Abstract
Coxsackievirus B3 (CVB3) is a primary cause of viral myocarditis, yet no effective therapeutic against CVB3 is available. Nucleic acid-based interventional strategies against various viruses, including CVB3, have shown promise experimentally, but limited stability and inefficient delivery in vivo remain as obstacles to their potential as therapeutics. We employed phosphorodiamidate morpholino oligomers (PMO) conjugated to a cell-penetrating arginine-rich peptide, P007 (to form PPMO), to address these issues. Eight CVB3-specific PPMO were evaluated with HeLa cells and HL-1 cardiomyocytes in culture and in a murine infection model. One of the PPMO (PPMO-6), designed to target a sequence in the 3′ portion of the CVB3 internal ribosomal entry site, was found to be especially potent against CVB3. Treatment of cells with PPMO-6 prior to CVB3 infection produced an approximately 3-log10 decrease in viral titer and largely protected cells from a virus-induced cytopathic effect. A similar antiviral effect was observed when PPMO-6 treatment began shortly after the virus infection period. A/J mice receiving intravenous administration of PPMO-6 once prior to and once after CVB3 infection showed an ∼2-log10-decreased viral titer in the myocardium at 7 days postinfection and a significantly decreased level of cardiac tissue damage, compared to the controls. Thus, PPMO-6 provided potent inhibition of CVB3 amplification both in cell cultures and in vivo and appears worthy of further evaluation as a candidate for clinical development.
Coxsackievirus B3 (CVB3), a member of the genus Enterovirus within the family Picornaviridae, is a primary causative agent of viral myocarditis (4, 17). In North America, viral myocarditis accounts for 20% of sudden heart failure in children and adolescents (9). The chronic sequela of CVB3-induced myocarditis, dilated cardiomyopathy, is responsible for approximately 50% of the cardiac transplants registered annually worldwide (1). Unfortunately, there is no specific therapeutic agent available to address CVB3-induced myocarditis, and as of this writing, no clinical trials for such are registered with the FDA (http://clinicaltrials.gov/).
CVB3 is a nonenveloped, single-stranded positive-polarity RNA virus containing a single open reading frame (ORF) flanked by 5′- and 3′-untranslated regions (5′- and 3′UTRs, respectively). The genomic RNA serves as a direct template for translation as well as for viral RNA replication through the transcription of a negative-strand intermediate. Like all picornaviruses, CVB3 possesses a long and highly structured 5′UTR. The initiation of CVB3 RNA translation is mediated through the binding of the 40S-ribosome-containing preinitiation complex to an internal ribosomal entry site (IRES) region within the viral 5′UTR (16).
Antisense oligonucleotides appear to have considerable potential as antiviral agents, with five having undergone clinical trials for the treatment of viral diseases and many more in ongoing studies (26). The successful application of antisense technology in vivo continues to be hampered by limited oligomer stability and inefficient delivery to RNA targets within relevant cells. An antisense phosphorothioate oligodeoxynucleotide (PS-ODN) targeting the 3′-terminal region of the CVB3 genome was shown to possess anti-CVB3 activity in cultured cells and in mouse hearts in vivo. However, the level of CVB3 inhibition was moderate and incapable of diminishing the severity of myocarditis, as evaluated histopathologically (33, 37).
Phosphorodiamidate morpholino oligomers (PMO) are a novel type of antisense compound in which each subunit contains a DNA base (A, C, G, or T) connected to a backbone consisting of a morpholine ring and phosphorodiamidate intersubunit linkage (29, 30). The uncharged and hydrophilic PMO are highly resistant to enzymes and biological fluids (14). The PMO mechanism of antisense action occurs via steric blocking of the cRNA sequence and thus differs from that of antisense agents, which induce RNase H-mediated cleavage of the RNA strand of an RNA-DNA duplex (28). There have been several recent reports of the antiviral efficacies of PMO compounds (7, 8, 15, 24, 32, 34). In this study, PMO were covalently conjugated to a cell-penetrating arginine-rich peptide (ARP), P007 (7, 23), in an attempt to enhance delivery to the heart. CVB3-specific ARP-PMO (PPMO) were designed based in part on previous reports identifying CVB3 RNA regions that are critical for efficient viral amplification. Previous work has identified a segment of the sequence in the CVB3 5′UTR spanning nucleotides (nt) 439 to 639 that is important and a 46-nt IRES “core sequence” that is indispensable for efficient viral translation (16, 36). The 5′-terminal region of the CVB3 5′UTR has been implicated in the translation initiation process (38), as well as in viral RNA synthesis (27). The CVB3 3′UTR contains sequence elements considered necessary for the regulation of viral RNA synthesis. Thus, we reasoned that blocking access to these critical sequences may interfere with virus particle production.
Cell culture experiments with a panel of PPMO showed that each of two PPMO targeting sequences in the IRES “core sequence” had high antiviral activities. Furthermore, two intravenously administered treatments with one of the IRES-targeted PPMO reduced levels of virus in the hearts of infected mice by over 95% and greatly reduced virus-induced heart tissue damage at a dose level assessed as nontoxic.
MATERIALS AND METHODS
PMO and PPMO.
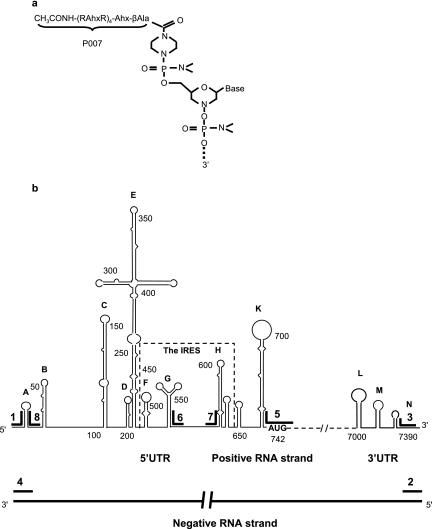
PMO were synthesized by methods previously described (30). An ARP [CH3CONH-(R-aminohexanoic acid-R)4-aminohexanoic acid-βAla, designated P007, where R is arginine and βAla is beta-alanine] was covalently conjugated to the 5′ end of each PMO through a noncleavable linker (Fig. 1a). PPMO compounds were synthesized, purified, and analyzed at AVI BioPharma, Inc. (Corvallis, OR) by methods described previously (21, 23). Eight PPMO, each of 17 to 21 bases in length, were designed to base pair with a complementary sequence in the genomic (positive) or antigenomic (negative) strand of CVB3 RNA. A 20-mer of random sequence with 50% G+C content was prepared as a PMO (PMO-C) and as a PPMO (PPMO-C) for use as negative controls. To monitor the delivery of PMO and PPMO into cells and tissues, fluorescein was conjugated to the 3′ ends of PMO-C and PPMO-C (creating Fl-PMO-C and Fl-PPMO-C, respectively). All PPMO base sequences were BLAST searched, and no unintentional matching with viral or host mRNAs was detected. PPMO names, base sequences, and target locations are listed in Table 1. Prior to their use, lyophilized PMO and PPMO were resuspended with filter-sterilized distilled water to a concentration of 1 to 2 mM and were stored at 4°C.
FIG. 1.
Structure of PPMO and the schematic locations in CVB3 RNA of PPMO target sequences. (a) PPMO is an antisense structural type in which each subunit consists of a purine or pyrimidine base attached to a morpholine ring, with the subunits being connected through phosphorodiamidate linkages. A cell-penetrating arginine-rich peptide (P007) (see Materials and Methods) is covalently conjugated to the 5′ end of each PMO through a noncleavable linker. (b) Six genomic (positive-strand) PPMO targets are indicated within the proposed secondary structures of the 5′- and 3′UTRs of CVB3 RNA. Two PPMO target the terminal regions of the viral antigenome (negative strand). PPMO base sequences are listed in Table 1.
TABLE 1.
PPMO sequences and target locations in CVB3 RNA
| PPMO | Sequence (5′-3′) | Target location in CVB3 RNAa |
|---|---|---|
| 1 | CAACCCACAGGCTGTTTTAA | +, 5′ terminus (1-21) |
| 2 | AAATTCTCCGCATTCGGTGCG | −, 5′ terminus |
| 3 | CGCACCGAATGCGGAGAATTT | +, 3′ terminus (7378-7398) |
| 4 | TTAAAACAGCCTGTGGGTTG | −, 3′ terminus |
| 5 | CCCATTTTGCTGTATTC | +, AUG region (730-746) |
| 6b | ATGAAACACGGACACCCAAAG | +, IRES core sequence (547-567) |
| 7 | TAAGCAGCCAGTATAGGAATA | +, IRES core sequence (570-590) |
| 8 | CAATGGGCCTGTGGGTGGG | + (23-41) |
| Cc | AGTCTCGACTTGCTACCTCA |
Based on the complete CVB3 genome sequence (GenBank accession no. M33854). Target locations and nucleotide positions (in parentheses) in the virus-positive (+) genomic or virus-negative (−) antigenomic strand are noted.
Prepared as both PMO and PPMO (see Materials and Methods).
C, negative control (random sequence). Prepared in four different forms: PMO-C, PPMO-C, Fl-PMO-C, and Fl-PPMO-C (see Materials and Methods).
Viruses and cells.
The CVB3 Kandolf strain was used for all cell cultures and in vivo experiments, unless otherwise specified. The CVB3 Gauntt strain was used in a single experiment, where stated. CVB3 strains were propagated in HeLa cells (American Type Culture Collection) maintained in virus growth medium (Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 100 μg/ml penicillin-streptomycin [Invitrogen]). Respiratory syncytial virus (RSV) (human Long strain type A) was a generous gift from Richard Hegele and was propagated in HEp-2 cells (American Type Culture Collection) as described previously (31). HL-1 cells, a cardiac muscle cell line established from an AT-1 mouse atrial cardiomyocyte tumor lineage (6), were a kind gift from William C. Claycomb and maintained in Claycomb medium (JRH Biosciences, Lenexa, KS) supplemented with 10% fetal bovine serum (JRH Bioscience), 100 μg/ml penicillin-streptomycin, 0.1 mM norepinephrine (Sigma), and 2 mM l-glutamine (Invitrogen).
Cell culture procedure and plaque assay.
For cell culture experiments, cells at ∼80% confluence were incubated with PPMO- or water-containing (as a mock treatment) growth medium for 4 h, unless otherwise specified. The PPMO or mock treatment was then removed, and the cells were rinsed with growth medium and infected with CVB3 at the multiplicity of infection (MOI) indicated below for 1 h or infected with RSV at a MOI of 0.1 for 1.5 h in a volume of 0.5 ml medium. After the viral-infection period, the inoculum was removed, and the cells were replenished with virus growth medium, in the absence of PPMO, and incubated at 37°C in 5% CO2. At various time points postinfection (p.i.), as described below, supernatants and cell lysates were collected and stored at −80°C. Viral titers were determined by plaque assay as described previously (37). Briefly, HeLa cells were seeded into six-well plates (8 × 105 cells/well) and incubated at 37°C. Cell monolayers at ∼90% confluence were washed with phosphate-buffered saline (PBS) and then overlaid with 500 μl of serial 10-fold dilutions of supernatants from cell cultures or heart lysates. The cells were incubated for 1 h and the supernatants removed. The cells were then overlaid with 2 ml of sterilized soft Bacto agar, incubated at 37°C for 72 h, fixed with Carnoy's fixative for 30 min, and stained with 1% crystal violet. The plaques were counted, and the numbers of viral PFU/ml were calculated.
Western immunoblotting.
Western blotting was performed by standard protocols as previously described (33). Briefly, equal amounts of protein were loaded into the lanes for 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat milk containing 0.1% Tween 20 and probed with either a monoclonal mouse antibody (Ab) against CVB3 capsid protein VP1 (DAKO), a polyclonal Ab against RSV (Biodesign), or a β-actin monoclonal Ab (Sigma), followed by incubation with horseradish peroxidase-conjugated secondary antibody. Signal was detected with ECL system reagents (Amersham).
Cell viability assay.
HeLa and HL-1 cells were subjected to PPMO or mock treatment for 4 h and to viral infection as described above. Cell viability was measured at 8 h p.i. (for HeLa cells) or 40 h p.i. (for HL-1 cells) using the CellTiter 96 AQueous one-solution cell proliferation assay {which uses MTS [3,4-(5-dimethylthiazol-2-yl)-5-(3-carboxymethoxy phenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt]} (Promega) according to the manufacturer's instructions. Cells were incubated with MTS solution for 2 h, and the absorbance was measured at 492 nm using an enzyme-linked immunosorbent assay reader. The absorbance values of PPMO- and mock-treated cells were converted to percentages by comparison to that of mock-treated noninfected samples, which was set at 100% survival. In addition, cells were exposed to PPMO-C treatment as described above but in the absence of virus, and cell viability analysis was carried out.
In vitro transcription and translation assay with monocistronic plasmid.
The protein-coding sequence for firefly luciferase, without the ATG initiator Met codon, was subcloned into the multiple cloning sites of the pCINeo plasmid (Promega) at the SalI and NotI sites (creating pCI-Neoluc). Bases 1 to 792 of CVB3 (GenBank accession number M33854), which correspond to bases −741 to +51 relative to the AUG translation start codon, were PCR amplified from pCMV5 (35) with primers CV5′-For (5′-ATCTCGAGTTAAAACAGCCTGTGGG-3′) and CV5′-Rev (5′-ATGTCGACCAGCCTGGTCTCATGTGCC-3′) and then restricted with XhoI and SalI and subcloned into pCI-Neoluc. This effectively replaced the start codon of the luciferase gene with a sequence representing the entire CVB3 5′UTR and the first 51 nucleotides of the coding sequence (creating pCV5′luc). The authentic AUG start codon in the CVB3 sequence “leader” is in frame with the coding sequence of luciferase in pCV5′luc. pCV5′luc was linearized with NotI, and in vitro-transcribed RNA was produced using a T7 Megascript kit (Ambion) according to the manufacturer's instructions. In vitro translations were carried out with transcribed RNA at a final concentration of 1 nM and PPMO or mock treatment as specified below. Luciferase-induced light emission was read on a model FLx800 microplate luminometer as previously described (25). The average light units produced by the set of reactions for each treatment were normalized to the mean light units of mock-treated control reactions and are expressed as a percentage of the inhibition of the luciferase mock-treated signal of the reaction mixture.
In vitro transcription and translation assay with bicistronic plasmid.
A bicistronic plasmid, pSPT18 (CAT+P1), containing chloramphenicol acetyltransferase (CAT), the CVB3 5′UTR, and the viral P1 polyprotein coding sequence, was used for in vitro transcription and translation, as described previously (36). Briefly, plasmid DNA was linearized with BglII and bicistronic RNA transcripts synthesized with SP6 RNA polymerase (Promega). Transcription products were treated with DNase I and extracted with phenol-chloroform. Equal amounts of RNA transcripts were translated in 25-μl reaction mixtures that contained rabbit reticulocyte lysate (Promega), [35S]methionine, an amino acid mixture minus methionine, and PMO or water, as used for mock treatment. A 100-fold molar excess of PMO over RNA transcripts was used. Aliquots of 35S-labeled translation products (CAT and P1 polyprotein) were subjected to SDS-PAGE, and the gels were then dried and exposed to X-ray film.
Biodistribution of PPMO and histology staining.
To determine the distribution of PMO or PPMO within cells and in various organs, 200 μg of Fl-PMO-C or Fl-PPMO-C was injected into one mouse each via the tail vein. Mice were sacrificed at 16 h after injection, and tissues were perfused with PBS and 4% paraformaldehyde-PBS. Frozen tissue sections were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) (Molecular Probes) and examined by confocal microscopy (Leica).
For histological analysis, formalin-fixed midventricular portions of cardiac tissue, spleen, liver, kidney, and pancreas were sectioned into 4-μm slices and stained with hematoxylin and eosin (H&E). H&E-stained sections were graded in a blind manner by two separate researchers for the extent of myocarditis, based on the intensity and character of tissue injury and inflammatory infiltration, as previously described (5). Histologic grades corresponded to the following scale: 0 for no or a questionable presence of foci, 1 for limited focal distribution, 2 to 3 for intermediate severity, and 4 to 5 for coalescent and extensive foci over the entirety of the transversely sectioned ventricular tissue.
PPMO treatment and viral infection of mice.
For PPMO treatment of mice in the absence of virus, 12 4-week-old A/J (H-2a) mice (Jackson Laboratory) were randomized into four groups and injected intravenously via the tail vein twice, at 0 and 48 h, with either 100, 150, or 200 μg PPMO-6 or an identical volume (200 μl) of PBS. All mice were observed for appearance and behavior and weighed daily for 7 days.
For the experiment with virus, 18 mice (same age and type as described above) were randomized to three groups (six per group) and infected intraperitoneally with 105 PFU of CVB3. Mice were administered a volume of 200 μl containing either 200 μg of PPMO in PBS or PBS alone intravenously via tail vein injection at 3 h before infection and then again on day 2 p.i. All mice were sacrificed on day 7 p.i., at which time the ventricular portions of the hearts were collected and transversely sectioned into apex, mid, and basal portions for analysis. Apex portions were weighed, homogenized in Dulbecco's modified Eagle's medium, and diluted to 1 mg of tissue per ml. Mid portions were fixed in 10% formalin and used for histopathology. Basal portions were stored at −80°C for possible future reference.
Statistical analysis.
Results are expressed as means ± standard deviations (SD). Statistical analyses were performed with Student's t test. P values less than 0.05 were considered statistically significant.
RESULTS
PPMO design.
Six antisense PPMO were designed to base pair with various sequences in the 5′- and 3′UTRs of positive-strand, genomic CVB3 RNA (Fig. 1b; see Table 1 for specifics on all PPMO). Several of these PPMO were designed to target sequences in cis- and trans-acting RNA translation control elements identified through previous mutational mapping (16, 36) and inhibition studies utilizing PS-ODNs (33, 37). Two PPMO targeting the 5′-terminal region of the 5′UTR (PPMO-1 and -8) were designed to obstruct viral sequences previously shown to be important in translation (18) and/or RNA synthesis (27). Two PPMO (PPMO-6 and -7) were designed against the IRES “core sequence” (nt 546 to 592) identified previously by mutational mapping (16, 36). A region spanning the AUG start codon at nt 742 to 744 of the ORF was also selected (PPMO-5), as the AUG codon translation initiation region is a site frequently targeted with this type of antisense technology (13, 22). Also targeted was the 3′-terminal end of the genome (PPMO-3), in an attempt to block the RNA-dependent RNA polymerase transcription initiation site utilized during negative-strand synthesis. A PS-ODN targeting this same site had shown antiviral efficacy in a previous study (33). As the CVB3 replication cycle is dependent on the utilization of an antigenomic negative strand, two PPMO (PPMO-2 and -4) were designed to target the 5′- and 3′-terminal regions of negative-strand CVB3 RNA. In addition, a random-sequence PMO and a random-sequence PPMO were utilized in both fluoresceinated and nonfluoresceinated forms (see Materials and Methods) to control for off-target effects and to explore the cellular uptake of their respective chemistries.
Conjugation to cell-penetrating peptide P007 enhances PMO delivery.
ARPs, including P007, have previously been shown to facilitate PMO delivery into cells in culture (7, 21, 23, 25). To investigate the ability of P007 to deliver CVB3-specific PMO into HeLa and HL-1 cells under our cell culture conditions, cells were incubated with 10 μM Fl-PMO-C or Fl-PPMO-C in virus growth medium. Confocal microscopy revealed that after 1 h of incubation with Fl-PPMO-C, nearly all cells of both types were fluorescence positive, with the majority of signal being apparent as a punctate pattern in the cytoplasm. In contrast, almost no signal was observed in cells treated with Fl-PMO-C (Fig. 2). This difference was found to be consistent with observations from a similar in vivo evaluation described below.
FIG. 2.
Conjugation to P007 facilitates delivery of PMO into cells. HeLa cells (a and b) and HL-1 cardiomyocytes (c and d) were incubated with 10 μM Fl-PMO-C (a and c) or Fl-PPMO-C (b and d). After 1 h of incubation, fluorescence images were photographed through a confocal microscope.
CVB3 replication is inhibited by PPMO targeting the IRES “core sequence.”
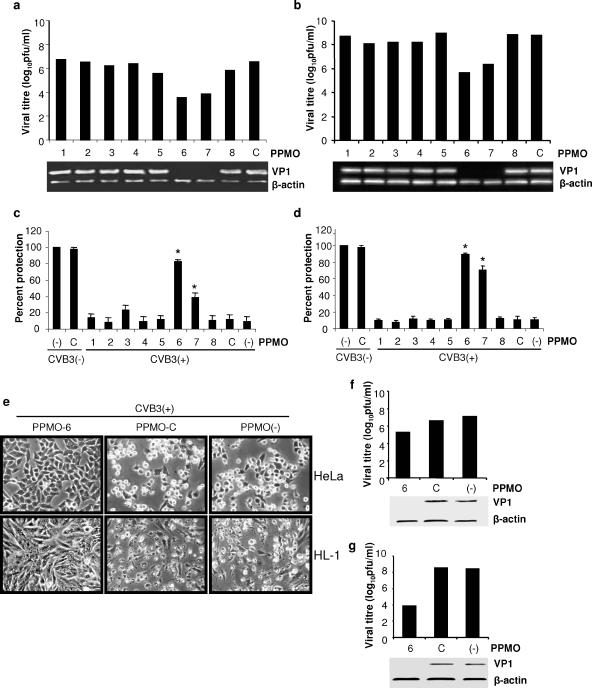
An initial evaluation of each PPMO at a single concentration was carried out to compare their antiviral efficacies. Two cell types were incubated with individual PPMO at a concentration of 10 μM for 4 h and then infected with CVB3 at an MOI of 10 for 1 h. The levels of viral particles were assessed by Western blotting (detecting the viral capsid protein VP1) and plaque assay from samples taken at 8 h (for HeLa cells) or 40 h (for HL-1 cells) p.i. As shown in Fig. 3a and b, VP1 protein was reduced to undetectable levels in cells treated with PPMO-6 or -7, yet there was no significant effect from the other antisense PPMO compared to that of the controls. Consistently, viral titers of PPMO-6- and -7-treated cells were around 3 log10 less than that of PPMO-C-treated cells. These results indicate that PPMO can effectively and specifically inhibit CVB3 replication in both HeLa and HL-1 cells and that the most potent CVB3 inhibitor of this panel of eight sequences is PPMO-6, followed by PPMO-7. The demonstration that PPMO-6 provided protection from the CVB3-induced cytopathic effect verified the observations described above. Ten micromolar PPMO-6-treated HeLa and HL-1 cells remained 83% and 89% viable, respectively, after CVB3 infection, compared to 11% and 10%, respectively, in the corresponding PPMO-C-treated cells (Fig. 3c, d, and e). The protection of cells from virus-induced cytopathic effect by PPMO-6 (Fig. 3e) is further confirmation that this PPMO itself at 10 μM was not cytopathic. Furthermore, standard cell viability assays in the absence of virus [CVB3(−) lanes in Fig. 3c and d] showed that PPMO-C generated almost no reduction in the viability of cells under the conditions of the above-described experiment, indicating an absence of generic PPMO cytotoxicity at the concentration used in this experiment.
FIG. 3.
CVB3-specific PPMO inhibit CVB3 replication in HeLa (a and c) and HL-1 (b and d) cells. (a and b) Plaque assays of infectious viral particles (upper panel) and Western blot analysis of CVB3 capsid protein VP1 (lower panel). Assays were conducted on HeLa and HL-1 cell monolayers as described in Materials and Methods. Cells were incubated with 10 μM PPMO or water (as a mock treatment) for 4 h, infected with CVB3 at an MOI of 10 for 1 h, and then incubated for 8 h (HeLa) or 40 h (HL-1) in the absence of PPMO, and samples were harvested for analysis. Supernatants were analyzed by plaque assay, and cell lysates were analyzed by Western blotting. β-Actin detection served as the protein loading control for Western blotting. Data shown are representative of two independent experiments that had equivalent outcomes. (c and d) PPMO protect cells from CVB3-induced cell death. MTS assays were performed on PPMO-treated and CVB3-infected [CVB3(+)] cells or PPMO-treated noninfected [CVB3(−)] cells, as described in Materials and Methods. Cell viability for all samples of infected or noninfected cells is expressed relative to that of the mock-treated, noninfected control, which is defined as 100% survival. Values shown here are means ± SD from three independent experiments. (*, P < 0.01). (e) Morphological changes of HeLa (top panels) and HL-1 (bottom panels) cells following PPMO treatment with either PPMO-6 or PPMO-C or mock treatment and infection, as described above. Photographs were taken at 8 and 40 h p.i. for HeLa and HL-1 cells, respectively. (f and g) PPMO-6 applied at 1 h p.i. inhibits ongoing CVB3 replication in HeLa (f) and HL-1 (g) cells. Cells were infected with CVB3 at an MOI of 10 and treated with 10 μM PPMO at 1 h p.i. Supernatants and cell lysates were collected and analyzed as described above (a and b). Data shown are representative of two independent experiments that had equivalent outcomes. For PPMO target locations in CVB3, see Table 1.
To investigate the effect of delaying PPMO treatment until after viral infection, cells were infected with CVB3 for 1 h, the inoculum was removed, and PPMO treatment was applied. This time point was selected because the kinetics of picornaviral replication are rapid. Cellular cap-dependent translation can start to shut down approximately 30 min after the initial infection and largely ceases by 2 h p.i. (12). Treatment with 10 μM PPMO-6 postinfection generated antiviral activity similar to that of the preinfection treatment protocol (Fig. 3f and g), implying a favorable characteristic with regard to potential therapeutic usefulness.
PPMO-6 inhibition of CVB3 is dose dependent and sequence specific.
To further characterize the potency of PPMO-6, a dose-response challenge of HeLa cells, followed by CVB3 infection, was performed. Treatment with increasing concentrations of PPMO-6 resulted in corresponding decreases in viral titer as measured by plaque assay and in VP1 expression as measured by Western blotting (Fig. 4a). These results indicate that PPMO-6 exerts anti-CVB3 activity in a dose-dependent manner.
FIG. 4.
PPMO-6 inhibits CVB3 amplification in a dose-dependent and sequence-specific manner. (a) Dose-dependent inhibition of CVB3 production in HeLa cells by PPMO-6 is shown. The experimental design is as described in the Fig. 3a legend. (b) The time course of inhibition of CVB3 replication by PPMO-6 is shown. HeLa cells were treated with PPMO-6 or -C and infected with CVB3, as described above. Supernatants and cell lysates were collected at the indicated time points p.i. for plaque assay and Western blotting. (c) Effective inhibition of viral production can be observed for at least 5 days after PPMO-6 treatment. HL-1 cells were treated with PPMO-6 or -C for 4 h and then infected 0, 1, 2, 3, 4, or 5 days (D0 to D5) later. Virus titers were determined by plaque assays from supernatants taken at 40 h p.i. The effect of PPMO-6 was calculated for each time point with respect to that of the negative-control PPMO-C, and those ratios were expressed relative to the PPMO-6 effect at day 0, which was defined as 100% inhibition. (d) CVB3-specific PPMO have no effect on RSV replication. HeLa cells were incubated with the indicated PPMO or mock treated (−) for 4 h, followed by RSV infection. Cell lysates were collected 2 days p.i. and used for Western blot detection of RSV proteins, as described in Materials and Methods. Data shown are representative of two independent experiments that had equivalent outcomes.
To investigate the kinetics of inhibition onset by PPMO-6, cells were pretreated with PPMO-6 or -C and then infected with virus. At various time points p.i., cell lysates and supernatants were collected for detecting VP1 expression levels and virus titers, respectively. Little difference between the effects of PPMO-6 or -C was detectable at 2 h and 4 h p.i. This is likely because the first replication cycle of CVB3 requires approximately 6 h, as indicated by the control lanes in the Western blot shown in Fig. 4b. We assume that the viral production apparent at 2 to 4 h in the titer graph of Fig. 4b is due largely to residual inoculum. Far fewer virus particles, and lower VP1 levels, were observed in PPMO-6-treated samples than in control treatment samples at 6 and 8 h p.i. (Fig. 4b).
We next sought to gain an indication of the length of time that a single treatment with PPMO-6 could protect cells from a future challenge by CVB3. HL-1 cells were treated for 4 h with PPMO-6 or PPMO-C and then infected with CVB3 for 1 h at day 0, 1, 2, 3, 4, or 5 after PPMO treatment. After the infection period, the cells were incubated for 40 h. The cytoprotective effect of PPMO-6 in relation to the period of time that elapsed between treatment and virus infection was measured by plaque assay at various time points. On days 4 and 5 posttreatment, PPMO-6 retained 78% and 47%, respectively, of the antiviral activity that it had displayed on day 0 posttreatment (Fig. 4c), indicating that PPMO-6 remains stable and able to access virus in the cells over time and can provide significant protection to HL-1 cells from CVB3 infection several days after application.
The sequence specificities of PPMO-6 and -7 were also investigated by the challenge of RSV infection under experimental conditions similar to those of the CVB3 experiments. HeLa cells were incubated with 10 μM of either PPMO-1, -3, -6, -7, or -C or water as mock treatment prior to RSV infection at an MOI of 0.1. Western immunoblotting showed that at 2 days p.i., RSV protein expression levels in all PPMO-treated samples were similar to that in the mock-treated sample (Fig. 4d). These results indicate that none of the PPMO were generically antiviral or cytotoxic and that PPMO-6 and -7 likely inhibit CVB3 replication through sequence-specific interference.
To test whether PPMO-6 could potentially inhibit multiple strains of CVB3, the CVB3 Gauntt strain was challenged by PPMO-6, PPMO-C, and mock treatment, as in the dose-response experiment described above. The Gauntt strain sequence is more than 99% identical at nt 300 to 599 of the 5′UTR and 100% identical in the PPMO-6 target region with the Kandolf strain. PPMO-6 generated levels of inhibition against the Gauntt strain that were similar to those generated against the Kandolf strain, as measured by plaque assay (data not shown). The nucleotide sequences of nt 300 to 599 from 14 CVB3 clinical isolates are available in GenBank (10). There is 89% to 99% sequence agreement between the Kandolf strain and the other CVB3 isolates within these 300 nucleotides, including some differences at the PPMO-6 target site (data not shown). Thus, although PPMO-6 appears to be imperfectly designed as a pan-CVB3 agent, it may well have considerable activity against most, if not all, other CVB3 strains.
In vitro inhibition of viral RNA translation.
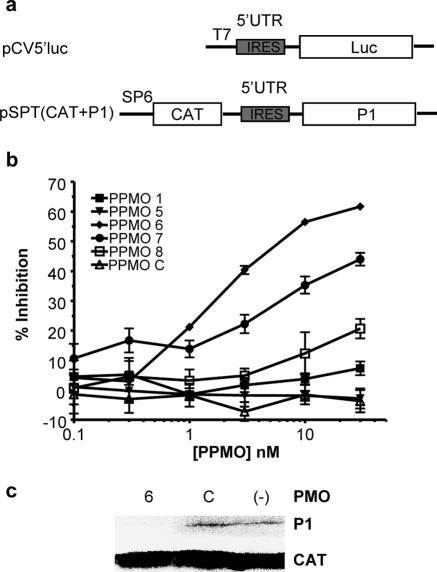
Based on our current understanding of the PPMO mechanism of action and the function of the picornaviral IRES region, we suspected that PPMO-6 and -7 inhibit viral production by interfering specifically with viral translation. To investigate the ability of these oligomers to act as translation inhibitors, we employed two in vitro translation systems. A monocistronic luciferase reporter assay system utilized plasmid-generated RNA comprising the entire CVB3 5′UTR and the first 51 nt of coding sequence, followed by the luciferase coding sequence (Fig. 5a). Rabbit reticulocyte lysate reaction mixtures containing in vitro-transcribed RNA were challenged by several PPMO. The results depicted in Fig. 5b show that PPMO-6 was a potent inhibitor of translation, producing a >50% reduction in translated luciferase at a concentration of under 10 nM. PPMO-7 was the next most effective PPMO tested. The relative levels of activity of the various PPMO used in this assay mirrored the results obtained in the single-dose survey depicted in Fig. 3a and b.
FIG. 5.
PPMO-6 inhibits viral RNA translation. (a) Map of the monocistronic pCV5′luc and bicistronic pSPT (CAT+P1) plasmids used to synthesize RNA by in vitro transcription. (b) In vitro-transcribed monocistronic RNA containing the CVB3 5′UTR and first 51 nt of coding sequence, followed by luciferase, was used in in vitro translation reaction mixtures with rabbit reticulocyte lysate and various PPMO (indicated in the key). Relative percentages of inhibition were calculated by comparison with results for mock treatment controls. Specifics of plasmid construction, reaction conditions, and luciferase determination are described in Materials and Methods. (c) Bicistronic RNA containing CAT as an internal control, followed by the CVB3 5′UTR and P1 polyprotein coding sequences, was translated with a 100-fold molar excess of PMO-6 (lane 6) or -C (lane C) or mock treated (lane −) in rabbit reticulocyte lysate with [35S]methionine, as described in Materials and Methods. Translation products were analyzed by SDS-PAGE.
A second in vitro translation assay system employed plasmid pSPT18 (CAT+P1), from which bicistronic RNA transcripts were synthesized (36). Translation of the downstream cistron (encoding the CVB3 polyprotein P1) present in the transcripts is mediated by the authentic CVB3 5′UTR, with the translation of CAT serving as an internal control (Fig. 5a). In the transcripts from this plasmid, the upstream cistron (CAT) serves to block the 5′ end of the CVB3 5′UTR to ensure that the downstream cistron (CVB3 P1) is translated through the utilization of IRES-mediated initiation. As shown in Fig. 5c, PMO-6 almost completely blocked the translation of P1, while PMO-C had little effect compared to that of the mock treatment. The results from these two experiments show that PMO-6 can interfere with the process of translation and provide further indication that the target sequence of PMO-6 is critical in CVB3 translation.
Inhibition of CVB3 replication and attenuation of myocarditis in mouse hearts.
Encouraged by highly positive cell culture results, we proceeded to attempt an initial evaluation of the antiviral viability of PPMO-6 in a mouse model. First, to investigate the ability of PMO or PPMO to achieve delivery into the cells of relevant tissues in vivo, a single bolus dose of 200 μg of Fl-PMO-C or Fl-PPMO-C was administered intravenously to one mouse apiece. Heart, pancreas, and kidney samples were harvested 16 h postinjection, and the tissues were perfused, fixed, and examined by confocal microscopy. Fluorescence was clearly visible in heart, kidney, and pancreas from the Fl-PPMO-C mouse (Fig. 6) but not from the mouse receiving Fl-PMO-C (only heart is shown in Fig. 6a).
FIG. 6.
Distribution of fluorescein-labeled PMO or PPMO in murine organs. Fl-PMO-C or Fl-PPMO-C was administered to A/J mice by intravenous tail vein injection. Sixteen hours later, tissues were harvested and processed for histological staining. Nuclei were counterstained with DAPI. The localizations of the fluorescein-labeled PMO or PPMO in the heart from the Fl-PMO-C-treated mouse (a) and the heart (b), kidney (c), and pancreas (d) from the Fl-PPMO-C-treated mouse were photographed with a confocal microscope.
To determine a robust yet nontoxic dose of PPMO-6 for use in an in vivo CVB3 challenge, groups of mice (four mice per group) were injected twice with 100 to 200 μg of PPMO-6 per dose (or PBS) 48 h apart, in the absence of virus. None of the dosages resulted in any evident toxicity throughout the ensuing 7-day monitoring period. Daily weighing of all animals revealed no loss of average body weight in any of the groups (data not shown). Mice were observed daily, and all displayed a normal appearance (e.g., no ruffled fur) and behavior (e.g., no obvious lethargy). Histopathologic examination of heart, liver, kidney, spleen, and pancreas revealed no abnormalities (data not shown).
Based on the preliminary dose-versus-toxicity experiment, mice were randomized into three groups (six mice per group), and each was injected with 200 μg of PPMO-6, -C, or PBS intravenously at 3 h prior to CVB3 infection and then again 48 h p.i. Seven days p.i., body weights were measured to determine virus-induced weight loss before euthanization. The average body weight loss was 17.44% for the PBS group, 15.82% for the PPMO-C-treated group, and 12.55% for the PPMO-6-treated group. None of these differences were statistically significant (P > 0.05). For the evaluation of the effect of PPMO treatment on CVB3 titers in the mouse hearts, plaque assays were performed on the pooled apex portions of the ventricles from each group. As shown in Fig. 7a, the amount of infectious virus particles in the tissues of the PPMO-6-treated group was approximately 2 log10 less than that in the control groups (treated with either PPMO-C or PBS). To examine whether this reduction of viral production in the myocardium was sufficient to attenuate the severity of myocarditis-associated tissue damage, the histopathology of stained midportion ventricular tissue sections was analyzed. The pathological grade of myocarditis in PPMO-6-treated mice was significantly lower than in PPMO-C- or PBS-treated mice (Fig. 7b). Extensive myocardial infiltration by inflammatory cells and myocyte necrosis (grades 4 to 5) were observed in tissues of the PPMO-C- and PBS-treated mice, whereas PPMO-6-treated mice had absent to mild (grade 0 to 1) tissue damage (Fig. 7c through f). In addition, the tissue damage to pancreas, liver, and spleen, as evaluated visually by microscopy of stained tissue slices, was far less in PPMO-6-treated mice than in the two control groups (data not shown).
FIG. 7.
PPMO-6 inhibits CVB3 replication in vivo and attenuates the severity of murine myocarditis. (a) CVB3 plaque assay of heart tissue after PPMO treatment. Mice (n = 6) subjected to intravenous treatments of 200 μg of PPMO-6, PPMO-C, or PBS at 3 h before and 2 days after infection with 105 PFU CVB3 were euthanized at day 7 p.i., and organs were harvested. The apical ventricular portions of hearts from each group were pooled and used for plaque assays. The titer difference between PPMO-6 and the control mice (PPMO-C and PBS) was statistically significant (*, P < 0.05). (b) Histopathology grading of heart tissue damage. The severity of myocarditis in mice treated with PPMO-6 is significantly lower than that of mice treated with PPMO-C or PBS (*, P < 0.05). (c to f) Hematoxylin-eosin-stained heart tissue sections (magnification, ×200). Heart tissue from noninfected mice (c) or infected mice treated with (d) PPMO-6, (e) PPMO-C, or (f) PBS shows marked differences in levels of CVB3-induced damage. Extensive inflammation and tissue damage are visible in PPMO-C- and PBS-treated mouse hearts compared to those of PPMO-6-treated and noninfected mice.
DISCUSSION
Coxsackievirus B3 can infect and injure multiple tissues in mice and humans, leading to myocarditis, pancreatitis, hepatitis, and meningitis (19). Although the specific mechanisms causing viral myocarditis remain somewhat unclear, it is known that CVB3 can cause direct damage to heart tissue and apoptosis of cardiomyocytes (18). Such injury to the heart can result in a weakened status or progression to heart failure. Therefore, directly reducing CVB3 replication in the heart is a rational treatment strategy against CVB3-induced myocarditis. Recently, several nucleic acid-based compounds, including PS-ODNs and small interfering RNAs, have been reported to have anti-CVB3 activity (20, 33, 37, 38). However, limitations in stability and in cellular entry in vivo appear to have impeded their further development as drugs. In this study, we investigated the effects of PMO, an enzymatically stable type of antisense oligomer, against CVB3 in both cell culture and a mouse model. PMO were conjugated to a cell-penetrating ARP to provide improved delivery into cells in culture and in vivo. PPMO were designed to base pair with sequence in the 5′- and 3′UTRs of CVB3 RNA that has been established previously as important in viral translation and/or RNA synthesis. One PPMO in particular, PPMO-6, demonstrated potent and specific anti-CVB3 activity, generating >100-fold reductions in viral titers in cell cultures and in mouse heart tissue in vivo.
PPMO-6 was over 200-fold more active in cell culture experiments than the most active anti-CVB3 PS-ODN previously reported (33, 37). Given the similar experimental conditions used in those previous studies and this study, we conclude that the greater specific antiviral activity of PPMO is due, at least partially, to its structural characteristics. The PPMO backbone confers stability against nuclease digestion, and this characteristic was evident in our study, which showed that 5 days after a single 4-h treatment, PPMO-6 retained 47% of the antiviral activity that it was capable of generating on the original day of the treatment of CVB-infected cardiomyocyte cultures.
A limitation of PS-ODN chemistry is that, likely due to an anionic character, they bind to certain cellular proteins, such as heparin-binding proteins, thereby producing cellular toxicity (2). Likewise, although the backbone of PPMO is neutral, the P007 peptide is highly positively charged, which could be expected to cause some toxicity. We did not investigate this issue in depth; however, viral titers were lowered over 99% in cell cultures at PPMO concentrations that resulted in a negligible loss of cell viability. Furthermore, highly significant antiviral activity in vivo was observed at a dose level (200 μg) that, after two intravenous treatments, resulted in no apparent ill effects to the mice.
In this study, only two (PPMO-6 and -7) of the eight PPMO designed against CVB3 exhibited antiviral activity. Other investigators have provided various explanations for why some PPMO may lack efficacy (7, 8, 15, 24), including conformational incompatibility between a PPMO and its RNA target, such as a severe RNA secondary structure, that may interfere with hybridization. Other explanations for low activity include the inaccessibility of an RNA target due to ultrastructural constraints or simply the inability of a PPMO that is successfully bound to viral RNA to significantly impact virus production. We made no attempt to gain mechanistic insight as to why some PPMO worked and others did not against whole virus in this study. Previous work has demonstrated that the RNA sequence that lies between loops G and H of the CVB3 IRES, spanning genomic nt 547 to 592, is critical to the viral life cycle, as deletions in this sequence region can abolish viral translation and infectivity (3, 16, 36). The targets of both PPMO-6 and -7 lie in this IRES “core sequence,” which includes the polypyrimidine tract region and Shine-Dalgarno-like sequence (35). We therefore assume that PPMO-6 and -7 probably prevent host factors involved in the preinitiation of translation, such as the 40S ribosomal subunit, from associating with critical IRES sequence elements. This assumption is supported by our in vitro translation inhibition assays using bicistronic RNA. Although in vitro translation systems have limited relevance to the molecular events that occur with whole virus in living cells, the results from Fig. 5c show that PMO-6 can act as a potent inhibitor of IRES-mediated translation. Interestingly, the PPMO targeting sequence in the 5′- and 3′-terminal regions of the viral genome and antigenome did not show nearly the level of CVB3 inhibition that corresponding PS-ODNs had in previous studies (33). Together, these differences may imply that PPMO are particularly effective at preventing the assembly or scanning of certain translation preinitiation complexes but that PS-ODNs are able to mediate RNase H-induced damage to viral RNA termini, a process that PPMO cannot engender. The high efficacy of IRES-targeted PPMO suggests that PPMO may prove useful as a reagent in the study of the molecular mechanics of IRES function in picornaviruses.
Various small positively charged peptides have been used to facilitate the transport of proteins or nucleic acid oligomers across cellular membranes. The human immunodeficiency virus Tat protein is probably the most extensively studied protein for this purpose. For instance, an 11-amino-acid transduction domain of Tat protein transduced an apoptosis repressor protein into cells of isolated perfused hearts (11). Various sequences of ARP conjugates have been utilized to facilitate PMO delivery in antiviral cell culture experiments (7, 15, 24, 32). An ARP with the sequence R9F2C increased the in vivo efficacy of a PMO against Ebola virus in a mouse model (8). In this study, we employed an ARP, P007, containing 6-aminohexanoic acid, to enhance the delivery of PMO into cells. Monitoring of fluoresceinated PMO and PPMO indicated that P007 was able to markedly improve the delivery of PMO into cells in culture and in vivo to a number of organs, including the heart, pancreas, and kidney, the major sites of CVB3 infection and replication. The pattern of distribution of Fl-PPMO-C in cultured cells and in mouse organs appeared punctate, indicating that much of the compound may be trapped in endosomes and/or lysosomes. The degree of sequence-specific antiviral activity, however, argues that a significant amount of PMO was able to access RNA targets in cytosol. The effective and nontoxic anti-CVB3 activity of PPMO-6 is likely attributable in part to this particular peptide.
The high efficacies of both PPMO-6 and -7, which target different segments of the IRES “core sequence,” suggest the possibility that other picornavirus infections could be approached with this strategy as well. The low toxicity and substantial antiviral efficacy observed when an intravenous route of administration of PPMO-6 was used are encouraging with respect to further preclinical development. However, the inability of PPMO-6 to more convincingly protect mice from virally induced weight loss is worrisome, and further attempts to improve outcome through improvements to the PPMO compound itself, or in dosing strategy, appear warranted. Future studies with PPMO-6 and -7 will likely include (i) further optimization of the PMO base sequence and delivery peptide, (ii) efforts to generate and characterize viral escape mutants resulting from PPMO use, (iii) examination of the effects of employing multiple PPMO, and (iv) exploration of the effects of dosing regimens on in vivo efficacy and toxicity.
Acknowledgments
We are grateful to the Chemistry Department at AVI BioPharma for expert synthesis, purification, and analysis of all PMO and PPMO used in this study and Reinhard Kandolf (University of Tubingen, Germany) and William C. Claycomb (Louisiana State University) for generously providing us with CVB3 cDNA and HL-1 cells, respectively. We also thank Brian Wong, Hon Leong, and Farnoosh Tayyari for their technical assistance.
This work was supported by grants from the Canadian Institutes of Health Research (MOP-14068) and Heart and Stroke Foundation of British Columbia and Yukon (20R20002).
Footnotes
Published ahead of print on 20 September 2006.
REFERENCES
- 1.Boucek, M. M., A. Faro, R. J. Novick, L. E. Bennett, B. M. Keck, and J. D. Hosenpud. 2001. The registry of the International Society for Heart and Lung Transplantation: fourth official pediatric report—2000. J. Heart Lung Transplant. 20:39-52. [DOI] [PubMed] [Google Scholar]
- 2.Brown, D. A., S. H. Kang, S. M. Gryaznov, L. DeDionisio, O. Heidenreich, S. Sullivan, X. Xu, and M. I. Nerenberg. 1994. Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J. Biol. Chem. 269:26801-26805. [PubMed] [Google Scholar]
- 3.Cheung, P., M. Zhang, J. Yuan, D. Chau, B. Yanagawa, B. McManus, and D. Yang. 2002. Specific interactions of HeLa cell proteins with Coxsackievirus B3 RNA: La autoantigen binds differentially to multiple sites within the 5′ untranslated region. Virus Res. 90:23-36. [DOI] [PubMed] [Google Scholar]
- 4.Chow, L. H., K. W. Beisel, and B. M. McManus. 1992. Enteroviral infection of mice with severe combined immunodeficiency. Evidence for direct viral pathogenesis of myocardial injury. Lab. Investig. 66:24-31. [PubMed] [Google Scholar]
- 5.Chow, L. H., C. J. Gauntt, and B. M. McManus. 1991. Differential effects of myocarditic variants of Coxsackievirus B3 in inbred mice. A pathologic characterization of heart tissue damage. Lab. Investig. 64:55-64. [PubMed] [Google Scholar]
- 6.Claycomb, W. C., N. A. Lanson, Jr., B. S. Stallworth, D. B. Egeland, J. B. Delcarpio, A. Bahinski, and N. J. Izzo, Jr. 1998. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA 95:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deas, T. S., I. Binduga-Gajewska, M. Tilgner, P. Ren, D. A. Stein, H. M. Moulton, P. L. Iversen, E. B. Kauffman, L. D. Kramer, and P.-Y. Shi. 2005. Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. J. Virol. 79:4599-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enterlein, S., K. L. Warfield, D. L. Swenson, D. A. Stein, J. L. Smith, C. S. Gamble, A. D. Kroeker, P. L. Iversen, S. Bavari, and E. Muhlberger. 2006. VP35 knockdown inhibits Ebola virus amplification and protects against lethal infection in mice. Antimicrob. Agents Chemother. 50:984-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldman, A. M., and D. McNamara. 2000. Myocarditis. N. Engl. J. Med. 343:1388-1398. [DOI] [PubMed] [Google Scholar]
- 10.Gauntt, C. J., and M. A. Pallansch. 1996. Coxsackievirus B3 clinical isolates and murine myocarditis. Virus Res. 41:89-99. [DOI] [PubMed] [Google Scholar]
- 11.Gustafsson, A. B., M. R. Sayen, S. D. Williams, M. T. Crow, and R. A. Gottlieb. 2002. TAT protein transduction into isolated perfused hearts: TAT-apoptosis repressor with caspase recruitment domain is cardioprotective. Circulation 106:735-739. [DOI] [PubMed] [Google Scholar]
- 12.Haller, A. A., and B. L. Semler. 1995. Translation and host cell shutoff in human enterovirus infection, p. 113-133. In H. A. Rotbart (ed.), Human enterovirus infections. ASM Press, Washington, D.C.
- 13.Heasman, J. 2002. Morpholino oligos: making sense of antisense? Dev. Biol. 243:209-214. [DOI] [PubMed] [Google Scholar]
- 14.Hudziak, R. M., E. Barofsky, D. F. Barofsky, D. L. Weller, S. B. Huang, and D. D. Weller. 1996. Resistance of morpholino phosphorodiamidate oligomers to enzymatic degradation. Antisense Nucleic Acid Drug Dev. 6:267-272. [DOI] [PubMed] [Google Scholar]
- 15.Kinney, R. M., C.-Y. Huang, B. C. Rose, A. D. Kroeker, T. W. Dreher, P. L. Iversen, and D. A. Stein. 2005. Inhibition of dengue virus serotypes 1 to 4 in Vero cell cultures with morpholino oligomers. J. Virol. 79:5116-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, Z., C. M. Carthy, P. Cheung, L. Bohunek, J. E. Wilson, B. M. McManus, and D. Yang. 1999. Structural and functional analysis of the 5′ untranslated region of coxsackievirus B3 RNA: in vivo translational and infectivity studies of full-length mutants. Virology 265:206-217. [DOI] [PubMed] [Google Scholar]
- 17.McManus, B. M., L. H. Chow, S. J. Radio, S. M. Tracy, M. A. Beck, N. M. Chapman, K. Klingel, and R. Kandolf. 1991. Progress and challenges in the pathological diagnosis of myocarditis. Eur. Heart J. 12(Suppl. D):18-21. [DOI] [PubMed] [Google Scholar]
- 18.McManus, B. M., L. H. Chow, J. E. Wilson, D. R. Anderson, J. M. Gulizia, C. J. Gauntt, K. E. Klingel, K. W. Beisel, and R. Kandolf. 1993. Direct myocardial injury by enterovirus: a central role in the evolution of murine myocarditis. Clin. Immunol. Immunopathol. 68:159-169. [DOI] [PubMed] [Google Scholar]
- 19.Melnick, J. L. 1996. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 655-712. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 20.Merl, S., C. Michaelis, B. Jaschke, M. Vorpahl, S. Seidl, and R. Wessely. 2005. Targeting 2A protease by RNA interference attenuates coxsackieviral cytopathogenicity and promotes survival in highly susceptible mice. Circulation 111:1583-1592. [DOI] [PubMed] [Google Scholar]
- 21.Moulton, H. M., M. H. Nelson, S. A. Hatlevig, M. T. Reddy, and P. L. Iversen. 2004. Cellular uptake of antisense morpholino oligomers conjugated to arginine-rich peptides. Bioconjug. Chem. 15:290-299. [DOI] [PubMed] [Google Scholar]
- 22.Nasevicius, A., and S. C. Ekker. 2000. Effective targeted gene “knockdown” in zebrafish. Nat. Genet. 26:216-220. [DOI] [PubMed] [Google Scholar]
- 23.Nelson, M. H., D. A. Stein, A. D. Kroeker, S. A. Hatlevig, P. L. Iversen, and H. M. Moulton. 2005. Arginine-rich peptide conjugation to morpholino oligomers: effects on antisense activity and specificity. Bioconjug. Chem. 16:959-966. [DOI] [PubMed] [Google Scholar]
- 24.Neuman, B. W., D. A. Stein, A. D. Kroeker, M. J. Churchill, A. M. Kim, P. Kuhn, P. Dawson, H. M. Moulton, R. K. Bestwick, P. L. Iversen, and M. J. Buchmeier. 2005. Inhibition, escape, and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense morpholino oligomers. J. Virol. 79:9665-9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neuman, B. W., D. A. Stein, A. D. Kroeker, A. D. Paulino, H. M. Moulton, P. L. Iversen, and M. J. Buchmeier. 2004. Antisense morpholino-oligomers directed against the 5′ end of the genome inhibit coronavirus proliferation and growth. J. Virol. 78:5891-5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Opalinska, J. B., and A. M. Gewirtz. 2002. Nucleic-acid therapeutics: basic principles and recent applications. Nat. Rev. Drug Discov. 1:503-514. [DOI] [PubMed] [Google Scholar]
- 27.Rohll, J. B., N. Percy, R. Ley, D. J. Evans, J. W. Almond, and W. S. Barclay. 1994. The 5′-untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J. Virol. 68:4384-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein, D., E. Foster, S. B. Huang, D. Weller, and J. Summerton. 1997. A specificity comparison of four antisense types: morpholino, 2′-O-methyl RNA, DNA, and phosphorothioate DNA. Antisense Nucleic Acid Drug Dev. 7:151-157. [DOI] [PubMed] [Google Scholar]
- 29.Summerton, J. 1999. Morpholino antisense oligomers: the case for an RNase H-independent structural type. Biochim. Biophys. Acta 1489:141-158. [DOI] [PubMed] [Google Scholar]
- 30.Summerton, J., and D. Weller. 1997. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 7:187-195. [DOI] [PubMed] [Google Scholar]
- 31.Tayyari, F., T. C. Sutton, H. E. Manson, and R. G. Hegele. 2005. CpG-oligodeoxynucleotides inhibit RSV-enhanced allergic sensitisation in guinea pigs. Eur. Respir. J. 25:295-302. [DOI] [PubMed] [Google Scholar]
- 32.van den Born, E., D. A. Stein, P. L. Iversen, and E. J. Snijder. 2005. Antiviral activity of morpholino oligomers designed to block various aspects of Equine arteritis virus amplification in cell culture. J. Gen. Virol. 86:3081-3090. [DOI] [PubMed] [Google Scholar]
- 33.Wang, A., P. K. M. Cheung, H. Zhang, C. M. Carthy, L. Bohunek, J. E. Wilson, B. M. McManus, and D. Yang. 2001. Specific inhibition of coxsackievirus B3 translation and replication by phosphorothioate antisense oligodeoxynucleotides. Antimicrob. Agents Chemother. 45:1043-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warfield, K. L., D. L. Swenson, G. G. Olinger, D. K. Nichols, W. D. Pratt, R. Blouch, D. A. Stein, M. J. Aman, P. L. Iversen, and S. Bavari. 2006. Gene-specific countermeasures against Ebola virus based on antisense phosphorodiamidate morpholino oligomers. PLoS Pathog. 2:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, D., P. Cheung, Y. Sun, J. Yuan, H. Zhang, C. M. Carthy, D. R. Anderson, L. Bohunek, J. E. Wilson, and B. M. McManus. 2003. A Shine-Dalgarno-like sequence mediates in vitro ribosomal internal entry and subsequent scanning for translation initiation of coxsackievirus B3 RNA. Virology 305:31-43. [DOI] [PubMed] [Google Scholar]
- 36.Yang, D., J. E. Wilson, D. R. Anderson, L. Bohunek, C. Cordeiro, R. Kandolf, and B. M. McManus. 1997. In vitro mutational and inhibitory analysis of the cis-acting translational elements within the 5′ untranslated region of coxsackievirus B3: potential targets for antiviral action of antisense oligomers. Virology 228:63-73. [DOI] [PubMed] [Google Scholar]
- 37.Yuan, J., P. K. Cheung, H. Zhang, D. Chau, B. Yanagawa, C. Cheung, H. Luo, Y. Wang, A. Suarez, B. M. McManus, and D. Yang. 2004. A phosphorothioate antisense oligodeoxynucleotide specifically inhibits coxsackievirus B3 replication in cardiomyocytes and mouse hearts. Lab. Investig. 84:703-714. [DOI] [PubMed] [Google Scholar]
- 38.Yuan, J., P. K. M. Cheung, H. M. Zhang, D. Chau, and D. Yang. 2005. Inhibition of coxsackievirus B3 replication by small interfering RNAs requires perfect sequence match in the central region of the viral positive strand. J. Virol. 79:2151-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]