies against proteins pB438L and p72 (loading control). The position of the detected proteins is indicated. (C) Protein synthesis in cells infected with vB438Li virus. Cells mock infected or infected as in panel B were pulse-labeled with 0.5 mCi/ml of [35S]methionine-cysteine from 18 to 20 hpi, lysed, and analyzed by SDS-PAGE. The positions of polyprotein pp220, the major capsid protein p72, and the reporter β-galactosidase (β-Gal) and β-glucuronidase (β-Gus) proteins are indicated. The electrophoretic mobilities of molecular mass markers are indicated at the left. (D) One-step growth curves of vB438Li virus. Vero cells were infected with 5 PFU of vB438Li virus per cell in the presence or absence of 250 μM IPTG. At the indicated times of infection, the total virus titer of each sample was determined by plaque assay on Vero cells in the presence of the inducer. Parental BA71V infections were also titrated as a control.
The IPTG dependence of the recombinant vB438Li virus was tested in a plaque assay performed in the presence and in the absence of 250 μM IPTG. In the presence of the inducer, the number of plaques was similar for both parental and recombinant viruses, while in its absence the plaque number of the recombinant was drastically reduced (data not shown).
To examine whether the reduction in plaque number of vB438Li in the absence of the inducer correlates with a decreased expression of protein pB438L, BA71V- or vB438Li-infected cells were analyzed by Western blotting using the anti-pB438L antibody. As shown in Fig. 2B, protein pB438L was barely detectable in infections with the recombinant virus under restrictive conditions, while in the presence of IPTG the expression of the protein was comparable to that with the parental virus. This finding shows that the expression of pB438L can be strongly repressed in the inducible virus. Repression of protein pB438L, however, did not significantly affect the overall viral gene expression, as can be seen in the pulse-labeling experiment presented in Fig. 2C.
In a further approach, we studied the effect of pB438L repression on virus multiplication. Figure 2D shows the one-step growth curves obtained under the different conditions. As can be seen, the virus titers of recombinant virus vB438Li under permissive conditions were essentially the same as those found with the parental BA71V. Under restrictive conditions, however, the vB438Li titers were significantly reduced, with a maximal difference of ca. 2 log units at 24 hpi. We also examined whether the release of B438L repression at different times postinfection led to normal virus growth. We found that the recovery of virus production was dependent on the time of IPTG addition. Thus, when the induction was performed at 12 hpi virus growth was fully recovered, but when IPTG was added at later times (18 and 24 hpi) the maximal titers were strongly reduced in comparison with those obtained under permissive conditions. The recovery observed at 12 hpi is probably due to de novo virus formation, since the assembling icosahedral particles seen by electron microscopy are apparently not derived from the tubular structures (not shown). Taken together, the results obtained indicate that recombinant virus vB438Li is an IPTG-dependent lethal-conditional mutant.
Polyprotein processing is impaired in infections with recombinant vB438Li under restrictive conditions.
It has been previously shown that the proteolytic processing of the ASFV polyproteins pp220 and pp62, an essential process for the maturation of the viral core (2, 3, 5), is inhibited under conditions that lead to alterations in the virus assembly process (3, 5, 35). Since pB438L protein is a structural component of the virus particle, it was possible that the defect in virus growth under restrictive conditions might be due to alterations in the morphogenetic process. We therefore determined the processing of the two viral polyproteins in cells infected with the recombinant vB438Li in the absence of the inducer. As can be seen in Fig. 3, the processing of polyprotein pp220 to its mature products p150 and p37, as well as that of pp62 to its product p35, is inhibited under restrictive conditions, suggesting that the virus assembly pathway may be altered in the absence of pB438L protein expression.
FIG. 3.
Polyprotein processing requires the expression of protein pB438L. Polyprotein processing was analyzed by immunoblotting with antibodies against polyprotein pp220 and its mature products p150 and p37 and polyprotein pp62 and its mature product p35 in the extracts used in the experiment for Fig. 2B. M, mock-infected cells; BA71V, cells infected with parental BA71V; + and −, cells infected with vB438Li virus in the presence and absence, respectively, of 250 μM IPTG. A sample of 2 μg of highly purified ASFV (V) was also analyzed. The position of the detected proteins is indicated.
Immunofluorescence microscopy of cells infected with recombinant vB438Li virus.
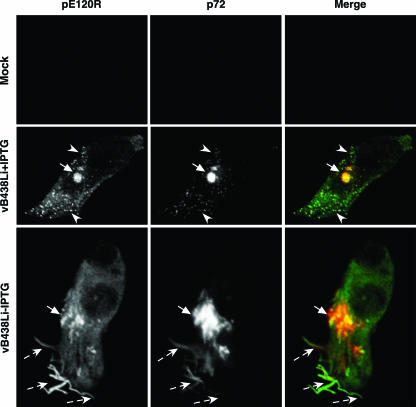
In a further approach to ascertain whether the ASFV assembly route is altered in the absence of pB438L, we examined by immunofluorescence microscopy the localization of several structural proteins, such as the capsid proteins p72 and pE120R and polyprotein pp62, in cells infected with the recombinant virus in the presence or in the absence of IPTG, using antibodies specific for these proteins. Figure 4 shows that in cells infected with the recombinant vB438Li virus under permissive conditions, the anti-p72 and anti-pE120R antibodies strongly labeled the viral factories, as well as virus particles scattered throughout the cytoplasm. Strikingly, in the absence of the inducer, the antibodies against these capsid proteins detected long filamentous forms, while the punctate labeling characteristic of the virus particles was not observed. The filiform structures were found at the viral factory, as well as proximal to the plasma membrane and even outside the cell. This suggests that these structures can be translocated from the assembly site to the cell membrane. Similar results were obtained in the case of polyprotein pp62 (not shown). These observations suggest that in the absence of pB438L protein the viral morphogenesis is severely altered with a blockage in the formation of normal viral particles.
FIG. 4.
Immunofluorescence microscopy analysis of vB438Li-infected cells. Vero cells mock infected or infected with recombinant vB438Li virus in the presence or absence of 250 μM IPTG were fixed at 14 hpi and double-labeled with rabbit anti-pE120R antibody and mouse anti-p72 monoclonal antibody. Labeling was revealed with Alexa 488-goat anti-rabbit immunoglobulin G and with Alexa 594-goat anti-mouse immunoglobulin G, respectively. Viral factories and virions are indicated by arrows and arrowheads, respectively. The filiform structures are indicated by dashed arrows.
Electron microscopy of vB438Li-infected cells.
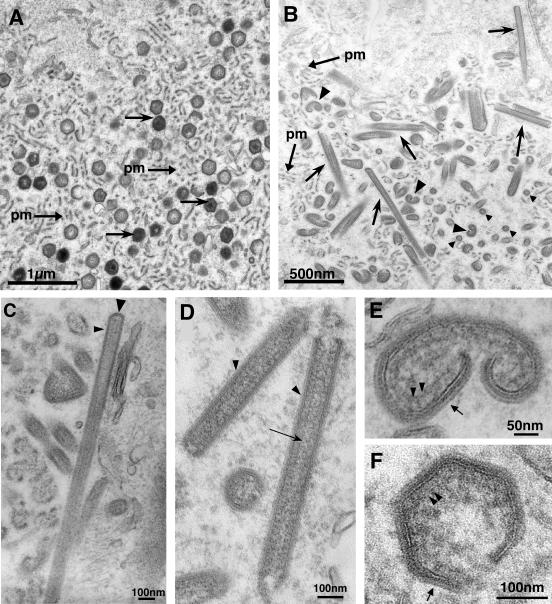
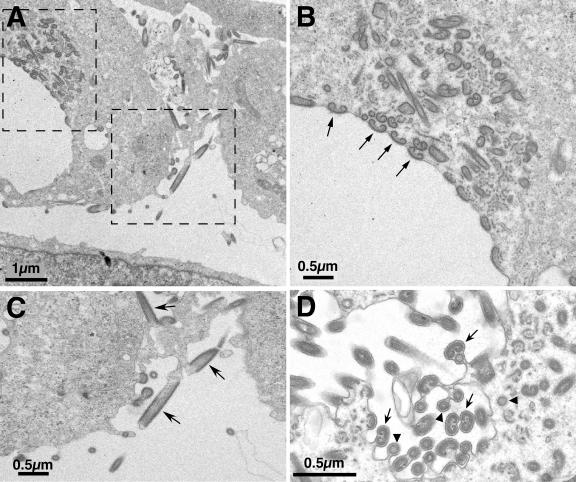
To establish more precisely the step at which virus morphogenesis is blocked in the absence of protein pB438L, cells infected with vB438Li virus in the presence or in the absence of IPTG were examined by electron microscopy. The viral factories in cells infected with the recombinant virus under permissive conditions were similar to those found in infections with the parental virus, with abundant precursor viral membranes as well as immature and mature viruses (Fig. 5A). In the absence of IPTG, the assembly sites contained the precursor membranes but not mature icosahedral particles. Instead, numerous tubular structures with a length of 1 μm or more were found (Fig. 5B). These large forms probably correspond to the filamentous structures detected by immunofluorescence microscopy. At higher magnification (Fig. 5C and D), it can be seen that these filiform particles, with a diameter of about 80 nm, contain a well-defined and electron-dense outer layer, as well as an inner layer reminiscent of the core shell of the normal particle and an apparent central channel with a diameter of about 25 nm. Frequently, the outer layer closes at least one of the ends of the tubules (Fig. 5C).
FIG. 5.
Electron microscopy of vB438Li-infected cells. Shown are ultrathin Epon sections of vB438Li-infected Vero cells incubated for 18 h in the presence (A) or in the absence (B, C, D, and E) of IPTG. In the presence of the inducer (A), the assembly sites contain large amounts of precursor viral membranes (pm), as well as immature and mature virions (arrows). Under restrictive conditions (B), the defective factories contain, in addition to precursor membranes, large numbers of aberrant tubular (arrows) and bilobulate (arrowheads) structures. Panels C and D show higher magnifications of the tubular structures. The outer electron-dense layer and the apparent central channel of these structures are indicated by arrowheads and an arrow, respectively. The closed end of the tubule in panel C is indicated by a large arrowhead. Panel E shows a higher magnification of a bilobulate structure. The outer and inner domains of these structures are indicated by an arrow and arrowheads, respectively. These layers resemble the capsid and the inner envelope (arrow) and the core shell domain (arrowheads) of a normal assembling particle (F).
In addition to the filamentous forms, the viral factories contain, under restrictive conditions, aberrant forms with a bilobulate structure (Fig. 5B). As shown in more detail in Fig. 5E, these bilobulate forms possess an outer layer that resembles the capsid and inner envelope of the normal particle, as well as an internal material similar to the core shell which is being constructed in a developing normal particle (Fig. 5F).
These different abnormal structures, like the parental virus, are released from the cell by budding through the plasma membrane (Fig. 6A to C) or are translocated, also by budding, into the lumen of intracellular vesicles (Fig. 6D). The circular structures clearly seen in this latter panel, where the central channel is very evident, most likely correspond to cross sections of the tubular forms. Also noteworthy is the arrangement of the bilobulate structures in close proximity to the plasma membrane, offering a larger surface of contact with the membrane in what probably represents a step immediately prior to the budding event itself (Fig. 6B). These observations suggest that the aberrant structures generated under restrictive conditions contain the viral proteins involved in the exit of the normal mature particle from the cell.
FIG. 6.
Budding of aberrant structures. (A) Cell infected with vB438Li virus in the absence of IPTG. (B and C) Higher magnifications of the regions delimited by the dashed lines in panel A. Arrows in panel B indicate bilobulate structures in apparent contact with the plasma membrane. (C) Tubular structures being released by budding (arrows). (D) Images of intracellular budding of the aberrant structures. Arrowheads point to circular structures, corresponding to cross sections of the tubules, while arrows indicate bilobulate structures.
Protein composition of the filamentous particles determined by immunoelectron microscopy.
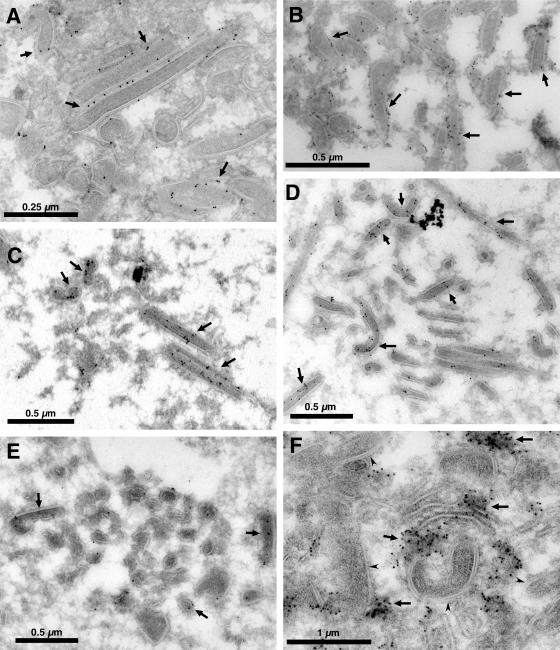
As mentioned above, the domains of the aberrant filamentous particles are reminiscent of several domains of the mature viral particle, such as the capsid, the inner envelope, and the core shell. To determine whether their protein composition is also similar to that of the mature virion, we analyzed by immunoelectron microscopy the presence in the aberrant tubules of several viral proteins that are markers of the domains of the normal particle. As can be seen in Fig. 7, the antibody against the major capsid protein p72 strongly labeled the outermost layer of the tubules (Fig. 7A), which also contain protein pE120R, another component of the normal viral capsid (Fig. 7B). However, in the absence of protein pB438L, the capsid-like outer domain of the aberrant particles does not acquire the icosahedral morphology of the mature virus. On the other hand, the presence of protein pE120R in the outer layer of the filiform structures could be expected, since these structures can be transported from the viral factory to the plasma membrane (Fig. 6A and C) and protein pE120R has been shown elsewhere to be involved in this process (7).
FIG. 7.
Immunoelectron microscopy of vB438Li-infected cells. Vero cells were infected with recombinant virus vB438Li in the absence of IPTG. At 24 hpi, the cells were fixed and prepared for cryosectioning. Cryosections were incubated with antibodies against the capsid proteins p72 (A) and pE120R (B), the inner envelope protein p17 (C), the core shell polyproteins pp220 (D) and pp62 (E), and a monoclonal anti-DNA antibody (F), followed by incubation with protein A-gold (10 nm). The arrows indicate representative labeling observed with antibodies against the capsid proteins, the inner envelope protein, and the core shell polyproteins in the aberrant structures. The anti-DNA antibody (F) labels foci dispersed throughout the factory (arrows), but the tubular structures are devoid of grains (arrowheads).
We then analyzed whether protein p17, a component of the inner envelope of the virus particle (37), is present in the abnormal structures. As shown in Fig. 7C, the anti-p17 antibody strongly labeled one of the outer domains of the tubules, suggesting that these structures contain, underneath the capsid, a membrane that might be similar to the inner envelope of the normal virus. On the other hand, the inner domain of these structures contains the polyproteins pp220 and pp62, as shown by the labeling with the corresponding antibodies (Fig. 7D and E).
Finally, we determined whether the filamentous particles contain the viral DNA, which is localized in the nucleoid of the normal virion. As could be expected from the observation that the aberrant forms are apparently devoid of this viral domain, the anti-DNA antibody did not label these forms, indicating that they are not able to incorporate the viral DNA, which is found forming dense clusters irregularly distributed throughout the factory (Fig. 7F).
DISCUSSION
Little is known at present about the mechanisms and protein interactions involved in the formation of the icosahedral capsid of the ASFV particle. As shown previously, capsid construction requires protein p72, the major structural component of this domain, as no virus particles are generated in its absence (24). The results presented here indicate that, in addition to protein p72, the ASFV structural protein pB438L also plays an essential role in the construction of the icosahedral capsid of the virus particle.
Large icosahedral viruses use identical or similarly folded proteins to establish different interactions while forming the capsid (42). Using structural and bioinformatic analysis, a similar “double-barreled” fold has been identified in the major capsid protein of different viruses, such as members of the Tectiviridae, Adenoviridae, Phycodnaviridae, Iridoviridae, and Asfarviridae and Mimivirus (10-12, 26, 30, 44). It has been suggested that this similarity might extend beyond the architecture of the major structural protein and that the assembly of the Tectivirus PRD1 capsid might be a scalable process that could apply to some of the largest known viruses (1, 44). In PRD1, the major component of the capsid is protein P3. Each P3 subunit is composed of two “jelly roll” domains (11, 12), giving the trimer a pseudohexagonal shape. Another necessary protein for capsid assembly is protein P30, which is proposed to be located between the adjacent facets of the icosahedral capsid and is required for stable capsid construction (38). The icosahedral vertices are occupied by pentamers of protein P31 that are associated with the trimeric P5 protein and the receptor-binding protein P2 to form flexible spikes (39). In the case of other larger viruses, like Chilo iridescent virus (Iridoviridae) and Paramecium bursaria Chlorella virus (Phycodnaviridae), it is known that the capsid is formed by hexavalent capsomers in the triangular facets and pentavalent capsomers in the 12 vertices of the icosahedron, but the composition of the pentavalent capsomers is unknown. However, at least for Paramecium bursaria Chlorella virus, it has been suggested that they could be formed by oligomers of a different structural protein (48).
Protein pB438L of ASFV is a structural protein that can associate with membranes during the infection, behaving as an integral membrane protein after treatment with sodium carbonate or Triton X-114. However, a hydrophobicity plot (27) of the pB438L protein sequence shows that it does not have any hydrophobic sequence typical of membrane-spanning proteins (data not shown). Also, no predicted motifs for posttranslational modifications, such as addition of fatty acids (myristic and palmitic) or of isoprenoids (geranyl and farnesyl) that could account for membrane targeting of protein pB438L, have been found. Additionally, in vitro-synthesized protein pB438L cannot associate with membranes. These data suggest that protein pB438L might associate with viral membranes possibly through a strong interaction with another viral or cellular integral membrane protein. This behavior might be similar to that observed with the product of the late gene A4L (p39) of vaccinia virus (18). This protein, which has a predominantly hydrophilic nature and is neither modified by fatty acids nor able to associate with membranes when synthesized in vitro (18), is, however, a membrane-associated protein during virus infection. Protein p39 is located between the core and the viral membranes and may form part of the spike-like structure on the core surface as a matrix-like linker protein (18, 32, 36). Possibly, vaccinia virus protein p39 attains its membrane association through interaction with another viral protein(s) in the membrane surrounding the intracellular mature virus (18). It is likewise possible that the ASFV structural protein pB438L might also associate with the inner envelope of the virion through its interaction with another protein of this membrane.
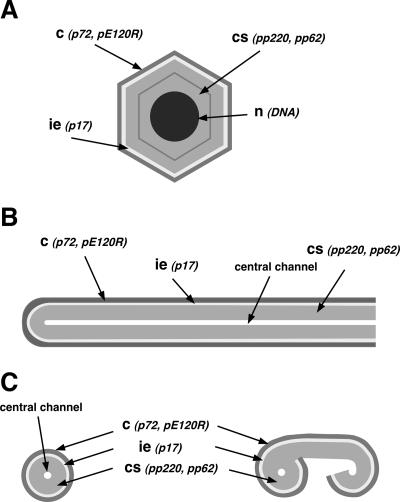
Using a recombinant ASFV that inducibly expresses protein pB438L, we found that this structural protein is essential for the formation of infectious virus particles. In the absence of protein pB438L, no viral icosahedral particles were observed within the infected cells by electron microscopy analysis. Instead, large aberrant tubular structures of viral origin, as well as bilobulate forms, were generated (diagrams in Fig. 8). The filamentous structures appeared to be covered by a capsid-like layer, which, however, did not present an icosahedral morphology. This layer was shown to contain the two capsid proteins described so far, namely, the major capsid protein p72 and protein pE120R, which is implicated in the movement of the particle from the viral factory to the budding sites at the plasma membrane (7). Indeed, this capsid is to some extent functional, as the filamentous structures can move from the virus assembly sites to the plasma membrane and exit the cell by budding. The tubules also possess an inner envelope, containing the viral protein p17, as well as an internal domain structurally similar to the core shell of the normal icosahedral particle. However, the core shell of the aberrant particles contains the two polyproteins pp220 and pp62 in an unprocessed state and does not therefore undergo the maturational event involving the proteolytic processing. A central electron-translucent channel lacking the viral DNA is found inside the tubular structures. In summary, our results indicate that, in the absence of protein pB438L, the viral capsid cannot acquire its icosahedral form which leads to the generation of tubules with an anomalous capsid-like layer.
FIG. 8.
Schematic representation of the structures generated during vB438Li infection. (A) Normal icosahedral particle. (B) Longitudinal section of a tubular particle. (C) Cross section of a tubular particle (left) and a bilobulate structure (right). c, capsid; ie, inner envelope; cs, core shell; n, nucleoid. Protein markers of each domain are indicated in parentheses. The DNA in the nucleoid of the normal particle is also indicated. The different structures are drawn approximately to scale.
The other anomalous structures generated under restrictive conditions, the bilobulate forms, present in cross section several layers resembling the domains of a developing normal particle. These aberrant forms are, like the tubular structures, massively translocated by budding into the lumen of intracellular vesicles and can apparently establish contacts with the plasma membrane. On the other hand, the morphological similarity observed in cross section between the tubular and bilobulate forms (Fig. 6D), together with the fact that the diameter of the lobules is the same as that of the tubules, suggests a morphogenetic relationship between the two structures.
The ASFV aberrant filamentous particles show a striking morphological resemblance with members of the Filoviridae family, such as Marburg and Ebola viruses (33, 41). Even the dimensions of the Marburg and Ebola virus filamentous virions, with a length of several micrometers and a diameter of 80 nm, as well as the width of the axial space (20 nm) are very similar to those of the ASFV tubules described here. This is particularly intriguing given the differences in domain organization and protein composition between the filoviruses and the ASFV filiform particles. The generation of ASFV tubules with the morphology and dimensions of the filovirus particles may reflect the existence of common architectural constraints in the construction of both kinds of particles, which, in the case of ASFV, would be imposed when the capsid layer, due to the absence of protein pB438L, cannot acquire its icosahedral symmetry.
It has been reported that the ASFV internal domains are not necessary for the correct assembly of the icosahedral capsid, since in the absence of polyprotein pp220 and hence of the core domain, empty, icosahedral particles consisting only of the inner envelope and the capsid are formed (5). In our view, this suggests that the failure to correctly assemble an icosahedral capsid in the absence of protein pB438L is related to a defect in one of the two external domains, the inner envelope or the capsid. The fact that protein pB438L has been shown to be membrane associated further supports a localization at these sites. Formation of aberrant tubular structures has been reported before in infections with other related icosahedral viruses, such as the iridovirus of the marine worm Nereis diversicolor (19). In the case of icosahedral viruses, it has been proposed that structures with tubular morphology may arise from the loss of the pentameric capsomers (or “pentasymmetrons”) (47). The fact that, in the absence of protein pB438L, the viral particles generated possess a tubular structure in which the icosahedral symmetry is lost suggests that the formation of the capsid pentasymmetrons might be compromised under these circumstances.
Our findings underscore the fact that a single viral protein, pB438L, can direct the morphogenetic process of ASFV towards the construction of an icosahedral capsid instead of a tubular structure. The elucidation of the mechanism by which protein pB438L accomplishes this will require a better knowledge of the organization and protein composition of the external domains of the virus particle.
.
Acknowledgments
This study was supported by grants from the Spanish Ministerio de Educación y Ciencia (BFU2004-00298), the European Commission (QLK2-CT-2001-02216), and the Wellcome Trust (075813/C/04/Z) and by an institutional grant from Fundación Ramón Areces. J. M. Rodríguez was supported by the Ramón y Cajal program of the Ministerio de Educación y Ciencia. C. Epifano was a predoctoral fellow of the Ministerio de Educación y Ciencia.
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Abrescia, N. G., J. J. Cockburn, J. M. Grimes, G. C. Sutton, J. M. Diprose, S. J. Butcher, S. D. Fuller, C. San Martin, R. M. Burnett, D. I. Stuart, D. H. Bamford, and J. K. Bamford. 2004. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 432:68-74. [DOI] [PubMed] [Google Scholar]
- 2.Alejo, A., G. Andrés, and M. L. Salas. 2003. African swine fever virus proteinase is essential for core maturation and infectivity. J. Virol. 77:5571-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrés, G., A. Alejo, J. Salas, and M. L. Salas. 2002. African swine fever virus polyproteins pp220 and pp62 assemble into the core shell. J. Virol. 76:12473-12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrés, G., A. Alejo, C. Simón-Mateo, and M. L. Salas. 2001. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 276:780-787. [DOI] [PubMed] [Google Scholar]
- 5.Andrés, G., R. García-Escudero, M. L. Salas, and J. M. Rodríguez. 2002. Repression of African swine fever virus polyprotein pp220-encoding gene leads to the assembly of icosahedral core-less particles. J. Virol. 76:2654-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrés, G., R. García-Escudero, C. Simón-Mateo, and E. Viñuela. 1998. African swine fever virus is enveloped by a two-membraned collapsed cisterna derived from the endoplasmic reticulum. J. Virol. 72:8988-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrés, G., R. García-Escudero, E. Viñuela, M. L. Salas, and J. M. Rodríguez. 2001. African swine fever virus structural protein pE120R is essential for virus transport from assembly sites to plasma membrane but not for infectivity. J. Virol. 75:6758-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrés, G., C. Simón-Mateo, and E. Viñuela. 1997. Assembly of African swine fever virus: role of polyprotein pp220. J. Virol. 71:2331-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angulo, A., E. Viñuela, and A. Alcamí. 1993. Inhibition of African swine fever virus binding and infectivity by purified recombinant virus attachment protein p12. J. Virol. 67:5463-5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16:673-685. [DOI] [PubMed] [Google Scholar]
- 11.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 1999. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98:825-833. [DOI] [PubMed] [Google Scholar]
- 12.Benson, S. D., J. K. Bamford, D. H. Bamford, and R. M. Burnett. 2002. The X-ray crystal structure of P3, the major coat protein of the lipid-containing bacteriophage PRD1, at 1.65 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 58:39-59. [DOI] [PubMed] [Google Scholar]
- 13.Breese, S. S., Jr., and C. J. DeBoer. 1966. Electron microscope observations of African swine fever virus in tissue culture cells. Virology 28:420-428. [DOI] [PubMed] [Google Scholar]
- 14.Brookes, S. M., A. D. Hyatt, T. Wise, and R. M. Parkhouse. 1998. Intracellular virus DNA distribution and the acquisition of the nucleoprotein core during African swine fever virus particle assembly: ultrastructural in situ hybridisation and DNase-gold labelling. Virology 249:175-188. [DOI] [PubMed] [Google Scholar]
- 15.Carrascosa, A. L., M. del Val, J. F. Santarén, and E. Viñuela. 1985. Purification and properties of African swine fever virus. J. Virol. 54:337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrascosa, J. L., J. M. Carazo, A. L. Carrascosa, N. García, A. Santisteban, and E. Viñuela. 1984. General morphology and capsid fine structure of African swine fever virus particles. Virology 132:160-172. [DOI] [PubMed] [Google Scholar]
- 17.Cobbold, C., M. Windsor, and T. Wileman. 2001. A virally encoded chaperone specialized for folding of the major capsid protein of African swine fever virus. J. Virol. 75:7221-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cudmore, S., R. Blasco, R. Vincentelli, M. Esteban, B. Sodeik, G. Griffiths, and J. Krijnse-Locker. 1996. A vaccinia virus core protein, p39, is membrane associated. J. Virol. 70:6909-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devauchelle, G. 1977. Ultrastructural characterization of an iridovirus from the marine worm Nereis diversicolor (O.F. Muller). Architecture of the virion and virus morphogenesis. Virology 81:237-246. [DOI] [PubMed] [Google Scholar]
- 20.Enjuanes, L., A. L. Carrascosa, M. A. Moreno, and E. Viñuela. 1976. Titration of African swine fever (ASF) virus. J. Gen. Virol. 32:471-477. [DOI] [PubMed] [Google Scholar]
- 21.Esteves, A., M. I. Marques, and J. V. Costa. 1986. Two-dimensional analysis of African swine fever virus proteins and proteins induced in infected cells. Virology 152:192-206. [DOI] [PubMed] [Google Scholar]
- 22.Eulalio, A., I. Nunes-Correia, A. L. Carvalho, C. Faro, V. Citovsky, J. Salas, M. L. Salas, S. Simoes, and M. C. Pedroso de Lima. 2006. Nuclear export of African swine fever virus p37 protein occurs through two distinct pathways and is mediated by three independent signals. J. Virol. 80:1393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galindo, I., E. Viñuela, and A. L. Carrascosa. 2000. Characterization of the African swine fever virus protein p49: a new late structural polypeptide. J. Gen. Virol. 81:59-65. [DOI] [PubMed] [Google Scholar]
- 24.García-Escudero, R., G. Andrés, F. Almazán, and E. Viñuela. 1998. Inducible gene expression from African swine fever virus recombinants: analysis of the major capsid protein p72. J. Virol. 72:3185-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffiths, G. 1993. Fine structure immunocytochemistry. Springer-Verlag, Heidelberg, Germany.
- 26.Iyer, L. M., L. Aravind, and E. V. Koonin. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720-11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 28.Laver, W. G., N. G. Wrigley, and H. G. Pereira. 1969. Removal of pentons from particles of adenovirus type 2. Virology 39:599-604. [DOI] [PubMed] [Google Scholar]
- 29.Martínez-Pomáres, L., C. Simón-Mateo, C. López-Otín, and E. Viñuela. 1997. Characterization of the African swine fever virus structural protein p14.5: a DNA binding protein. Virology 229:201-211. [DOI] [PubMed] [Google Scholar]
- 30.Nandhagopal, N., A. A. Simpson, J. R. Gurnon, X. Yan, T. S. Baker, M. V. Graves, J. L. Van Etten, and M. G. Rossmann. 2002. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. USA 99:14758-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newcomb, W. W., and J. C. Brown. 1991. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J. Virol. 65:613-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pedersen, K., E. J. Snijder, S. Schleich, N. Roos, G. Griffiths, and J. K. Locker. 2000. Characterization of vaccinia virus intracellular cores: implications for viral uncoating and core structure. J. Virol. 74:3525-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peters, C. L. 1996. Viral hemorrhagic fevers. In N. Nathanson, R. Ahmed, F. González-Scarano, D. E. Griffin, K. V. Holmes, F. A. Murphy, and H. L. Robinson (ed.), Viral pathogenesis. Lippincott-Raven, Philadelphia, Pa.
- 34.Rodríguez, J. M., F. Almazán, E. Viñuela, and J. F. Rodríguez. 1992. Genetic manipulation of African swine fever virus: construction of recombinant viruses expressing the beta-galactosidase gene. Virology 188:67-76. [DOI] [PubMed] [Google Scholar]
- 35.Rodríguez, J. M., R. García-Escudero, M. L. Salas, and G. Andrés. 2004. African swine fever virus structural protein p54 is essential for the recruitment of envelope precursors to assembly sites. J. Virol. 78:4299-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roos, N., M. Cyrklaff, S. Cudmore, R. Blasco, J. Krijnse-Locker, and G. Griffiths. 1996. A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J. 15:2343-2355. [PMC free article] [PubMed] [Google Scholar]
- 37.Rouiller, I., S. M. Brookes, A. D. Hyatt, M. Windsor, and T. Wileman. 1998. African swine fever virus is wrapped by the endoplasmic reticulum. J. Virol. 72:2373-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rydman, P. S., J. K. Bamford, and D. H. Bamford. 2001. A minor capsid protein P30 is essential for bacteriophage PRD1 capsid assembly. J. Mol. Biol. 313:785-795. [DOI] [PubMed] [Google Scholar]
- 39.Rydman, P. S., J. Caldentey, S. J. Butcher, S. D. Fuller, T. Rutten, and D. H. Bamford. 1999. Bacteriophage PRD1 contains a labile receptor-binding structure at each vertex. J. Mol. Biol. 291:575-587. [DOI] [PubMed] [Google Scholar]
- 40.Salas, J., M. L. Salas, and E. Viñuela. 1999. African swine fever virus: a missing link between poxviruses and iridoviruses?, p. 467-480. In E. Domingo, R. G. Webster, and J. J. Holland (ed.), Origin and evolution of viruses. Academic Press, New York, N.Y.
- 41.Sánchez, A., A. S. Khan, S. R. Zaki, C. J. Nabel, T. G. Ksiazek, and C. J. Peters. 2001. Filoviridae, p. 1279-1304. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Virology, vol. 1. Lippincott Williams & Wilkins, Hagerstown, Md. [Google Scholar]
- 42.San Martín, C., J. T. Huiskonen, J. K. Bamford, S. J. Butcher, S. D. Fuller, D. H. Bamford, and R. M. Burnett. 2002. Minor proteins, mobile arms and membrane-capsid interactions in the bacteriophage PRD1 capsid. Nat. Struct. Biol. 9:756-763. [DOI] [PubMed] [Google Scholar]
- 43.Sanz, A., B. García-Barreno, M. L. Nogal, E. Viñuela, and L. Enjuanes. 1985. Monoclonal antibodies specific for African swine fever virus proteins. J. Virol. 54:199-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saren, A. M., J. J. Ravantti, S. D. Benson, R. M. Burnett, L. Paulin, D. H. Bamford, and J. K. Bamford. 2005. A snapshot of viral evolution from genome analysis of the tectiviridae family. J. Mol. Biol. 350:427-440. [DOI] [PubMed] [Google Scholar]
- 45.Simón-Mateo, C., G. Andrés, F. Almazán, and E. Viñuela. 1997. Proteolytic processing in African swine fever virus: evidence for a new structural polyprotein, pp62. J. Virol. 71:5799-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simón-Mateo, C., G. Andrés, and E. Viñuela. 1993. Polyprotein processing in African swine fever virus: a novel gene expression strategy for a DNA virus. EMBO J. 12:2977-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wrigley, N. G. 1969. An electron microscope study of the structure of Sericesthis iridescent virus. J. Gen. Virol. 5:123-134. [DOI] [PubMed] [Google Scholar]
- 48.Yan, X., N. H. Olson, J. L. Van Etten, M. Bergoin, M. G. Rossmann, and T. S. Baker. 2000. Structure and assembly of large lipid-containing dsDNA viruses. Nat. Struct. Biol. 7:101-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yáñez, R. J., J. M. Rodríguez, M. L. Nogal, L. Yuste, C. Enriquez, J. F. Rodríguez, and E. Viñuela. 1995. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 208:249-278. [DOI] [PubMed] [Google Scholar]