Abstract
In addition to the phosphoprotein, the P gene of measles virus (MV) also encodes the V and C proteins by an RNA editing process and by alternative initiation of translation in a different reading frame, respectively. Although the MV C protein is required for efficient MV replication in vivo and in some cultured cells, its exact functions in virus infection are currently unclear. Here, we report that a recombinant MV lacking the C protein (MVΔC) grew poorly in a human cell line possessing the intact interferon (IFN) pathway and that this growth defect was associated with reduced viral translation and genome replication. The translational inhibition was correlated with phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Moreover, increased IFN induction was observed in MVΔC-infected cells. The NS1 protein of influenza virus, which binds to double-stranded RNA (dsRNA) and consequently inhibits IFN induction and dsRNA-dependent protein kinase activation, complemented the growth defect of MVΔC. These results indicate that the MV C protein inhibits IFN induction and modulates host antiviral responses, thereby ensuring MV growth in host cells.
Measles virus (MV), a member of the genus Morbillivirus of the family Paramyxoviridae, is an enveloped virus with a nonsegmented negative-sense RNA as a genome. The MV genome has six genes that encode the nucleocapsid (N), phospho- (P), matrix (M), fusion (F), hemagglutinin (H), and large (L) proteins. In addition to the P protein, the P gene encodes two other proteins, the V and C proteins, by an RNA editing process and by alternative initiation of translation in a different reading frame, respectively (20). A recombinant MV lacking the C protein propagates efficiently in certain cultured cell lines (14, 40), but not in other cell lines (14, 45, 51), human peripheral blood mononuclear cells (PBMCs) (14, 45, 51), or animal models, such as CD46 transgenic mice, severe combined immunodeficiency mice engrafted with human thymus/liver implants, and macaques (38, 51, 55). The cause of its growth defect in vitro and in vivo is currently unknown. In a minigenome system, the MV C protein was found to inhibit viral transcription and genome replication (4, 42). The C protein is also known to play a role in virus assembly (11). Furthermore, one study reported that the C protein inhibits the interferon (IFN) response (45), although other studies failed to confirm this observation (36, 51). The contributions of these activities of the C protein to MV growth and pathogenicity remain to be determined.
Host cells initiate antiviral responses by detecting pathogen-associated molecular patterns, such as double-stranded RNA (dsRNA), highly structured single-stranded RNA, and envelope glycoproteins in the case of viruses (1). Cytoplasmic dsRNAs are detected by two intracellular RNA helicases, RIG-I and mda-5 (1). These helicases transmit signals to a downstream adapter molecule, IPS1, which in turn activates specific kinases (1). Although it has been difficult to obtain direct evidence that dsRNA is produced in negative-stranded RNA virus-infected cells (57), RIG-I was shown to play a major role in IFN induction in these cells (1, 27). The activated kinases phosphorylate IFN-regulatory factors (IRFs), leading to the production of IFN-α/β (28). Following secretion, IFN-α/β bind to IFN-α/β receptors on the surface of adjacent cells, thereby activating the Jak/Stat signaling pathway (28). Phosphorylated Stats activate the transcription of specific genes, referred to as IFN-stimulated genes, containing an IFN-stimulated response element in their transcriptional enhancer sequences (28). The products of the IFN-stimulated genes usually possess activities that inhibit viral growth at various steps of the viral replication cycle. Among the IFN-inducible antiviral proteins, the most well-characterized are the dsRNA-dependent protein kinase R (PKR), 2′,5′-oligoadenylate synthetase, and Mx protein, which play roles in inhibiting virus translation, cleaving viral RNA, and sequestering viral N proteins, respectively (19, 28, 30). PKR, a serine/threonine kinase, is normally inactive but becomes activated by binding dsRNA. Activated PKR phosphorylates the alpha subunit of eukaryotic translation initiation factor 2 (eIF2α), thereby preventing the recycling of initiation factors and consequently inhibiting translation (28).
In order to ensure their growth, many viruses encode proteins in their genome that regulate host antiviral responses (17, 21, 28). These proteins often inhibit molecules involved in the induction, signal transduction, or effector activity of the IFN system. For example, the nonstructural NS1 protein of influenza virus binds dsRNA, thereby inhibiting IFN induction and PKR activation (12, 23). Accordingly, an NS1 protein-deficient influenza virus grows poorly in IFN-competent cell lines (5, 18).
In the present study, we demonstrate that a recombinant MV lacking the C protein was unable to grow efficiently in a human cell line possessing the intact IFN pathway and that this growth defect was linked to reduced viral translation and genome replication. The translational inhibition was correlated with the phosphorylation of eIF2α. Moreover, enhanced IFN induction was observed in the infected cells. The NS1 protein of influenza virus complemented the growth defect of the C protein-deficient MV. Taken together, these results indicate that the MV C protein can modulate host antiviral responses.
MATERIALS AND METHODS
Cells and viruses.
Vero cells constitutively expressing human signaling lymphocyte activation molecule (SLAM), an MV receptor (54), designated Vero/hSLAM cells (37), were maintained in Dulbecco's modified Eagle's medium (DMEM; ICN Biomedicals, Aurora, Ohio) supplemented with 7.5% fetal bovine serum (FBS) and 500 μg/ml Geneticin (G418; Nacalai Tesque, Tokyo, Japan). A549 cells constitutively expressing human SLAM, designated A549/hSLAM cells (49), and Chinese hamster ovary (CHO) cells constitutively expressing human SLAM, designated CHO/hSLAM cells (54), were maintained in RPMI 1640 medium (ICN Biomedicals) supplemented with 10% FBS and 500 μg/ml G418. B95a cells (29) were maintained in RPMI 1640 medium supplemented with 7.5% FBS. HeLa cells constitutively expressing the NS1 protein of the influenza A virus PR8 strain (HeLa/NS1) or the neomycin resistance gene alone (HeLa/neo) were provided by K. Takeda. These cells were generated by transfecting HeLa cells with pCXN2-FLAG-PR8-NS1, encoding the FLAG-tagged PR8-NS1 protein, or pCXN2 (35), respectively, and maintained in DMEM supplemented with 7.5% FBS and 500 μg/ml G418. Recombinant MVs were generated as described previously (33, 47, 48). Briefly, CHO/hSLAM cells were infected with vTF7-3 (15), a vaccinia virus encoding the T7 RNA polymerase (a gift from B. Moss), and transfected with a full-length genome plasmid that encodes the antigenome of MV under the control of the T7 promoter and three support plasmids, pCAG-T7-IC-N, pCAG-T7-IC-PΔC, and pGEMCR-9301B-L (48). The cells were then incubated in the above-described medium containing 50 μg/ml z-Asp-CH2-DCB (Peptide Institute Inc., Osaka, Japan), a caspase inhibitor (33, 47, 48). On the next day, the transfected CHO/hSLAM cells were cocultured with B95a cells to amplify the recombinant MV rescued from the full-length genome plasmid. Since B95a is an Epstein-Barr virus-transformed marmoset B-lymphoid cell line, the viruses underwent one passage in Vero/hSLAM cells, which are nonsusceptible to the Epstein-Barr virus. All the recombinant MVs used in the present study had a transcriptional unit of enhanced green fluorescent protein (EGFP) at the first 3′ locus of the virus genome (22). Throughout the study, multiplicities of infection (MOIs) were calculated using the virus titers determined on Vero/hSLAM cells. Since A549- and HeLa-derived cell lines show lower infectivity titers after MV infection than Vero-derived cell lines, the MOIs for the A549- and HeLa-derived cell lines may be overestimated.
Reagents and antibodies.
A fusion blocking peptide, Z-D-Phe-Phe-Gly (Peptide Institute Inc.) (43), was used at 100 μg/ml in appropriate experiments. The serum of a patient with subacute sclerosing panencephalitis (59), a cocktail of mouse monoclonal antibodies against the MV M protein (Chemicon, Temecula, CA), a rabbit polyclonal antibody against the MV H protein (a gift from T. Kohama), and a rabbit polyclonal antibody against the MV F protein (a gift from R. Cattaneo) (8) were used to detect the N, M, H, and F proteins, respectively. Antibodies for detecting the P, V, and C proteins were raised against the peptides (C)PPNPSRASTSETPIKKGT, CRTDTGVDTRIWYHNLPEIPE, and (C)WPSRKPWQHGQKYQTTQDRTE, respectively. These peptides correspond to 18 amino acids at positions 215 to 232 of the P protein, 21 amino acids at positions 279 to 299 of the V protein, and 21 amino acids at positions 20 to 40 of the C protein, respectively. The cysteines (C) at the termini of the peptides were used for coupling to keyhole limpet hemocyanin for immunization of rabbits. A monoclonal antibody, JL-8 (BD Biosciences/Clontech, Palo Alto, CA), was used for detecting EGFP. A horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin (Ig) antibody (Amersham Biosciences, Piscataway, NJ), a goat anti-human Ig antibody (EY Laboratories, San Mateo, CA), and a sheep anti-mouse Ig antibody (Amersham Biosciences) were used as secondary antibodies. For indirect immunofluorescence labeling, a mouse monoclonal antibody against human IRF3 (SL-12.1; BD PharMingen, Heidelberg, Germany) and a rabbit polyclonal antibody against eIF2α phosphorylated at Ser51 (Cell Signaling Technology, Beverly, MA) were used in combination with Alexa Fluor 594-conjugated donkey anti-mouse and anti-rabbit IgG (H+L) antibodies (Molecular Probes, Eugene, OR), respectively.
Construction of full-length genome plasmids.
All full-length genome plasmids were derived from p(+)MV323, which encodes the full-length antigenomic cDNA of the virulent IC-B strain of MV (50). The plasmid p(+)MV323-EGFP, containing an additional transcriptional unit of EGFP, was described previously (22). p(+)MVΔC-EGFP was generated by introducing an opal mutation and an amber mutation in the reading frame of the C protein at amino acid positions 2 and 6, respectively. These mutations were synonymous in the reading frame of the P and V proteins. The viruses rescued from p(+)MV323-EGFP and p(+)MVΔC-EGFP are referred to as MV wild type (MVwt) and MVΔC, respectively. MVΔC was incapable of producing the C protein, as reported previously (51). The plasmid p(+)MV323/EdH-EGFP, which encodes the Edmonston H protein in place of the IC-B H protein, was described previously (22). The plasmid p(+)MVΔC/EdH-EGFP was generated by replacing the PacI-SpeI fragment containing the open reading frame of the H protein of p(+)MVΔC-EGFP with the corresponding fragment of p(+)MV2A (a gift from M. A. Billeter) encoding the full-length antigenomic cDNA of the Edmonston strain (41). The viruses rescued from p(+)MV323/EdH-EGFP and p(+)MVΔC/EdH-EGFP are referred to as MVwt-EdH and MVΔC-EdH, respectively.
Virus titration.
Monolayers of Vero/hSLAM cells on six-well plates were incubated with serially diluted virus samples for 1 h at 37°C. The monolayers were then washed with phosphate-buffered saline (PBS) and overlaid with DMEM containing 5% FBS and 0.5% agarose. At 3 days postinfection (p.i.), the PFU were determined by counting the numbers of plaques under a fluorescence microscope.
Reverse transcription-quantitative PCR (RT-QPCR).
Total RNA was extracted from virus-infected cells with the TRIzol reagent (Life Technologies, Gaithersburg, MD), treated with DNase I, and reverse transcribed into cDNAs using SuperScript II reverse transcriptase (Invitrogen Life Technologies, Carlsbad, CA). The cDNAs of the viral RNA genome, viral mRNAs, and cellular IFN-β and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNAs were quantified using SYBR Premix Ex Taq (TaKaRa Bio Inc., Shiga, Japan) and a LightCycler (Roche Diagnostics, Indianapolis, IN) as described previously (47, 49). For quantification of the cDNA of the IFN-β mRNA, we used the primer pair 5′-CTCCTGGCTAATGTCTATCA-3′ and 5′-GCAGAATCCTCCCATAATAT-3′. Data were analyzed with the LightCycler software (version 3.5; Roche Diagnostics).
ELISA.
Monolayers of A549/hSLAM cells were infected with MVwt or MVΔC at an MOI of 0.5. At 48 h p.i., the amount of IFN-β in the culture medium was determined by enzyme-linked immunosorbent assay (ELISA) using a HuIFN-β ELISA kit (Fujirebio Inc., Tokyo, Japan).
Immunoblotting.
Monolayers of A549/hSLAM, HeLa/neo, and HeLa/NS1 cells were infected with each virus. At 24 h p.i., the cells were lysed in passive lysis buffer (Promega, Madison, WI). The polypeptides in the cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted onto polyvinylidene difluoride membranes (Hybond-P; Amersham Biosciences), and incubated with primary antibodies against the MV proteins diluted in Tris-buffered saline and 0.05% Tween 20 (TBST) containing 5% nonfat dry milk at 4°C for 16 h. The membranes were then washed five times with TBST and incubated with peroxidase-conjugated secondary antibodies for 1 h at room temperature. After five washes with TBST, the membranes were treated with the ECL Plus reagent (Amersham Biosciences), and chemiluminescent signals on the membranes were detected and visualized using a VersaDoc 3000 imager (Bio-Rad, Hercules, CA). The relative amounts of the proteins were determined by densitometry using the Quantity One software (Bio-Rad).
Immunofluorescence staining.
Cells were seeded on coverslips in six-well plates and infected with each virus in the presence of the fusion blocking peptide. At 36 h p.i., the cells were fixed and permeabilized with PBS containing 2.5% formaldehyde and 0.5% Triton X-100. The fixed cells were washed with PBS and then sequentially incubated with a primary antibody against IRF3 (SL-12.1) or phosphorylated eIF2α for 1 h at room temperature, followed by an Alexa Fluor 594-conjugated secondary antibody for 1 h at room temperature. Nuclear DNA was stained with 4′,6′-diamidino-2-phenylindole (DAPI; Nacalai Tesque) at 0.2 μg/ml. Cells were observed using a confocal microscope (Radiance 2100; Bio-Rad).
RESULTS
Characterization of a recombinant MV lacking the C protein.
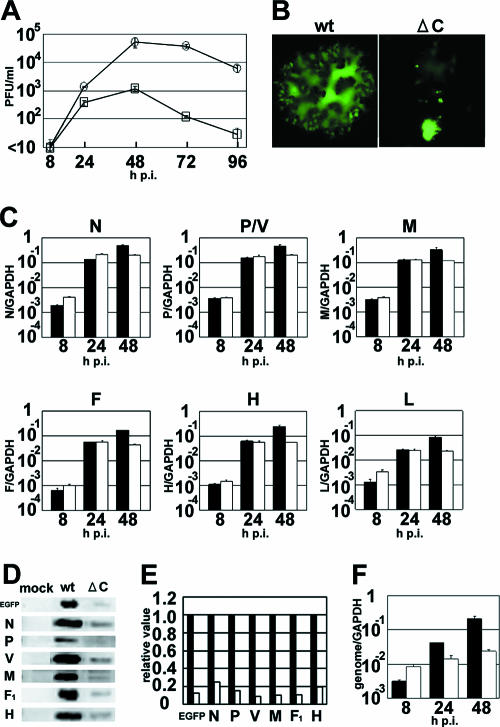
A C protein-deficient MV has been reported to exhibit a growth defect in vitro and in vivo (14, 38, 45, 51, 55). In an attempt to determine the cause of this defect, we examined whether MVΔC, a C protein-deficient recombinant MV based on the wild-type IC-B strain, could grow in A549 cells constitutively expressing human SLAM (A549/hSLAM cells) (Fig. 1A). A549 cells have functional IFN induction and signal transduction pathways and are commonly used to study interactions between the IFN system and negative-stranded RNA viruses (46, 56). All the recombinant MVs used in the present study expressed EGFP. MVΔC replicated poorly in A549/hSLAM cells, and its peak titer was almost 2 logs lower than that of the parental MVwt (Fig. 1A). Moreover, the cytopathic effects in MVΔC-infected cells were different from those in MVwt-infected cells. At 48 h p.i., cells infected with MVΔC at an MOI of 0.01 had rounded up and detached from the culture dish, while those infected with MVwt at the same MOI formed large syncytia (Fig. 1B). These results for A549/hSLAM cells were similar to previous observations for 293 cells expressing human SLAM (51), confirming that this type of cytopathic effect is characteristic of MVΔC-infected cells.
FIG. 1.
Characterization of the recombinant C protein-deficient MV during multiple steps of growth. (A) Replication kinetics. A549/hSLAM cells were infected with MVwt or MVΔC at an MOI of 0.01. At various intervals, cells were scraped into the culture medium and the virus titers were determined on Vero/hSLAM cells. Circles, MVwt; squares, MVΔC. The data represent the means ± standard deviations of triplicate samples. (B) Syncytium formation. A549/hSLAM cells were infected with MVwt or MVΔC at an MOI of 0.01. At 48 h p.i., the cells were observed under a fluorescence microscope. (C) Quantification of viral transcripts. A549/hSLAM cells were infected with MVwt or MVΔC at an MOI of 0.5. At 8, 24, and 48 h p.i., the N, P/V, M, F, H, and L mRNAs in the virus-infected cells were quantified by RT-QPCR. All data were normalized by the corresponding level of GAPDH mRNA. Filled bars, MVwt; open bars, MVΔC. The data represent the means ± standard deviations of triplicate samples. (D) Detection of viral proteins by immunoblotting. A549/hSLAM cells were infected with MVwt or MVΔC at an MOI of 0.5. At 24 h p.i., EGFP and the N, P, V, M, F (F1 subunit), and H proteins in the virus-infected cells were detected by immunoblotting using appropriate primary and secondary antibodies. (E) Quantification of the chemiluminescence signals of the immunoblotting shown in panel D. Filled bars, MVwt; open bars, MVΔC. (F) Quantification of the viral genome by RT-QPCR. A549/hSLAM cells were infected with MVwt or MVΔC at an MOI of 0.5. Total RNA samples extracted from the infected cells were reverse transcribed into cDNAs with genome-specific primers, and the amounts of the cDNA for the genome were quantified by RT-QPCR. Filled bars, MVwt; open bars, MVΔC. The data represent the means ± standard deviations of triplicate samples.
To determine the defective step or steps of the infectious cycle of MVΔC, we used RT-QPCR to examine the amounts of viral mRNAs in cells infected with MVwt or MVΔC at an MOI of 0.5 (Fig. 1C). Similar amounts of all the viral mRNAs were detected in cells infected with either MVΔC or MVwt at 8 and 24 h after infection. However, at 48 h p.i., the amounts of all the viral mRNAs were severalfold lower in MVΔC-infected cells than in MVwt-infected cells. Next, we performed Western blot analyses on the amounts of viral proteins at 24 h after infection at an MOI of 0.5, when all the viral mRNAs were present at similar amounts in MVwt- and MVΔC-infected cells. The amounts of viral proteins and virus-encoded EGFP were lower in MVΔC-infected cells than in MVwt-infected cells (Fig. 1D). Quantitative analyses revealed that the N and H protein levels were ∼5-fold lower in MVΔC-infected cells than in MVwt-infected cells, while the relative amounts of EGFP and the P, V, M, and F (F1 subunit) proteins were even lower in MVΔC-infected cells (Fig. 1E). Furthermore, we carried out RT-QPCR to analyze the amounts of genomic RNAs in cells infected with MVwt or MVΔC at several time points after infection at an MOI of 0.5 (Fig. 1F). Although the amount of genomic RNA increased with time in MVwt-infected cells, it hardly changed in MVΔC-infected cells from 8 to 48 h after infection. These results suggest that MVΔC cannot replicate efficiently in A549/hSLAM cells, at least partly because it produces much lower amounts of viral proteins and genomic RNA in infected cells than MVwt, despite the fact that it expresses almost equal amounts of viral mRNAs to MVwt.
Viral transcription and translation in MVΔC-infected cells in the presence of a fusion blocking peptide.
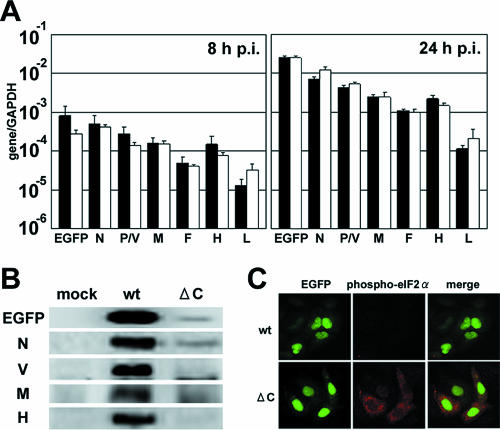
The above experiments were performed under conditions in which multiple steps of MV growth occurred. To exclude the possible effect of multiple rounds of infection on the findings, we repeated the experiments in the presence of a fusion-blocking peptide that can prevent secondary infections from taking place. The amounts of viral mRNAs in MVΔC-infected cells were comparable to those in MVwt-infected cells at 8 h and 24 h p.i. under these experimental conditions (Fig. 2A). The amounts of viral proteins and virus-encoded EGFP in MVΔC-infected cells at 24 h p.i. were again drastically reduced compared with those in MVwt-infected cells (Fig. 2B). These results confirmed that MVΔC growth is inhibited at the step of viral protein production in infected cells.
FIG. 2.
Translational inhibition in MVΔC-infected cells. (A) Quantification of viral transcripts in a single step of growth. A549/hSLAM cells were infected with MVwt or MVΔC at an MOI of 0.5 and incubated in the presence of a fusion-blocking peptide. At 8 and 24 h p.i., the EGFP, N, P/V, M, F, H, and L mRNAs in the virus-infected cells were quantified by RT-QPCR. All data were normalized by the corresponding level of GAPDH mRNA. Filled bars, MVwt; open bars, MVΔC. The data represent the means ± standard deviations of triplicate samples. (B) Detection of viral proteins by immunoblotting. A549/hSLAM cells were infected with MVwt or MVΔC at an MOI of 0.5 and incubated in the presence of a fusion-blocking peptide. At 24 h p.i., EGFP and the N, V, M, and H proteins in the virus-infected cells were detected by immunoblotting using appropriate primary and secondary antibodies. (C) Immunofluorescence staining of phospho-eIF2α in MV-infected cells. A549/hSLAM cells were infected with MVwt or MVΔC at an MOI of 0.5 and incubated in the presence of a fusion-blocking peptide. At 36 h p.i., the cells were fixed and permeabilized, and phospho-eIF2α was detected by indirect immunofluorescence staining using a phospho-eIF2α-specific antibody. Green and red fluorescence indicates EGFP and phospho-eIF2α, respectively.
Next, we examined a possible cause of the translational inhibition. eIF2α is a component of the translation initiation complex, and virus infections are generally known to induce phosphorylation of eIF2α, which in turn inhibits translation (16, 24). Immunofluorescence labeling revealed that ∼50% of the MVΔC-infected cells showed stronger signals for phosphorylated eIF2α than the surrounding uninfected cells, while the signals of most MVwt-infected cells remained as weak as those of uninfected cells (Fig. 2C). These data suggest that enhanced phosphorylation of eIF2α accounts, at least partly, for the translational inhibition observed in MVΔC-infected cells.
Increased IFN induction and production in MVΔC-infected cells.
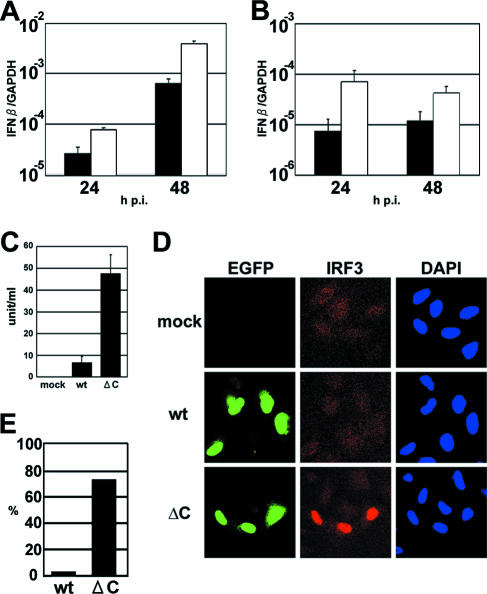
A C protein-deficient MV has been reported to grow normally in Vero cells (14, 40, 45), which have a defect in IFN production (32). Moreover, eIF2α is phosphorylated by the IFN-inducible PKR (3, 58). Therefore, we considered that the growth defect of MVΔC may be related to the IFN response and examined the levels of IFN induction in MVΔC- and MVwt-infected cells (Fig. 3A and B). Regardless of the presence of the fusion-blocking peptide, the amounts of IFN-β mRNA were 5- to 10-fold higher in MVΔC-infected cells than in MVwt-infected cells at all the time points examined. The amounts of secreted IFN-β were analyzed by ELISA and found to be higher in medium samples from MVΔC-infected cells than in those from MVwt-infected cells (Fig. 3C). The distribution of IRF3, a transcription factor for the IFN-β gene, in the infected cells was analyzed by immunofluorescence staining (Fig. 3D). The cytoplasm of all the cells was homogenously stained and weakly positive. Nuclear translocation of IRF3 was detected in ∼75% of MVΔC-infected cells, compared to only ∼5% of MVwt-infected cells (Fig. 3E). These results indicate that MVΔC activates IRF3 and induces IFN-β more strongly than MVwt.
FIG. 3.
Increased IFN induction in MVΔC-infected cells. (A) Quantification of IFN-β mRNA in MVΔC-infected cells. A549/hSLAM cells were infected with MVwt or MVΔC at an MOI of 0.5. At 24 and 48 h p.i., total RNAs were extracted from the cells, and the levels of IFN-β mRNA were quantified by RT-QPCR. All data were normalized by the corresponding level of GAPDH mRNA. Filled bars, MVwt; open bars, MVΔC. The data represent the means ± standard deviations of triplicate samples. (B) IFN-β induction in a single step of growth. The experiments shown in panel A were repeated in the presence of a fusion-blocking peptide. (C) Monolayers of A549/hSLAM cells were infected with MVwt or MVΔC at an MOI of 0.5. At 48 h p.i., the amounts of IFN-β in the culture medium were determined by ELISA. (D) Immunofluorescence staining of IRF3 in virus-infected cells. A549/hSLAM cells were infected with MVwt or MVΔC at an MOI of 0.5 and incubated in the presence of a fusion-blocking peptide. At 36 h p.i., the cells were fixed and permeabilized, and IRF3 was detected by indirect immunofluorescence staining. Green and red fluorescence indicates EGFP and IRF3, respectively. Nuclear DNA was stained with DAPI (blue). (E) The percentages of cells with nuclear localization of IRF3 relative to the total number of infected cells were determined using a fluorescence microscope.
Restoration of MVΔC growth by the influenza virus NS1 protein.
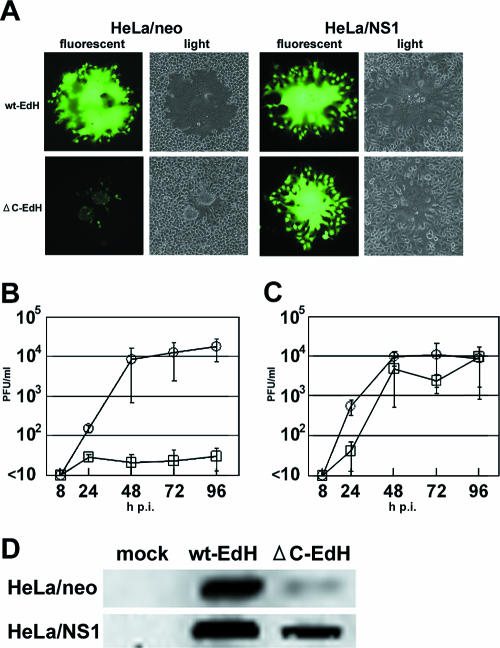
Since the influenza virus NS1 protein is known to counteract the host antiviral responses mediated by the IFN and PKR systems, we next examined whether the NS1 protein can complement the growth defect of MVΔC. To this end, we used HeLa cells constitutively expressing the NS1 protein of the influenza virus PR8 strain (HeLa/NS1 cells). Since MVΔC cannot infect HeLa cells due to their lack of SLAM expression, we prepared MVΔC-EdH containing the H protein of the Edmonston strain instead of the IC-B H protein. This virus can use the ubiquitously expressed CD46 as an alternative receptor (13, 34, 54).
We infected HeLa/neo and HeLa/NS1 cells with MVwt-EdH or MVΔC-EdH at an MOI of 0.01 (Fig. 4A). HeLa/neo cells infected with MVΔC-EdH rounded up and detached from the culture dish, similar to MVΔC-infected A549/hSLAM cells (Fig. 1B), while MVwt-EdH induced large syncytia in the cells. In HeLa/NS1 cells, MVΔC-EdH caused large syncytia similar to those induced by MVwt-EdH. We next examined the growth of MVΔC-EdH in HeLa/neo and HeLa/NS1 cells infected at an MOI of 0.01. MVΔC-EdH hardly replicated in HeLa/neo cells, unlike MVwt-EdH (Fig. 4B), consistent with a previous report (45). On the other hand, MVΔC-EdH grew much more efficiently in HeLa/NS1 cells, although its growth was not as efficient as that of MVwt-EdH (Fig. 4C). The amount of the N protein in HeLa/NS1 cells infected with MVΔC-EdH at 48 h p.i. was partially restored, compared with the amount in HeLa/neo cells (Fig. 4D). A similar result was obtained for the H protein (data not shown). These results indicate that the influenza virus NS1 protein can complement the growth defect of MVΔC in IFN-competent cells.
FIG. 4.
Restoration of the growth of MVΔC by the influenza virus NS1 protein. (A) Syncytium formation. HeLa/neo and HeLa/NS1 cells were infected with MVwt-EdH or MVΔC-EdH at an MOI of 0.01. At 48 h p.i., the cells were observed under a light or fluorescence microscope. (B and C) Replication kinetics. HeLa/neo (B) and HeLa/NS1 (C) cells were infected with MVwt-EdH or MVΔC-EdH at an MOI of 0.01. At various intervals, the cells were scraped into the culture medium, and the virus titers were determined on Vero/hSLAM cells. Circles, MVwt-EdH; squares, MVΔC-EdH. The data represent the means ± standard deviations of triplicate samples. (D) Detection of viral proteins by immunoblotting. HeLa/neo or HeLa/NS1 cells were infected with MVwt-EdH or MVΔC-EdH at an MOI of 0.5 and then incubated in the presence of a fusion-blocking peptide. At 48 h p.i., the N protein levels in the virus-infected cells were detected by immunoblotting.
DISCUSSION
A C protein-deficient MV grows well in some cell lines but not in other cell lines, such as HeLa and 293 cells, human peripheral blood mononuclear cells, or cynomolgus monkeys (14, 38, 40, 45, 51). Although these observations suggest that the C protein of MV is a cell-type-specific virulence factor, the mechanism by which it supports viral growth in certain cells remains unclear. In the present study, we found that MVΔC grew poorly in A549/hSLAM cells, due to reduced production of viral proteins and genomic RNA, compared with MVwt. Since the N protein of MV converts the viral RNA-dependent RNA polymerase from the transcription mode to the genome replication mode (39), it is likely that the decreased genomic RNA synthesis in MVΔC-infected cells results from a reduction in the level of newly synthesized N protein.
Previously, the MV C protein was found to inhibit viral transcription and genome replication in a minigenome system (4, 42). We also observed a similar inhibitory effect of the C protein of the IC-B strain in our minigenome assay system and virus rescue system (48) (unpublished observation). These findings are somewhat in conflict with the finding that MVΔC-infected cells produced almost the same amounts of viral mRNAs as MVwt-infected cells. We speculate that the C protein expression level may be controlled such that it does not inhibit viral RNA synthesis in infected cells. The MV C protein was also reported to play a role in virus assembly (11). However, we found that MVΔC replicated well in HeLa/NS1 cells, as well as in Vero/hSLAM cells (data not shown). Thus, the role of the MV C protein in virus assembly may not be critical for MV replication in cultured cells.
In the present study, the level of phosphorylated eIF2α was higher in MVΔC-infected cells than in MVwt-infected cells, accounting for the reduced translation in the former cells. It will be of great interest to clarify whether the translational inhibition in MVΔC-infected cells is viral protein specific or generally found for all viral and cellular proteins. At present, we are unable to answer this question because the low titers of our MVΔC stocks have prevented us from performing experiments at high MOIs. We observed that MVΔC induced stronger nuclear translocation of IRF3 and higher IFN-β production than MVwt. Shaffer et al. showed that neutralizing antibodies against IFN-α/β could restore the growth defect of a C protein-deficient MV (45), whereas Takeuchi et al. (51) and ourselves observed only small effects of anti-IFN-α/β antibodies on the growth of MVΔC (data not shown). These observations suggest that viral stress-inducible genes other than IFNs (44, 53) may also contribute to the growth defect of MVΔC. Furthermore, constitutively expressed PKR may be activated by dsRNA after virus infection and subsequently phosphorylate eIF2α, thereby inhibiting translation even in the presence of anti-IFN antibodies. Nevertheless, our data suggest that the translational inhibition and growth defect of MVΔC are at least partly related to increased IFN induction in MVΔC-infected cells.
A C protein-deficient MV has been reported to grow well in Vero, B95a, and human neuroblastoma SK-N-MC cells (14, 38, 40, 45, 51). We also observed that MVΔC replicated well in Vero/hSLAM cells, although the plaque sizes were smaller than those of MVwt (data not shown). Importantly, these cell lines have a defect in the IFN and/or PKR system (10, 26, 32). Thus, the C protein of MV is likely to be able to modulate the IFN and PKR systems, since MVΔC showed growth in cells with defects in these systems. The C protein of the MV Edmonston strain, a laboratory strain that has adapted to a variety of cultured cells, inhibits the Jak-Stat pathway of IFN-α/β signaling (45). However, we and others have failed to detect any inhibition of this pathway by the C protein of the wild-type IC-B strain (viruses derived from this strain were used in the present study) or that of the Edmonston strain (36, 51). Therefore, we consider that the C protein of the MV IC-B strain inhibits the induction of IFN but probably not the Jak-Stat pathway. The V proteins of many paramyxoviruses counteract mda-5, which detects cytoplasmic dsRNA and transmits a signal for IFN induction (2). In a reporter assay using expression plasmids, the V protein of the MV IC-B strain did indeed block the signal of mda-5 (unpublished observation). Since MVΔC produces the V protein, the insufficient blockage of IFN induction by MVΔC may indicate that MV needs both the V and C proteins to effectively inhibit IFN induction and ensure its replication in cells.
Many viruses produce some proteins to evade host antiviral responses (17, 21). We examined whether the NS1 protein of influenza virus could complement the growth of MVΔC in HeLa cells, since a previous study indicated that MV lacking the C protein is unable to grow efficiently in this cell line (45). We found that MVΔC grew and translated viral proteins almost as efficiently as MVwt in HeLa cells constitutively expressing the NS1 protein, indicating that the NS1 protein can counteract the translational inhibition in MVΔC-infected cells and support efficient growth of MVΔC. Thus, the C protein of MV appears to have a similar function to the NS1 protein of influenza virus. However, unlike the influenza virus NS1 protein, the MV C protein did not block IFN induction in a standard reporter assay using poly(I-C) as an inducer (unpublished observation). Therefore, these two proteins may differ somewhat in their exact functions.
Some viral proteins that modulate the PKR pathway can also regulate viral RNA synthesis (7, 25, 31, 52). These viral proteins generally have the capacity to bind RNA (6, 9, 25). The NS1 protein of influenza virus, Tat protein of human immunodeficiency virus type 1, NS5A protein of hepatitis C virus, and E3L protein of vaccinia virus can bind to RNA specifically or nonspecifically and modulate IFN induction, PKR activity, and/or viral RNA synthesis (6, 7, 9, 17, 25). The C protein of MV may have similar properties to these proteins. In fact, previous studies have indicated that the C protein of MV can regulate viral RNA synthesis (4, 42), and our present study showed that it can modulate the IFN and PKR systems. It will be of interest to examine the RNA-binding activity of the MV C protein, since it has abundant basic amino acids, a property found in other RNA-binding proteins. Studies are currently in progress to elucidate the precise mechanism by which the C protein of MV modulates the host innate immunity.
Acknowledgments
We thank H. Minagawa and H. Kawaguchi for helpful discussions and T. Kohama, R. Cattaneo, B. Moss, M. A. Billeter, and K. Takeda for providing the anti-MV H antibody, anti-MV-F antibody, recombinant vaccinia virus, p(+)MV-2A plasmid, and cell lines (HeLa/neo and HeLa/NS1), respectively.
This work was supported by grants from the Ministry of Education, Science and Culture of Japan, the Ministry of Health, Labor and Welfare of Japan, the Japan Society for the Promotion of Science, and the Uehara Memorial Foundation.
Footnotes
Published ahead of print on 20 September 2006.
REFERENCES
- 1.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124:783-801. [DOI] [PubMed] [Google Scholar]
- 2.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balachandran, S., P. C. Roberts, L. E. Brown, H. Truong, A. K. Pattnaik, D. R. Archer, and G. N. Barber. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13:129-141. [DOI] [PubMed] [Google Scholar]
- 4.Bankamp, B., J. Wilson, W. J. Bellini, and P. A. Rota. 2005. Identification of naturally occurring amino acid variations that affect the ability of the measles virus C protein to regulate genome replication and transcription. Virology 336:120-129. [DOI] [PubMed] [Google Scholar]
- 5.Bergmann, M., A. Garcia-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burd, C. G., and G. Dreyfuss. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265:615-621. [DOI] [PubMed] [Google Scholar]
- 7.Cai, R., B. Carpick, R. F. Chun, K. T. Jeang, and B. R. Williams. 2000. HIV-I TAT inhibits PKR activity by both RNA-dependent and RNA-independent mechanisms. Arch. Biochem. Biophys. 373:361-367. [DOI] [PubMed] [Google Scholar]
- 8.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien, C. Y., R. Tejero, Y. Huang, D. E. Zimmerman, C. B. Rios, R. M. Krug, and G. T. Montelione. 1997. A novel RNA-binding motif in influenza A virus non-structural protein 1. Nat. Struct. Biol. 4:891-895. [DOI] [PubMed] [Google Scholar]
- 10.Clarke, P. A., N. A. Sharp, and M. J. Clemens. 1990. Translational control by the Epstein-Barr virus small RNA EBER-1. Reversal of the double-stranded RNA-induced inhibition of protein synthesis in reticulocyte lysates. Eur. J. Biochem. 193:635-641. [DOI] [PubMed] [Google Scholar]
- 11.Devaux, P., and R. Cattaneo. 2004. Measles virus phosphoprotein gene products: conformational flexibility of the P/V protein amino-terminal domain and C protein infectivity factor function. J. Virol. 78:11632-11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donelan, N. R., C. F. Basler, and A. Garcia-Sastre. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 77:13257-13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorig, R. E., A. Marcil, A. Chopra, and C. D. Richardson. 1993. The human CD46 molecule is a receptor for measles virus (Edmonston strain). Cell 75:295-305. [DOI] [PubMed] [Google Scholar]
- 14.Escoffier, C., S. Manie, S. Vincent, C. P. Muller, M. Billeter, and D. Gerlier. 1999. Nonstructural C protein is required for efficient measles virus replication in human peripheral blood cells. J. Virol. 73:1695-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gale, M., Jr., S. L. Tan, and M. G. Katze. 2000. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 64:239-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Sastre, A. 2002. Mechanisms of inhibition of the host interferon alpha/beta-mediated antiviral responses by viruses. Microbes Infect. 4:647-655. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 19.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 20.Griffin, D. E. 2001. Measles virus, p. 1401-1441. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 21.Haller, O., G. Kochs, and F. Weber. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimoto, K., N. Ono, H. Tatsuo, H. Minagawa, M. Takeda, K. Takeuchi, and Y. Yanagi. 2002. SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein. J. Virol. 76:6743-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsiao, J. C., C. S. Chung, R. Drillien, and W. Chang. 2004. The cowpox virus host range gene, CP77, affects phosphorylation of eIF2 alpha and vaccinia viral translation in apoptotic HeLa cells. Virology 329:199-212. [DOI] [PubMed] [Google Scholar]
- 25.Huang, L., J. Hwang, S. D. Sharma, M. R. Hargittai, Y. Chen, J. J. Arnold, K. D. Raney, and C. E. Cameron. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 280:36417-36428. [DOI] [PubMed] [Google Scholar]
- 26.Ida-Hosonuma, M., T. Iwasaki, T. Yoshikawa, N. Nagata, Y. Sato, T. Sata, M. Yoneyama, T. Fujita, C. Taya, H. Yonekawa, and S. Koike. 2005. The alpha/beta interferon response controls tissue tropism and pathogenicity of poliovirus. J. Virol. 79:4460-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 28.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 29.Kobune, F., H. Sakata, and A. Sugiura. 1990. Marmoset lymphoblastoid cells as a sensitive host for isolation of measles virus. J. Virol. 64:700-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacMicking, J. D. 2004. IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 25:601-609. [DOI] [PubMed] [Google Scholar]
- 31.Marciniak, R. A., B. J. Calnan, A. D. Frankel, and P. A. Sharp. 1990. HIV-1 Tat protein trans-activates transcription in vitro. Cell 63:791-802. [DOI] [PubMed] [Google Scholar]
- 32.Mosca, J. D., and P. M. Pitha. 1986. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol. Cell. Biol. 6:2279-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakatsu, Y., M. Takeda, M. Kidokoro, M. Kohara, and Y. Yanagi. 2006. Rescue system for measles virus from cloned cDNA driven by vaccinia virus Lister vaccine strain. J. Virol. Methods 137:152-155. [DOI] [PubMed] [Google Scholar]
- 34.Naniche, D., G. Varior-Krishnan, F. Cervoni, T. F. Wild, B. Rossi, C. Rabourdin-Combe, and D. Gerlier. 1993. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 67:6025-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 36.Ohno, S., N. Ono, M. Takeda, K. Takeuchi, and Y. Yanagi. 2004. Dissection of measles virus V protein in relation to its ability to block alpha/beta interferon signal transduction. J. Gen. Virol. 85:2991-2999. [DOI] [PubMed] [Google Scholar]
- 37.Ono, N., H. Tatsuo, Y. Hidaka, T. Aoki, H. Minagawa, and Y. Yanagi. 2001. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J. Virol. 75:4399-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patterson, J. B., D. Thomas, H. Lewicki, M. A. Billeter, and M. B. Oldstone. 2000. V and C proteins of measles virus function as virulence factors in vivo. Virology 267:80-89. [DOI] [PubMed] [Google Scholar]
- 39.Plumet, S., W. P. Duprex, and D. Gerlier. 2005. Dynamics of viral RNA synthesis during measles virus infection. J. Virol. 79:6900-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radecke, F., and M. A. Billeter. 1996. The nonstructural C protein is not essential for multiplication of Edmonston B strain measles virus in cultured cells. Virology 217:418-421. [DOI] [PubMed] [Google Scholar]
- 41.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dotsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reutter, G. L., C. Cortese-Grogan, J. Wilson, and S. A. Moyer. 2001. Mutations in the measles virus C protein that up regulate viral RNA synthesis. Virology 285:100-109. [DOI] [PubMed] [Google Scholar]
- 43.Richardson, C. D., A. Scheid, and P. W. Choppin. 1980. Specific inhibition of paramyxovirus and myxovirus replication by oligopeptides with amino acid sequences similar to those at the N-termini of the F1 or HA2 viral polypeptides. Virology 105:205-222. [DOI] [PubMed] [Google Scholar]
- 44.Sarkar, S. N., and G. C. Sen. 2004. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol. Ther. 103:245-259. [DOI] [PubMed] [Google Scholar]
- 45.Shaffer, J. A., W. J. Bellini, and P. A. Rota. 2003. The C protein of measles virus inhibits the type I interferon response. Virology 315:389-397. [DOI] [PubMed] [Google Scholar]
- 46.Spann, K. M., K. C. Tran, B. Chi, R. L. Rabin, and P. L. Collins. 2004. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 78:4363-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takeda, M., Y. Nakatsu, S. Ohno, F. Seki, M. Tahara, T. Hashiguchi, and Y. Yanagi. 2006. Generation of measles virus with a segmented RNA genome. J. Virol. 80:4242-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeda, M., S. Ohno, F. Seki, K. Hashimoto, N. Miyajima, K. Takeuchi, and Y. Yanagi. 2005. Efficient rescue of measles virus from cloned cDNA using SLAM-expressing Chinese hamster ovary cells. Virus Res. 108:161-165. [DOI] [PubMed] [Google Scholar]
- 49.Takeda, M., S. Ohno, F. Seki, Y. Nakatsu, M. Tahara, and Y. Yanagi. 2005. Long untranslated regions of the measles virus M and F genes control virus replication and cytopathogenicity. J. Virol. 79:14346-14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeda, M., K. Takeuchi, N. Miyajima, F. Kobune, Y. Ami, N. Nagata, Y. Suzaki, Y. Nagai, and M. Tashiro. 2000. Recovery of pathogenic measles virus from cloned cDNA. J. Virol. 74:6643-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeuchi, K., M. Takeda, N. Miyajima, Y. Ami, N. Nagata, Y. Suzaki, J. Shahnewaz, S. Kadota, and K. Nagata. 2005. Stringent requirement for the C protein of wild-type measles virus for growth both in vitro and in macaques. J. Virol. 79:7838-7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan, S. L., and M. G. Katze. 2001. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology 284:1-12. [DOI] [PubMed] [Google Scholar]
- 53.Taniguchi, T., and A. Takaoka. 2002. The interferon-alpha/beta system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr. Opin. Immunol. 14:111-116. [DOI] [PubMed] [Google Scholar]
- 54.Tatsuo, H., N. Ono, K. Tanaka, and Y. Yanagi. 2000. SLAM (CDw150) is a cellular receptor for measles virus. Nature 406:893-897. [DOI] [PubMed] [Google Scholar]
- 55.Valsamakis, A., H. Schneider, P. G. Auwaerter, H. Kaneshima, M. A. Billeter, and D. E. Griffin. 1998. Recombinant measles viruses with mutations in the C, V, or F gene have altered growth phenotypes in vivo. J. Virol. 72:7754-7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wansley, E. K., P. J. Dillon, M. D. Gainey, J. Tam, S. D. Cramer, and G. D. Parks. 2005. Growth sensitivity of a recombinant simian virus 5 P/V mutant to type I interferon differs between tumor cell lines and normal primary cells. Virology 335:131-144. [DOI] [PubMed] [Google Scholar]
- 57.Weber, F., V. Wagner, S. B. Rasmussen, R. Hartmann, and S. R. Paludan. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams, B. R. 1999. PKR; a sentinel kinase for cellular stress. Oncogene 18:6112-6120. [DOI] [PubMed] [Google Scholar]
- 59.Yanagi, Y., B. A. Cubitt, and M. B. A. Oldstone. 1992. Measles virus inhibits mitogen-induced T cell proliferation but does not directly perturb the T cell activation process inside the cell. Virology 187:280-289. [DOI] [PubMed] [Google Scholar]