Abstract
Epstein-Barr virus (EBV) establishes a persistent infection in the human host and is associated with a variety of human cancers. Persistent infection results from a balance between the host immune response and viral immune evasion mechanisms. EBV infection is controlled initially by the innate immune response and later by T-cell-mediated adaptive immunity. EBV has evolved mechanisms to evade the host immune response so that it can persist for the lifetime of the host. Latent membrane protein 1 (LMP-1) is the EBV oncoprotein essential for B-cell immortalization by EBV. We show here that LMP-1 interacts with Tyk2, a signaling intermediate in the alpha interferon (IFN-α) signaling pathway, via a previously uncharacterized LMP-1 signaling domain. LMP-1 prevents Tyk2 phosphorylation and inhibits IFN-α-stimulated STAT2 nuclear translocation and interferon-stimulated response element transcriptional activity. Long-term culture of EBV+ lymphoblastoid cells in IFN-α is associated with outgrowth of a population expressing elevated LMP-1 protein levels, suggesting that cells expressing higher levels of LMP-1 survive the antiproliferative selective pressure imposed by IFN-α. These results show that LMP-1 can protect EBV+ cells from the IFN-α-stimulated antiviral/antiproliferative response and suggest that chronic IFN-α treatment may encourage the outgrowth of cells expressing elevated, and therefore potentially oncogenic, LMP-1 levels in EBV+ individuals.
Epstein-Barr virus (EBV) is a gammaherpesvirus that infects 90% of the world population. EBV infects naive B lymphocytes and establishes a latent, persistent infection via the expression of up to nine viral proteins. Latently infected human B cells are rendered immortal through the combined action of several of these latent gene products (16). EBV's efficient immortalization of human B cells makes this virus a risk factor for development of neoplastic disease. EBV is associated with many human cancers, including Burkitt's lymphoma (BL), Hodgkin's disease, and nasopharyngeal carcinoma ((29). Immunocompromised individuals are particularly at risk for the development of EBV-dependent lymphoproliferative diseases.
Latent membrane protein-1 (LMP-1) is an EBV oncoprotein expressed during latency and required for EBV immortalization of B cells (2, 15). The LMP-1 protein has three major domains defined by structure and function: an N-terminal cytoplasmic domain essential for proper membrane insertion of the polytopic transmembrane domain (3), a multispanning transmembrane domain required for constitutive activation of C-terminal signaling and cytostasis induction, and a C-terminal cytoplasmic signaling domain (20). LMP-1 functions in part as a constitutively active tumor necrosis factor receptor analog. LMP-1 activation is believed to occur as a result of its constitutive oligomerization and lipid raft association. LMP-1's C terminus encodes consensus sequences for tumor necrosis factor receptor (TNFR)-associated factor (TRAF) binding, referred to as C-terminal activating regions 1 and 2 (CTAR1 and -2). CTAR1 and -2 interact with TRAF1, -2, -3, and -5, TNFR-associated death domain, and phosphatidylinositol 3-kinase to activate the NF-κB, AP-1, mitogen-activated protein kinase (MAPK), and Akt signaling pathways (21). LMP-1 interacts with JAK3 via a third CTAR domain in the C terminus (CTAR3) (9). LMP-1 signaling most closely resembles that of CD40, a TNFR superfamily member that plays a key role in proliferation, survival, and differentiation of normal B cells to immunoglobulin-secreting plasma cells. While LMP-1 shares no overt homology with CD40, LMP-1 signaling can complement some CD40 null phenotypes and vice versa (17). LMP-1 signaling can be distinguished from CD40 signaling in that LMP-1 signaling is constitutive (10, 27), LMP-1 negatively regulates proliferation via signals from its transmembrane domain, and LMP-1 is oncogenic (20).
Interferons are a family of cytokines known to function in defense against infection by viral pathogens. Type I interferons, such as IFN-α, bind to alpha interferon receptors 1 and 2 (IFNAR1 and -2), resulting in receptor dimerization. In the classical IFN-α pathway, the receptor-bound Janus kinases (Jaks) nonreceptor tyrosine kinase 2 (Tyk2) and Jak1 are activated and cross-phosphorylate each other. Activated Jaks phosphorylate IFNAR1 and -2, which then serve as Src homology 2 domain docking sites for signal transducers of activated transcription 1 and 2 (STAT1 and -2). STAT1 and -2 are subsequently phosphorylated by activated Jak1 and Tyk2. Phosphorylated STATs heterodimerize and interact with interferon regulatory factor 9 (IRF9) to form the active transcriptional factor complex interferon-stimulated gene factor 3 (ISGF3), which regulates the expression of interferon-stimulated genes (ISGs) (35). Activation of this pathway by type 1 interferons establishes an antiviral and antiproliferative state in the cell. While IFN-α signaling generally occurs via the pathway described above, specific details vary depending on the signaling system in question (35). Nonclassical signaling pathways are also activated by IFN-α. These Jak-dependent but STAT-independent pathways include the phosphatidylinositol 3-kinase (PI3K) pathway, the Cbl-Crk pathway, and the Rac1/p38 map kinase signaling cascade (48).
A novel LMP-1 interactor, Tyk2, was identified in our efforts to characterize the native LMP-1 signaling complex. We show that LMP-1 interacts with Tyk2 and inhibits its tyrosine phosphorylation. STAT2 tyrosine phosphorylation and nuclear translocation are inhibited in cells expressing LMP-1, and LMP-1 blocks IFN-α activation of ISRE-mediated transcription. Culture of a lymphoblastoid cell line (LCL) in IFN-α results in the outgrowth of a population expressing significantly higher levels of the LMP-1 protein than the parent population. LMP-1-mediated inhibition of type I IFN signaling is compatible with LMP-1's role in EBV-driven B-cell proliferation and may contribute to the oncogenic properties of LMP-1 in vivo.
MATERIALS AND METHODS
Cells.
721 is a human, latently infected, EBV+ LCL. DG75 is an EBV− BL cell line. HH514 is an EBV+ BL cell line that does not express LMP-1 due to a deletion of the viral EBNA2 gene (36). All B-cell lines were grown in RPMI 1640 medium supplemented with 10% calf serum (R10C). 293 cells are an embryonic kidney carcinoma cell line grown in Dulbecco's modified Eagle's medium supplemented with 10% calf serum (D10C). All cells were grown at 37°C in high humidity with 5% CO2.
Plasmids.
pRSV-LMP-1, pRSV-lyLMP-1, and pRSV-LacZ are pRC-RSV-based vectors (Invitrogen) and have been described previously (4). pRSV-LMP-1/CΔ55 was designed by inserting the 700-bp Blp1-BstG1 fragment of pCMV-CΔ55 (28) into Blp1-BstG1 of pRSV-LMP-1. pRSV-LMP-1/CΔ55ΔCTAR1 (encoding LMP-1CΔ1,2) and pRSV-LMP-1/ΔCTAR1 (encoding LMP-1CΔ1) were constructed from pRSV-LMP-1CΔ55 and pRSV-LMP-1, respectively, using the QuikChange site-directed mutagenesis kit (Stratagene) to generate substitutions of proline 204 and glutamine 206 in LMP-1's CTAR1 to alanines. The sequence of each point mutant was confirmed by sequencing. pISRE-Luc and pGAS-Luc are luciferase reporters under the control of five tandem ISREs and four tandem gamma interferon (IFN-γ)-activated site (GAS) elements, respectively, upstream of minimal promoter elements (Stratagene). pMT2T-Tyk2 was provided by John Krolewski (Columbia University) and contains the human Tyk2 cDNA cloned into the pMT2T expression vector. The IRF9 and IRF9-S2C expression vectors, gifts from Curt Horvath (Northwestern University), encode IRF9 and IRF9 fused to the 104 C-terminal amino acids of STAT2, respectively, in pcDNA3 and have been described previously (18). pCMV-CD4 is a pcDNA3-based expression vector encoding human CD4 and was provided by Luwen Zhang (University of Nebraska).
Antibodies.
Anti-LMP-1 antiserum is an affinity-purified rabbit polyclonal serum raised against LMP-1's C terminus (residues 188 to 352) fused to glutathione-S-transferase. Anti-N-terminal LMP-1 antiserum is a rabbit polyclonal serum raised against LMP-1's cytoplasmic N terminus (gift of David Thorley-Lawson). CS1-4 is a pool of four mouse monoclonal antibodies to LMP-1 (Dako). Polyclonal rabbit antibodies to Tyk2 (C20), STAT2 (C20), STAT1 (C24), and mouse monoclonal antibody to phosphotyrosine (PY99) are from Santa Cruz Biotechnology. Rabbit anti-phospho-STAT2 antibody (07-224) is from Upstate Cell Signaling Solutions. Rabbit anti-phospho-STAT1 (9171) is from Cell Signaling Technology. Horseradish peroxidase-labeled goat antirabbit (W401B) and goat antimouse (W402B) secondary antibodies are from Promega. Alexa Fluor 488-conjugated goat antirabbit (A11008) and Alexa Flour 594-conjugated goat antimouse (A21203) secondary antibodies are from Molecular Probes. Working antibody dilutions for protein staining of Western blots were the following: anti-LMP-1 polyclonal (1:2,000), anti-N-terminal LMP-1 (1:500), CS1-4 (1:500), anti-Tyk2 (C20,1:400), antiphosphotyrosine (PY99; 1:1,000), anti-STAT2 (C20; 1:200), anti-phospho-STAT2 (07-224; 1:500), horseradish peroxidase-labeled secondary antibodies (1:5,000). All antibodies were diluted 1:500 for immunofluorescence analysis.
Immunoaffinity purification of LMP-1.
721 cells (1.5 × 109) were homogenized in 50 ml of 0.25 M sucrose and 1 mM phenylmethylsulfonyl fluoride (PMSF) at 4°C with 20 strokes of a dounce homogenizer and then centrifuged at 1,000 × g for 10 min. The supernatant was saved, and the pellet was resuspended in 50 ml of 0.25 M sucrose-1 mM PMSF and recentrifuged. The two supernatants were pooled and centrifuged at 105,000 × g for 1 h. The resulting pellet was resuspended in low-salt buffer (LSB) (50 mM HEPES-KOH, pH 7.4; 100 mM B-glycerolphosphate, 25 mM NaF, 1 mM MgCl2, 1 mM EGTA, 5% glycerol) containing 2× PMSF at a concentration of 20 mg/ml protein, an equal volume of LSB-10% Triton X-100 was added, and the solution was incubated on ice for 30 min. The detergent lysate was microcentrifuged at 13,000 × g for 15 min, and the supernatant was centrifuged at 100,000 × g for 1 h (Triton X-100 soluble starting material). Rabbit anti-LMP-1 antibodies (directed against LMP-1's cytoplasmic carboxy terminus, residues 188 to 352), covalently coupled to NHS-activated Sepharose beads (Amersham), were mixed overnight at 4°C with the 100,000 × g-centrifuged supernatant. Beads were packed into a column and washed with LSB-0.035% Triton X-100 without PMSF. The column was eluted with 100 mM glycine (pH 2.5). Fractions (30 μl) were either diluted in sodium dodecyl sulfate (SDS) sample buffer for further analysis by SDS-polyacrylamide gel electrophoresis (PAGE), silver staining, and Western blotting or prepared for liquid chromatography tandem mass spectrometry (LC/MS/MS).
LC/MS/MS.
LC/MS/MS was carried out by the University of Colorado—Boulder mass spectrometry facility using a 500-μm high-pressure liquid chromography column packed with C18 reverse-phase resin interfaced to an LCQ ion trap mass spectrometer (ThermoQuest, San Jose, CA).
Native PAGE.
Cells were resuspended in hypotonic lysis buffer (10 mM HEPES-KOH, pH 7.9, 0.5 mM KCl, 0.5 mM MgCl2, 0.1 mM EGTA, 0.5 mM dithiothreitol), incubated on ice for 30 min, and triturated 10 times through a 26.5-gauge needle. The lysate was microcentrifuged at 13,000 × g for 10 min. The pellet was resuspended in LSB with 2× PMSF, diluted with an equal volume of LSB-10% Triton X-100 (final concentration of 108 cell equivalents/ml), and incubated on ice for 30 min. The solubilized pellet was microcentrifuged at 13,000 × g for 15 min, and the supernatant was centrifuged at 100,000 × g for 1 h at 4°C. The final supernatant was loaded on a native acrylamide gradient gel (43). Briefly, 1.5-mm-thick continuous 4 to 29% polyacrylamide-1× Tris-borate-EDTA gels were cast for the Miniprotein 2 apparatus (Bio-Rad). Gels were prerun at 70 V for 20 min. Loaded gels were run for 20 min at 70 V, followed by 150 V for 18 h at 4°C. Gels were soaked in 50 mM Tris base, 40 mM glycine, 0.25% SDS for 20 min at 70°C. Gels were transferred as described in “SDS-PAGE and Western analysis”. High-molecular-weight native markers were from Amersham.
Transfections.
Cells (107/cuvette) were transfected by electroporation in 0.4-cm gapped cuvettes using a Bio-Rad gene pulser. DG75 cells and HH514 cells were electroporated in 0.35 ml R10C at 0.250 kV and 960 μF. 293 cells were electroporated in 0.5 ml D10C at 0.26 kV and 960 μF. Transfected cells were assayed 48 h posttransfection.
SDS-PAGE and Western analysis.
Samples were analyzed by SDS-PAGE and Western blotting as described previously (4) with the following exception: antiphosphotyrosine and anti-N-terminal LMP-1 blots were blocked and stained in 3% bovine serum albumin rather than in 5% milk. Whole-cell lysates (WCLs) analyzed by Western blotting were solubilized in 4× SDS sample buffer. Unless otherwise noted, lanes contain 105 cell equivalents. Where noted, blots from two to four independent experiments were quantified using ImageJ software. Molecular weight markers (Benchmark Pre-stained) were from Invitrogen. The identity of the 120-kDa phosphorylated Tyk2 band on blots visualized with antiphosphotyrosine antibodies was confirmed by including extracts from cells transfected with or without the Tyk2 expression vector as well as purified Tyk2 protein (not shown).
Coimmunoprecipitation.
Cells were resuspended in hypotonic lysis buffer, incubated on ice for 30 min, and triturated 10 times through a 26.5-gauge needle. The lysate was microcentrifuged at 13,000 × g for 10 min. The pellet was resuspended in 200 μl LSB containing 2× PMSF, diluted with an equal volume of LSB-10% Triton X-100, and incubated on ice for 30 min. The solubilized pellet was microcentrifuged at 13,000 × g for 15 min, and the resulting supernatant was centrifuged at 100,000 × g for 1 h at 4°C. The 100,000 × g-centrifuged supernatant served as the “lysate” for immunoprecipitation. The appropriate antibody (3 μl) was added to the starting material and incubated on ice for 30 min. Immune complexes were recovered by incubation with protein G-agarose (Roche Diagnostics) for 90 min. The beads were washed five times with LSB-0.035% Triton X-100 without PMSF and boiled in SDS-sample buffer.
Enrichment of CD4+ cells.
DG75 cells (5 × 107) were transfected with 5 μg of pCMV-CD4 and 10 μg of pRSV-LMP-1 and treated with 1,000 U/ml of IFN-α 48 h posttransfection. Cells were washed two times in ice-cold phosphate-buffered saline (PBS)-2% fetal bovine serum, and CD4+ cells were isolated using magnetic CD4-coupled Dynal beads (product no. 113.03) as described by the manufacturer. Enriched cells were collected in the following buffers: 4× SDS sample buffer for Western blot analysis, 5× RIPA buffer for immunoprecipitation, or MNE buffer (25 mM morpholineethanesulfonic acid, pH 6.5, 150 mM NaCl, 5 mM EDTA) containing 0.2% Triton X-100 for subcellular fractionation in sucrose gradients.
Tyk2 Immunoprecipitation.
Cell pellets were resuspended in 100 μl of 5× RIPA buffer and incubated on ice for 10 min. Samples were diluted with H2O to 1× RIPA, sonicated, and boiled for 10 min. Ten microliters of each sample was mixed 1:1 with 4× sample buffer for analysis as a WCL. The rest of each sample was incubated with 2 μl of anti-Tyk2 antibody for 30 min at 4°C and then incubated with protein G-agarose for 90 min. The beads were washed four times with 1× RIPA and once with PBS and boiled in SDS-sample buffer.
Subcellular fractionation.
For CD4+ sorted cells, sucrose gradients were analyzed as described previously (4), with volumes adjusted to compensate for lower cell numbers. Briefly, cells were homogenized on ice with a Dounce homogenizer in 0.35 ml of MNE buffer and 0.2% Triton X-100. Extracts were diluted 1:1 in MNE-80% sucrose, overlaid with 1.4 ml MNE-30% sucrose and 0.7 ml of MNE-5% sucrose, and centrifuged at 4°C for 18 h at 200,000 × g. Gradient fractions (100 μl) were collected for SDS-PAGE analysis. The Triton-insoluble material formed a tight pellet in the bottom of the tube, the Triton-soluble material was distributed between the pellet and across the 40%/30% interface, and the Triton-resistant buoyant material was found at the 30%/5% interface. Approximately 10 to 20% of STAT1 and -2 were found in the Triton-insoluble pellet, and the remainder was found in the soluble fraction.
ISRE and GAS reporter assays.
DG75, HH514, and 293 cells were transfected by electroporation in duplicate with LMP-1 and Tyk2 expression vectors, pRSV-LacZ, and either pISRE-Luc or pGAS-Luc. All transfections were performed with equivalent μg of DNA by addition of an expression vector control plasmid (pRC-RSV). Forty hours posttransfection, cells were treated with or without IFN-α (for ISRE activation) or IFN-γ (for GAS activation) for 6 to 7 h, and extracts were assayed for luciferase and β-galactosidase accumulation using the Dual Light assay kit from Tropix. Luciferase activity was normalized within each transfection with β-galactosidase activity.
Immunofluorescence.
721 cells were treated with IFN-α for 30 min, fixed for 20 min in 4% paraformaldehyde, and made permeable with 0.1% Triton X-100. Cells were then blocked for 10 min with PBS and 2% fetal calf serum and stained with anti-LMP-1 (CS1-4) and anti-STAT2 antibodies followed by Alexa Fluor 488-conjugated antirabbit and Alexa Fluor 594-conjugated antimouse secondary antibodies at 4°C for 1 h. Cells were resuspended in 4′,6′-diamidino-2-phenylindole mounting medium and mounted on slides. Slides were visualized under magnification ×60 with a Nikon Eclipse E800 fluorescence microscope, and images were captured with a Cooke SensiCam digital camera using SlideBook software. Cells were scored first for STAT2 nuclear localization and second for intensity of LMP-1 staining. LMP-1 staining intensity for each cell was scored as “+” for very low (but detectable) relative intensity or “++++” for very high relative intensity (cells with intermediate LMP-1 staining intensity were not included). A total of 58 cells expressing low LMP-1 levels and 69 cells expressing high LMP-1 levels were counted. Data are expressed as percentages of IFN-α-treated cells with nuclear STAT2 versus the LMP-1 level (“+” or “++++”). The relationship between LMP-1 levels and IFN-stimulated nuclear STAT2 in this cell population were also quantified by comparing mean STAT2 levels within the nucleus to mean STAT2 levels within the cytoplasm using ImageJ software. Each cell and its nucleus were outlined using ImageJ, and ImageJ calculated the area and mean light value for the outlined region. Data are expressed as a ratio of mean analyzed nuclear staining to mean analyzed cytoplasmic staining, [Mn(At − An)]/(MtAt − MnAn), where Mt is the mean light value of the total cell; Mn is the mean light value of the nuclear area; At is the area of the total cell; and An is the area of the nucleus. Ratios from each of the two populations were compared using Student's t test.
Long-term IFN-α treatment.
721 cells were incubated at 105 cells/ml in R10C with either IFN-α or IFN-γ and counted daily. Cells were fed with R10C and IFN every 3 to 4 days. Cultures were diluted 1:10 when they reached a density of ∼106 cells/ml. IFN-α was removed from a fraction of the plates after 14 days of growth, and cells were grown for another 27 days in the absence of IFN-α.
RESULTS
LMP-1 and Tyk2 copurify from B cells.
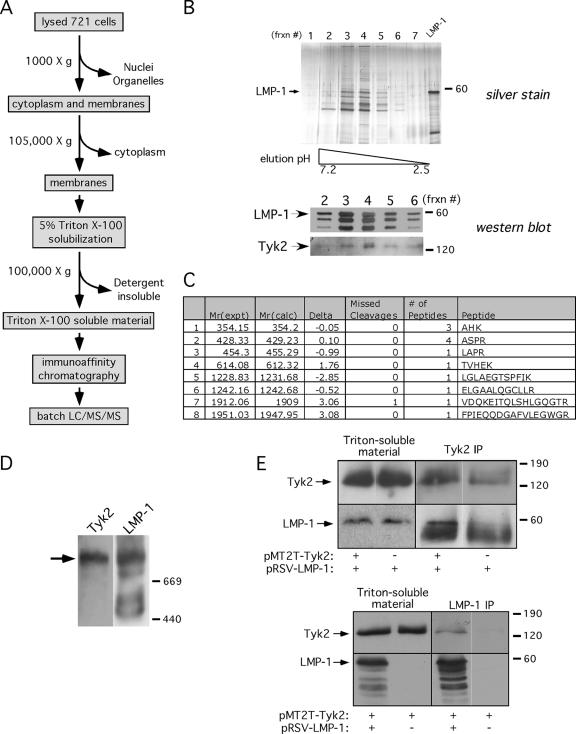
We have developed an affinity purification and mass spectrometric approach to identify novel interactors associated with native LMP-1 signaling complexes from EBV-immortalized cells (Fig. 1A). Membranes from EBV-immortalized lymphoblastoid cells (721 cells) were enriched by centrifugation and solubilized in Triton X-100. Detergent-soluble material was applied to an anti-LMP-1 antibody resin, and bound LMP-1 complexes were eluted at a low pH. The SDS-PAGE profile of the column elution analyzed by silver staining for total protein and Western blotting for LMP-1 is shown in Fig. 1B. Fraction 4 from the elution shown in Fig. 1 was analyzed in batch by LC/MS/MS. Tyk2 was among the interacting proteins identified by mass spectrometry and was shown by Western analysis to coelute with LMP-1 (Fig. 1B and C). TRAF3 and the p85 alpha subunit of PI3K (known direct LMP-1 interactors) and p38-MAPK (a known indirect LMP-1 interactor) were also identified in purified samples, either by mass spectrometry (p85 and p38-MAPK) or by Western analysis (TRAF3) (not shown). LMP-1 peptides were not identified, presumably because of the paucity of trypsin sites in the LMP-1 protein. To determine if Tyk2 was present in LMP-1 complexes prior to purification, detergent-soluble complexes were resolved by native PAGE (4 to 29%). Consistent with the copurification of Tyk2 with LMP-1, Tyk2 comigrated with the slowest-migrating solubilized native LMP-1 complex of >669 kDa (Fig. 1D). We chose to characterize the presumptive interaction between LMP-1 and Tyk2 further because of Tyk2's key role in type I IFN signaling.
FIG. 1.
Tyk2 copurifies with LMP-1. (A) LMP-1 immunoaffinity purification procedure. (B) LMP-1 complexes eluted from anti-LMP-1 antibody affinity column by decreasing pH, analyzed by silver staining (top gel) or Western blotting for LMP-1 and Tyk2 (bottom blots). The pH gradient is indicated below the silver-stained gel. A glutathione S-transferase-LMP-1 marker, comigrating with LMP-1, was loaded in the lane marked “LMP-1” (silver-stained gel), and the arrows to the left of the Western blots point to LMP-1 and Tyk2. Frxn #, fraction no. (C) Tyk2 peptides identified by tandem MS of LMP-1 complexes in fraction 4; Mr(expt) and Mr(calc), experimental and calculated peptide weights, respectively; Delta, difference between Mr (experimental) and Mr (calculated); Missed Cleavages, missed arginine/tryptophan cleavage sites; # of Peptides, detection frequency of peptide; Peptide, peptide sequence. (D) Triton-soluble material from 721 cell membranes analyzed by native PAGE (4 to 29%) and Western blotting for LMP-1 and Tyk2. Blots were first stained for Tyk2 (left) and then stripped and reprobed for LMP-1 (right). The primary antibody used for staining is shown above each blot, the migration of high-molecular-mass native markers is shown on the right (in kDa), and the arrow points to the predominant high-molecular-mass LMP-1 complex with which Tyk2 comigrates. (E) Tyk2 and LMP-1 immunoprecipitates from Triton X-100-soluble extracts of DG75 cells transfected with pRSV-LMP-1 with or without pMT2T-Tyk2 (top panel) or with pMT2T-Tyk2 with or without pRSV-LMP-1 (bottom panel). Triton-soluble starting material and immunoprecipitates were analyzed by Western blotting for Tyk2 or LMP-1. Expression vectors are shown below, precipitating antibodies are shown above, and blotting antibodies are shown to the left of each blot. Arrows point to LMP-1 and Tyk2; the band below LMP-1 in the top panel is the heavy chain from the precipitating anti-Tyk2 antibody. Migration of molecular mass markers is shown in kDa to the right of each blot. Images within black boxes are from a single scanned blot and are separated within the box if individual lanes have been rearranged to maintain consistency between figures (this applies to blots in all figures).
Coimmunoprecipitation assays were performed to confirm the LMP-1 and Tyk2 interaction. DG75 cells were transfected with LMP-1 and Tyk2 expression vectors, and reciprocal coimmunoprecipitations were performed 48 h later. LMP-1 coprecipitated with Tyk2 more efficiently when Tyk2 was ectopically expressed (compare LMP-1 levels in ± pMT2T-Tyk2 lanes), and Tyk2 coprecipitated with LMP-1 only when LMP-1 was expressed (Fig. 1E). These results demonstrate that LMP-1 and Tyk2 interact in EBV-immortalized lymphoblastoid cells and B lymphoma cells.
Tyk2 is underphosphorylated in B cells expressing LMP-1.
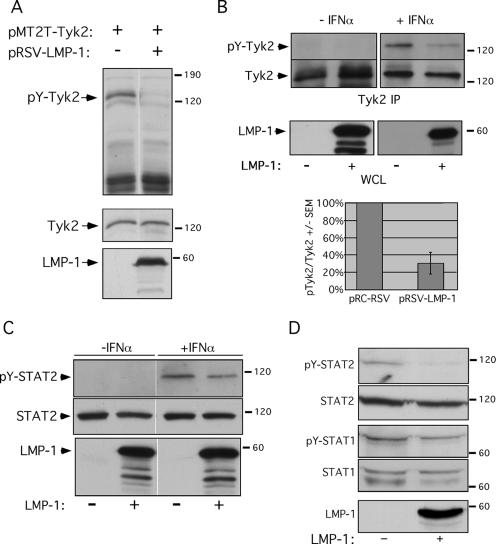
Tyk2 tyrosine phosphorylation was assessed to ask if the LMP-1/Tyk2 interaction affected Tyk2 activity. LMP-1 and Tyk2 were coexpressed in DG75 cells, and Tyk2 expression and tyrosine phosphorylation were examined by Western blotting (Fig. 2A). Consistent with previous reports (6), ectopically expressed Tyk2 was tyrosine phosphorylated in the absence of IFN-α treatment. LMP-1 expression efficiently inhibited tyrosine phosphorylation of ectopic Tyk2. IFN-α-stimulated Tyk2 phosphorylation in DG75 and HH514 cells was similarly inhibited by LMP-1 (not shown), indicating that LMP-1's effect on Tyk2 phosphorylation occurred in a variety of human B-cell backgrounds.
FIG. 2.
LMP-1 negatively regulates Tyk2 and STAT2 phosphorylation. (A) WCLs from DG75 cells transfected with pRSV-LMP-1, pMT2T-Tyk2, or empty vector, analyzed by Western blotting for phosphotyrosine (pY) (top blot), Tyk2 (middle blot), or LMP-1 (lower blot). Expression vectors are shown at the top of the blot. (B) DG75 cells were cotransfected with pCMV-CD4 and either pRC-RSV or pRSV-LMP-1, treated with or without IFN-α (1,000 U/ml, 30 min) 2 days posttransfection, and then sorted for CD4 to enrich for transfected cells. Tyk2 immunoprecipitates from LMP-1− and LMP-1+ cells solubilized in RIPA buffer were analyzed by Western blotting for pY (top blots) or Tyk2 (middle blots); corresponding WCLs were analyzed by Western blotting for LMP-1 expression (lower blots). The “−” and “+” signs below the blots mark the CD4-enriched LMP-1− and LMP-1+ populations, respectively. IFN-α treatment is noted above the blots. (Graph in B) Phosphorylated Tyk2 levels were normalized to total Tyk2 protein levels (from four independent experiments) and expressed as “pY-Tyk2/Tyk2” ± standard deviation (pTyk2/Tyk2 +/− SEM) (pY-Tyk2/Tyk2 values from LMP-1− extracts from CD4-sorted cells were set to 100%) (y axis). Introduced expression vectors are indicated (x axis). (C) WCLs of IFN-α-treated (1,000 U/ml, 15 min), CD4+-sorted LMP-1− and LMP-1+ DG75 cells, analyzed by SDS-PAGE and Western blotting for pY-STAT2 (top blot), STAT2 (middle blot), or LMP-1 (bottom blot). The “−” and “+” signs below the blots and “−/+ IFN-α” noted above the blots have the same meaning as in part B. The average decrease in IFN-α-stimulated STAT2 phosphorylation (pY-STAT2/STAT2) in LMP-1+ cells was 23%. Shown is one of two independent experiments. (D) Western analysis of pY-STAT2, STAT2, pY-STAT1, STAT1, and LMP-1 in the Triton-insoluble fraction of DG75 cells cotransfected with pCMV-CD4 and either pRC-RSV or pRSV-LMP-1, treated with 1,000 U/ml IFN-α for 30 min, and sorted for CD4 to enrich for transfected cells. Blots were stained for pY-STAT2, STAT2, pY-STAT1, STAT1, or LMP-1 as indicated. LMP-1 status is denoted below the blots. These data are representative of two independent experiments; blots were quantified as described in Material and Methods.
To determine if LMP-1 negatively regulated phosphorylation of endogenous Tyk2, DG75 cells were transfected with a human CD4 expression vector, with or without pRSV-LMP-1. The CD4 expression vector was included to allow isolation of transfected cells by sorting for cell surface CD4. This was necessary because the majority of cells in the bulk-transfected population will not express LMP-1 (the transfection efficiency of DG75 cells in our experiments is ∼5%), and thus, the negative regulation of Tyk2 phosphorylation by LMP-1 would likely be masked by the IFN-stimulated Tyk2 phosphorylation signal contributed by the untransfected cells in the population. Cells were treated with IFN-α for 30 min and sorted with anti-CD4 Dynal beads to isolate the transfected CD4+ subpopulation. Tyk2 immunoprecipitates from sorted cell extracts were analyzed by Western blotting for phosphotyrosine and Tyk2 (Fig. 2B). Basal (no IFN-α treatment) Tyk2 phosphorylation was undetectable. IFN-α-stimulated Tyk2 phosphorylation was diminished by 70% in the presence of LMP-1. Thus, LMP-1 expression inhibits tyrosine phosphorylation of endogenous as well as ectopically expressed Tyk2.
LMP-1 inhibits STAT2 phosphorylation.
We next determined if LMP-1's effect on Tyk2 phosphorylation was associated with a diminution in IFN-α-stimulated STAT activation. DG75 cells transiently expressing CD4 and LMP-1 were treated with IFN-α for 15 min and magnetically sorted to isolate CD4+ transfected subpopulations. STAT protein levels and tyrosine phosphorylation were analyzed in extracts of LMP-1− and LMP-1+ cell populations. IFN-α-stimulated cellular STAT2 tyrosine phosphorylation (pY-STAT2/total STAT2) in LMP-1+ cells was diminished 23% relative to that in LMP-1− cells (Fig. 2C), whereas cellular STAT1 phosphorylation was minimally affected by LMP-1 expression (not shown). These results suggest that tyrosine phosphorylation of STAT2 but not STAT1 is inhibited by LMP-1 expression, and they are consistent with those of previous studies demonstrating STAT2 but not STAT1 tyrosine phosphorylation is significantly reduced for Tyk2 null mice (14).
Activation of STAT proteins by extracellular signals results in their rapid subcellular redistribution from the cytoplasm to the plasma membrane and ultimately to the nucleus. Thus, STAT proteins are found in distinct and biochemically resolvable pools in both unstimulated and IFN-α-treated cells. Such pools include free dimeric STATs in the cytosol, receptor-associated STATs at the plasma membrane, and STATs bound to DNA or nucleoporins within the nucleus. Cytosolic STAT pools can be distinguished from IFNAR-associated and nuclear STATs by their differential solubility in Triton X-100. Importantly, approximately 20 to 30% of cellular LMP-1 is insoluble in Triton X-100 and, like the IFNAR, is presumed to be associated with elements of the cytoskeleton (23, 26). In this context, we next tested the hypothesis that LMP-1's effect on STAT2 phosphorylation targeted the Triton-insoluble STAT pool. Detergent-insoluble material from Triton X-100-solubilized whole-cell homogenates of sorted LMP-1− and LMP-1+ cells were analyzed by Western blotting for STAT expression and tyrosine phosphorylation (Fig. 2D). The amount of STAT protein recovered in the insoluble fraction was unaffected by LMP-1 expression (not shown). Levels of IFN-α-stimulated STAT1 phosphorylation were minimally affected by LMP-1. In contrast, IFN-α-stimulated phosphorylation of Triton-insoluble STAT2 (pY-STAT2/STAT2) from LMP-1+ cells was diminished 90% relative to that with LMP-1− cells (Fig. 2D). Thus, the effect of LMP-1 on STAT phosphorylation is specific for STAT2 and specifically for the detergent-insoluble pool of cellular STAT2.
LMP-1 inhibits IFN-α activation of ISRE elements.
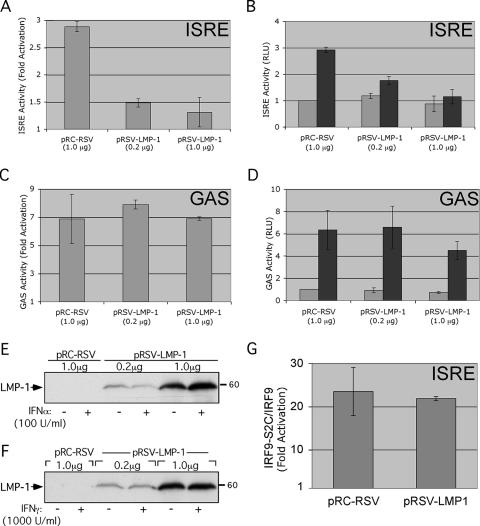
IFN-α stimulation triggers formation of ISGF3, a transcriptional regulatory complex that binds to ISREs and regulates ISG expression. Considering Tyk2's essential role in type I IFN signaling, IFN-α-stimulated ISRE activity should be diminished in cells expressing LMP-1. This prediction was tested by reporter assay. DG75 cells were transfected with a luciferase reporter under the control of five tandem ISRE elements from the ISG54 gene, with or without pRSV-LMP-1, and cultured for 48 h. Cells were treated with 100 U/ml of IFN-α (a concentration of IFN-α determined to be sufficient for near-maximal ISRE activation [Table 1 ]) 6 h prior to harvest, and cell extracts were assayed for luciferase accumulation and LMP-1 expression (Fig. 3A, B, and E). IFN-α treatment resulted in a threefold activation of the ISRE reporter (Fig. 3A). As predicted, LMP-1 inhibited IFN-α-stimulated, but not basal, ISRE activation (Fig. 3A and B).
TABLE 1.
LMP-1 inhibits IFN-α-stimulated ISRE activation in multiple cell linesa
| Cell type | pRSV-LMP-1 (μg)b | Ectopic Tyk2 | Relative ISRE activity at IFN-α level (U/ml) of:
|
Fold activation (100 U/ml) | ||
|---|---|---|---|---|---|---|
| 0 | 100 | 1,000 | ||||
| DG75 | − | − | 1.00 | 2.92 | 2.27 | 2.88 ± 0.09** |
| 0.2 | − | 1.18 | 1.76 | 1.51 | 1.48 ± 0.08** | |
| 1.0 | − | 0.88 | 1.15 | 1.16 | 1.30 ± 0.27** | |
| DG75 | − | + | 1.00 | 4.80 | 4.09 | 4.80 ± 1.34* |
| 0.2 | + | 1.21 | 2.28 | 1.77 | 1.90 ± 0.36* | |
| 1.0 | + | 0.78 | 1.14 | 1.02 | 1.43 ± 0.34* | |
| HH514 | − | + | 1.00 | 10.05 | ND | 10.05 ± 0.66* |
| 1.0 | + | 0.29 | 1.81 | ND | 6.28 ± 1.39* | |
| 293 | − | − | 1.00 | 10.55 | ND | 10.55 ± 0.72** |
| 2.0 | − | 0.79 | 5.83 | ND | 7.36 ± 0.52** | |
| 5.0 | − | 0.74 | 4.97 | ND | 6.65 ± 0.34** | |
Reporter assays were performed with DG75, HH514, and 293 cell lines as described in the legend to Fig. 3. pMT2T-Tyk2 (5 μg) was introduced into cells where indicated (−, no ectopic Tyk2; +, ectopic Tyk2). Equivalent amounts (μg) of Rous sarcoma virus expression vector were introduced in all cases. Data are expressed as “relative ISRE activity” (normalized ISRE activity relative to that of the untreated pRC-RSV control, which was set to 1.0, for each cell line transfected with pMT2T-Tyk2 or left untransfected) or “fold activation” (ISRE activity of IFN-α-treated samples (100 U/ml) relative to that of untreated samples). Asterisk, standard deviation; double asterisk, standard error of the mean from three independent experiments.
−, no pRSV-LMP-1.
FIG. 3.
LMP-1 inhibits IFN-α-stimulated ISRE activation. DG75 cells were transfected with 6 μg pISRE-Luc (A, B, E) or pGAS-Luc (C, D, F), 1 μg pRSV-LacZ, and the indicated amounts of pRSV-LMP-1 or pRC-RSV vector (x axis). Two days later, cells were treated with 100 U/ml IFN-α (A, B, E) or 1,000 U/ml IFN-γ (C, D, F) for 6 h prior to harvest, and cell lysates were assayed for LMP-1 expression (E, F), luciferase activity, and β-galactosidase activity (A to D). Luciferase activity was normalized within each transfection to β-galactosidase activity and is expressed as either “fold activation” (normalized ISRE [A] or GAS [C]) reporter activity of IFN-treated samples relative to that of untreated samples) or “RLU” (relative light units) (normalized ISRE [B] or GAS ([D] reporter activity relative to that of the untreated pRC-RSV control). Gray and black bars in B and D represent activity from untreated and IFN-treated samples, respectively. Results shown in A to D are the averages ± standard errors of the means from three independent experiments. (E and F) Western analysis of LMP-1 expression from one of the three experiments shown in A and B (E) or C and D (F). Introduced expression vectors and IFN concentrations used are shown above and below the blots, respectively. (G) LMP-1 has no effect on ISRE activation by constitutively active ISGF3. DG75 cells were transfected with pISRE-Luc, pRSV-LacZ, and pRSV-LMP-1 (as in A, B, and E) plus 3 μg pIRF9-S2C or pIRF9, and extracts were assayed for normalized luciferase activity. Data are expressed as IRF9-S2C/IRF9 “fold activation” (ISRE activation by IRF9-S2C relative to ISRE activation by IRF9 alone). Results are the averages ± standard errors of the means from three independent experiments.
LMP-1's effect on IFN-α-stimulated ISRE activity also was observed in HH514 and 293 cells, as well as in cells expressing ectopic Tyk2 (Table 1). Ectopic Tyk2 expression enhanced the observed degree of ISRE stimulation by IFN-α in the absence of LMP-1, suggesting that endogenous Tyk2 levels may be limiting in these cells. The magnitude of the IFN-α-stimulated ISRE response (fold activation) in cells expressing LMP-1 was independent of ectopic Tyk2 expression (Table 1). Together, these results show that LMP-1 inhibits IFN-α-stimulated ISRE activity in a variety of human cell lines.
LMP-1 does not affect IFN-γ-induced GAS activation.
The specificity of LMP-1's regulation of interferon signaling was tested by examining LMP-1's effect on IFN-γ-mediated type II IFN signaling. Although similar in overall design, the type I and type II IFN pathways differ with respect to associated signaling proteins mediating downstream gene regulation. IFN-γ binds to JAK1 and JAK2-associated receptor subunits that are distinct from the IFN-α-specific IFNAR1 and -2 subunits. Activated JAKs phosphorylate the receptor to recruit STAT1 proteins, which are subsequently phosphorylated. Phosphorylated STAT1 monomers homodimerize, translocate to the nucleus, and bind GAS elements to activate transcription (35). LMP-1's effect on IFN-γ regulation of GAS elements was tested to ask if LMP-1's regulation of type I IFN signaling extended to type II IFN signaling. LMP-1's ability to regulate type II IFN signaling was tested by reporter assay. DG75 cells were transfected with a luciferase reporter containing four tandem copies of the GAS element from the GBP1 gene upstream of a minimal promoter, with or without pRSV-LMP-1. LMP-1 expression had no significant effect on either basal or IFN-γ-stimulated GAS activity (Fig. 3C and D). LMP-1's effect on ISRE and GAS activity was routinely assayed within the same experiment and consistently resulted in inhibition of ISRE but not GAS activity despite similar LMP-1 expression levels (compare Fig. 3E and F). Thus, LMP-1's inhibition of IFN signaling is specific for the type I IFN pathway and is consistent with LMP-1's inhibition of Tyk2 phosphorylation.
Constitutively active ISGF3 is unaffected by LMP-1.
Our results thus far suggest that LMP-1 affects ISRE activity via inhibition of Tyk2 phosphorylation. If so, signals through the type I IFN pathway initiated downstream of Tyk2 should be independent of LMP-1 expression. Therefore, LMP-1's effect on constitutive ISGF3 activity was tested. ISGF3 activity can be rendered constitutive by fusing the DNA binding component (IRF9) to the C-terminal transcriptional activating domain of STAT2 (S2C) (18).
DG75 cells were transfected with pISRE-Luc, with or without pRSV-LMP-1, and with either the IRF9-S2C or IRF9 expression vector. ISRE activity was measured 2 days later (Fig. 3G). As described previously, IRF9-S2C but not IRF9 alone activated the ISRE reporter at least 20-fold (Fig. 3G) and was independent of IFN-α (not shown). Importantly, LMP-1 expression had no effect on ISRE activation by IRF9-S2C. This suggests that LMP-1 acts upstream of ISGF3, presumably at the level of Tyk2 activation, to inhibit ISRE activation. Furthermore, Fig. 3C, D, and G demonstrate that inhibition of IFN-α-stimulated ISRE activity does not result from the LMP-1-mediated general downregulation of transcription (37).
The LMP-1/Tyk2 interaction is independent of CTAR1, -2, and -3.
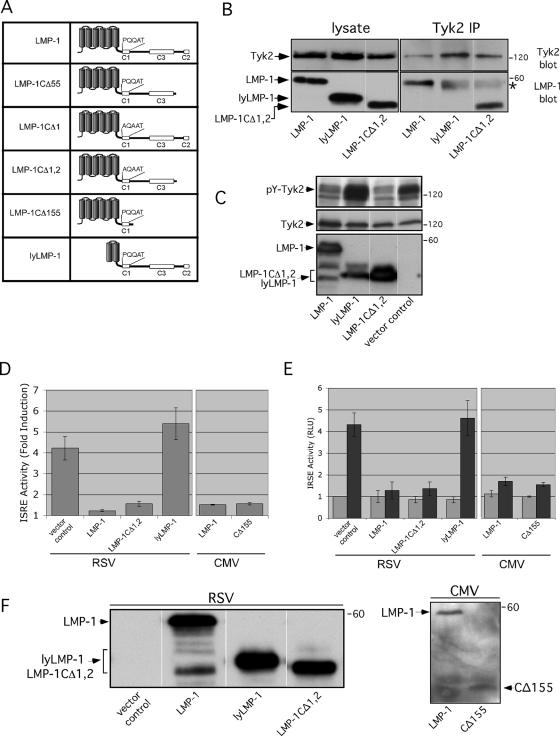
LMP-1 regulates signaling by binding to cellular proteins (i.e., TRAFs) via CTAR domains located in LMP-1's cytoplasmic C terminus. A series of LMP-1 deletion and/or substitution mutants was utilized to identify the LMP-1 domain required for regulation of IFN-α signaling. C-terminal mutants included LMP-1CΔ55 (deletion of C-terminal 55 residues that include CTAR2), LMP-1CΔ155 (deletion of C-terminal 155 residues that include CTAR2 and -3), LMP-1CΔ1 (substitution of P204A and Q206A to eliminate the PXQXT motif in CTAR1 [abolishes CTAR1 activity]) (5), and LMP-1CΔ1,2 (deletion of C-terminal 55 residues in LMP-1CΔ1 to eliminate CTAR2). An EBV-encoded N-terminal LMP-1 truncation called lyLMP-1 (7), which lacks the first four of six predicted membrane-spanning segments of LMP-1's transmembrane domain, was used to assess the contribution of LMP-1's transmembrane domain to regulation of IFN-α signaling. Experiments using deletion mutants lacking the entire C-terminal cytoplasmic domain were not included because their expression proved difficult to assess quantitatively relative to that of full-length LMP-1 by Western blotting (unpublished data).
Regions of LMP-1 required for interaction with Tyk2 and inhibition of Tyk2 tyrosine phosphorylation were assessed by introduction of LMP-1 mutant expression vectors into cells, followed by LMP-1/Tyk2 coimmunoprecipitation (Fig. 4B) and antiphosphotyrosine Western analysis (Fig. 4C). DG75 cells were transfected with Tyk2 and LMP-1 expression vectors, and reciprocal coimmunoprecipitations were performed. LMP-1 and LMP-1CΔ1,2 were detectable in Tyk2 immunoprecipitates, while lyLMP-1 was not (Fig. 4B). LMP-1CΔ155, LMP-1CΔ55, and LMP-1CΔ1 were also detectable in Tyk2 immunoprecipitations (not shown).
FIG. 4.
LMP-1 targets Tyk2 and IFN-α signaling in a CTAR-independent manner. (A) LMP-1 mutants. (B) Western analysis of anti-Tyk2 immunoprecipitates from lysates of DG75 cells expressing the indicated LMP-1 mutants (shown below blots). LMP-1 and mutants were expressed from pRC-RSV. The top and bottom blots are stained for Tyk2 and LMP-1 (using the mouse CS1-4 antibody), respectively. Starting material for immunoprecipitates (lysate) is shown on the left, and corresponding Tyk2 immunoprecipitates are shown on the right. The asterisk marks cross-reactive secondary antibody staining of the precipitating anti-Tyk2 antibody; cross-reaction of the rabbit precipitating antibody with the antimouse secondary antibody is variably observed but resolvable from the 60-kDa LMP-1 band. (C) Western analysis of WCLs from DG75 cells expressing ectopic Tyk2 and the indicated LMP-1 mutants expressed from pRC-RSV; blots stained for phosphotyrosine (pY) (top blot), Tyk2 (middle blot), or LMP-1 (lower blot). (D, E, F) DG75 cells were transfected with 6 μg pISRE-Luc, 1 μg pRSV-LacZ, and either pRC-RSV or the indicated LMP-1 expression vector (x axis). Two days later, cells were treated for 7 h with 100 U/ml IFN-α prior to harvest and assayed for ISRE activity. Results are displayed as described in the legend for Fig. 3. The expressed LMP-1 mutant is shown below the graph, and the promoter used to drive expression (pRSV or pCMV) is shown at the bottom. The graphs in D and E are “split” to emphasize that results from experiments employing cytomegalovirus promoter-driven LMP-1 expression vectors were assessed independently from those employing Rous sarcoma virus promoter-driven LMP-1 expression vectors. Results shown in D and E are the averages ± standard errors of the means from three independent experiments. (F) Western analysis of LMP-1 expression in untreated cells from one of the experiments shown in D and E. The blot on the right was stained with an anti-N-terminal LMP-1 antibody because CΔ155 is undetectable with anti-C-terminal LMP-1 antibodies.
If LMP-1's effect on Tyk2 phosphorylation results from the observed LMP-1/Tyk2 interaction, Tyk2-interacting LMP-1 mutants should inhibit Tyk2 phosphorylation. Tyk2 phosphorylation in cells expressing ectopic Tyk2 and LMP-1 mutants was analyzed by Western blotting. Consistent with the interaction data, Tyk2 phosphorylation was diminished in the presence of LMP-1 and LMP-1CΔ1,2 but not lyLMP-1 (Fig. 4C).
The ability of LMP-1 mutants to negatively regulate IFN-α signaling was tested by reporter assay. Reporter assays were conducted as described previously except that DG75 cells were treated with IFN-α for 7 h prior to harvest. Consistent with Tyk2 interaction and phosphorylation data (Fig. 4B and C), LMP-1CΔ1,2 and LMP-1CΔ155 but not lyLMP-1 suppressed IFN-α-stimulated ISRE activity to the same extent as LMP-1 (Fig. 4D and E). LMP-1CΔ1 and LMP-1CΔ55 also retained wild-type LMP-1 activity in this assay (not shown). Western analysis showed that similar levels of LMP-1 mutants were expressed (Fig. 4F).
These structure/function studies show that LMP-1's inhibition of IFN-α-stimulated ISRE activity correlates with Tyk2 binding and inhibition of Tyk2 phosphorylation. These results also demonstrate that the LMP-1/Tyk2 interaction is independent of CTAR1 and the C-terminal 155 residues of LMP-1, which include CTAR2 and -3.
IFN-α resistance correlates with LMP-1 protein levels in LCLs.
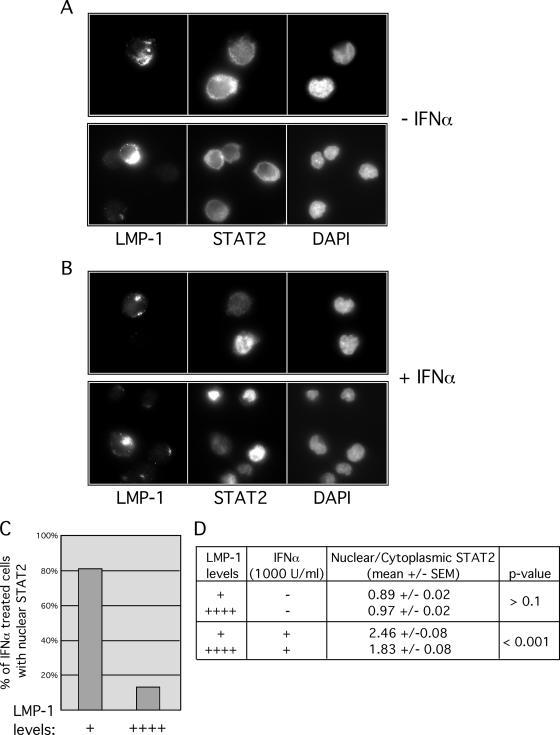
LMP-1 expression in a variety of human B-cell and non-B-cell lines resulted in inhibition of IFN-α-stimulated ISRE activity (Table 1; Fig. 3 and 4). We next investigated LMP-1 regulation of IFN-α signaling in EBV+ LCLs. LCLs (such as 721 cells) derived from in vitro EBV infection of human peripheral B cells require continuous LMP-1 expression for persistent proliferation (17). We predicted, based on results in Fig. 1 to 4, that 721 cells would be resistant to the effects of IFN-α. Cellular LMP-1 protein levels range over several logs within an LCL population (19). Therefore, the relationship between LMP-1 expression levels and STAT2 activation was determined using a single-cell assay. Relative nuclear and cytoplasmic STAT2 localization in 721 cells expressing either “high” or “low” levels of LMP-1 was assessed (Fig. 5). For untreated cells, no significant difference in STAT2 nuclear localization in cells expressing either high or low levels of LMP-1 was observed (Fig. 5A and D). However, IFN-α-stimulated STAT2 nuclear localization was diminished in cells expressing high levels of LMP-1 relative to that in cells expressing low LMP-1 levels (Fig. 5B, C, and D). Thus, IFN-α sensitivity correlates inversely with levels of LMP-1 protein expressed in 721 cells.
FIG. 5.
IFN-α-stimulated STAT2 nuclear localization is inhibited in 721 cells expressing high levels of LMP-1. (A, B) LMP-1 and STAT2 staining of 721 cells, untreated (A) or treated with IFN-α for 30 min (B). Cells were fixed, made permeable, and stained for LMP-1 and STAT2, and nuclei were stained with 4′,6′-diamidino-2-phenylindole as indicated below each panel. Two representative frames are shown (each) for untreated (A) and treated (B) cells. (C, D) The relationship between LMP-1 levels and IFN-stimulated STAT2 nuclear localization were quantified by two methods. (C) Stained cells were scored by visual inspection for STAT2 nuclear localization (“−” or “+”) and intensity of LMP-1 staining (“+”, very low but detectable; “++++”, very high). For some cells within a frame, the intensity of LMP-1 staining, although detectable, was too low to be captured without overexposing the adjacent cell. The percentage of IFN-α-treated cells with nuclear STAT2 was determined for cells expressing either high (“++++”) or low (“+”) levels of LMP-1. Data are expressed as percent IFN-α-treated cells with nuclear STAT2 versus level of LMP-1 expression. (D) The relationship between LMP-1 levels and STAT2 nuclear localization (−/+ IFN stimulation) was also quantified by determining the ratio of mean nuclear localization of STAT2/mean cytoplasmic localization in cells expressing either high (++++) or low (+) levels of LMP-1. Data are expressed as mean nuclear/cytoplasmic staining ± standard errors of the means. In untreated cells and in IFN-α-treated cells, statistical significance of ratios obtained from cells expressing high versus low LMP-1 levels was analyzed using Student's t test. Images shown in A and B are representative images of cells analyzed in C and D.
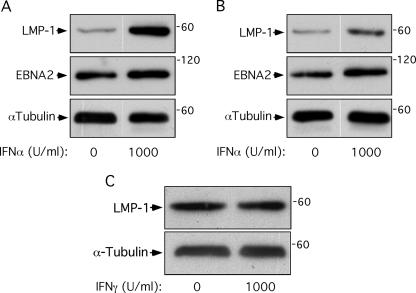
LCLs expressing LMP-1 survive prolonged growth in IFN-α.
IFN-α can trigger antiproliferative as well as antiviral responses in cells. IFN-α-stimulated STAT2 nuclear localization was diminished in 721 cells expressing high levels of LMP-1 (Fig. 5), suggesting cells expressing high levels of LMP-1 within a 721 cell population might be resistant to the antiproliferative effects of IFN-α. This hypothesis predicts that prolonged culture of 721 cells in IFN-α will select for outgrowth of a population expressing high levels of LMP-1 relative to cells grown without IFN-α. To test this hypothesis, 721 cells were plated in various concentrations of IFN-α and IFN-γ, and changes in LMP-1 protein levels in response to prolonged culture in IFN were analyzed by Western blotting (Fig. 6A and B). LMP-1 levels were similar in all cell populations after 24 h of growth in IFN-α but were detectably elevated by 72 h of treatment with 1,000 U/ml of IFN-α (not shown). IFN-α-treated cell populations were split after 14 days and cultured in the presence or absence of IFN-α for another 27 days. LMP-1 protein levels were higher in cells grown in 1,000 U/ml IFN-α for 41 days than in cells grown without IFN-α (Fig. 6A). LMP-1 protein levels remained elevated after removal of IFN-α and continued culture for 27 days (Fig. 6B). Levels of the EBV latent protein EBNA2 remained fairly constant under these conditions (Fig. 6A and B), suggesting that that the effects of IFN-α on LMP-1 were not secondary to selection for cells with a higher viral genome copy number but rather were specific for selection of cells expressing higher levels of LMP-1. Importantly, selection in IFN-γ had no effect on LMP-1 protein levels (Fig. 6C). The observation that LMP-1 protein levels remain elevated following removal of IFN-α is consistent with selection for IFN-α-resistant cells expressing high LMP-1 levels rather than IFN-α-mediated transcriptional or posttranslational regulation of LMP-1.
FIG. 6.
LMP-1 protein levels in EBV+ LCLs are elevated following prolonged growth in IFN-α. 721 cells were treated with IFN-α for 41 days (A) or treated with IFN-α for 14 days, washed, and then cultured in the absence of IFN-α for 27 days (B) or treated with IFN-γ for 21 days (C). Cells were then harvested and analyzed by Western blotting for LMP-1 and α-tubulin as a loading control. IFN treatment conditions are shown below the blots. These results are representative of three independent experiments.
DISCUSSION
Early reports describing antiviral and antiproliferative effects of IFN-α on EBV infection of B cells are consistent with LMP-1's anti-IFN-α activity. IFN-α treatment of unfractionated or T-cell-depleted cord blood mononuclear cells (8), adult T-cell-depleted mononuclear cells (8), or adult B cells (46) prior to, or at the time of, addition of B95-8 virus prevents EBV-mediated B cell outgrowth. This presumably occurs through the establishment of an antiviral/antiproliferative state in the targeted cells. In contrast, newly EBV-infected B cells become resistant to the effects of IFN-α within a few days postinfection (8, 46). The time course of acquisition of IFN-α resistance (25) correlates generally with the kinetics of LMP-1 protein expression in newly infected B cells (7, 42).
Several EBV gene products and promoters have been linked to regulation of, or by, the type I IFN pathway. The EBNA1 promoter Qp can be regulated by IRF1 and IRF2 (38) and repressed by IRF2 and IRF7 (32, 50, 53). EBNA2 can stimulate beta interferon expression and ISGF3 activity in BL cells (13), whereas Daudi cells, which represent a BL cell line harboring an EBNA2 deletion EBV strain that is exquisitely sensitive to IFN-α's antiproliferative effects, become IFN-α resistant upon ectopic expression of EBNA2 (12). Importantly, EBNA2 is a positive regulator of the LMP-1 promoter. Whether IFN-α resistance in EBNA2-positive Daudi cells is a consequence of EBNA2 expression itself or is due to EBNA2's induction of LMP-1 expression is unknown. Epstein-Barr virus-encoded small RNAs prevent IFN-α-induced apoptosis in Akata, Daudi, and Mutu1 cell lines (31) but are unlikely to confer IFN resistance, because they are highly expressed in both IFN-α-resistant and IFN-α-sensitive Daudi cell lines (44).
Tyk2 was identified in our initial screen to isolate proteins that interact with LMP-1's transmembrane domain. We focused on the physical and functional interaction between LMP-1 and Tyk2 because of Tyk2's essential role in antiviral signaling triggered by IFN-α. Tyk2 is present in the native LMP-1 complex isolated from EBV-immortalized cells, and its phosphorylation on tyrosine is greatly diminished in cells expressing LMP-1, suggesting LMP-1 either prevents Tyk2 phosphorylation or increases the rate of Tyk2 dephosphorylation (Fig. 1 and 2). LMP-1 suppresses both STAT1 and -2 phosphorylation, although the effect of LMP-1 on STAT2 phosphorylation is of greater magnitude (Fig. 2). This preferential suppression of STAT2 tyrosine phosphorylation by LMP-1 is consistent with the finding that some IFN-α-activated signaling pathways are Tyk2 independent (14, 41). Whereas both STAT1 and STAT2 are phosphorylated in response to IFN-α in mouse embryonic fibroblasts and bone marrow macrophages (Tyk2+/+), IFN-α-stimulated STAT2 but not STAT1 phosphorylation in Tyk2−/− mice is diminished (14). Notably, Tyk2 plays a key role in IFN-α-mediated but STAT1-independent inhibition of interleukin 7-dependent growth and survival of B- and T-cell lymphoid progenitors (40). Thus, IFN-α, via Tyk2, can be coupled to downstream signaling pathways via alternative mechanisms that do not involve STAT1.
The relatively modest effect on IFN-α-stimulated cellular STAT2 phosphorylation (23% reduction) is consistent with LMP-1's apparent specific targeting of the detergent-insoluble STAT2 pool (Fig. 2C and D). We found 10 to 20% of cellular STAT proteins in the detergent-insoluble pellet. This fraction contains IFNAR, IFNAR-associated STATs, nuclear STATs, and 20 to 30% of cellular LMP-1 (23, 26, 34, 39). The Triton X-100 insolubility of IFNAR and LMP-1 results from their association with the cytoskeleton (23, 26, 34). The differential effect of LMP-1 on the insoluble STAT2 pool is consistent with a model in which LMP-1 associates with the IFNAR/Jak/STAT signaling complex at the membrane to regulate its activity.
LMP-1's inhibition of Tyk2 phosphorylation correlates with a diminution in ISRE activation by IFN-α in a wide range of human cell lines (Fig. 3 and 4; Table 1). LMP-1's regulation of IFN signaling is upstream of ISGF3 and is specific for the type I IFN pathway (Fig. 3). LMP-1's interaction with the IFN-α pathway does not require CTAR1 or the last 155 residues of the C terminus, which encode CTAR2 and -3. Thus, it is likely that the LMP-1/Tyk2 interaction is mediated by LMP-1's transmembrane domain and/or the membrane-proximal 30 to 40 residues of the C terminus. This CTAR independence suggests strongly that LMP-1's effect on IFN signaling is not a secondary effect of C-terminal TRAF-dependent NF-κB or Jun N-terminal protein kinase activation.
We found the sensitivity of EBV-immortalized 721 cells to IFN-α to depend upon the level of LMP-1 protein expressed in the cell (Fig. 5). LMP-1 protein levels span a 2-log range in populations of EBV-immortalized LCLs, including 721 cells (19), and IFN-α-stimulated STAT2 nuclear translocation was blocked in 721 cell subpopulations expressing high levels of LMP-1 (Fig. 5). Consistent with this observation, prolonged growth of 721 cells in 1,000 U/ml IFN-α results in the outgrowth of cells stably expressing higher levels of the LMP-1 protein than those grown without IFN-α (Fig. 6). Our results do not distinguish whether the levels of LMP-1 in the entire population are elevated following IFN selection or if a subset of cells within the selected population are expressing extremely high levels of LMP-1. We think the latter possibility is unlikely given LMP-1's profound negative effect on proliferation when expressed at very high levels in LCLs (19).
Our interpretation of these results is that 721 cells expressing high enough levels of LMP-1 to block IFN-α signaling but not so high as to inhibit proliferation (11) emerge following selection in IFN-α. Such a model may have implications for chronic IFN-α treatment of EBV+ patients. Current therapeutic applications of IFN-α include treatments for hematological malignancies, such as multiple myeloma, solid tumors, such as malignant melanoma, and viral diseases, such as hepatitis B and C (35). EBV+ patients undergoing long-term IFN-α treatment may be at risk for IFN-α-induced selection of EBV+ B cells expressing elevated, and therefore potentially more oncogenic, levels of LMP-1.
Tyk2 plays key roles in additional IFN-α and cytokine-stimulated pathways. Tyk2 is involved in the direct activation of PI3K and NF-κB by IFN-α (35), interleukin 12 signaling (1), and IFN-γ signaling (35). To our knowledge, viral targeting of IFN-α signaling via Tyk2 is limited to human papillomavirus (22), Japanese encephalitis virus (24), and now EBV. The widespread involvement of Tyk2 in cytokine signaling raises the possibility of viral regulation of as-of-yet-unappreciated Tyk2-dependent pathways. Understanding LMP-1's effect on other Tyk2-dependent signaling events may provide insight into how viruses interfere with Tyk2 signaling.
Ectopic LMP-1 stimulates basal expression of a subset of ISGs through CTAR-mediated NF-κB activation of IRF7 expression (51, 52, 54). LMP-1's effect on IFN-α-stimulated ISG regulation was not explored in these studies, and our observed CTAR-independent negative regulation of IFN-α signaling by LMP-1 is not inconsistent with these reports. Expression of either or both of these mechanistically distinct LMP-1 activities could occur in a manner dictated by the signaling environment of the infected cell. For example, IFN-β, one of the subset of LMP-1/IRF7-stimulated ISGs, is secreted by latently infected B cells (30, 49). IFN secretion by the latently infected cell would be predicted to exert an autocrine-mediated antiproliferative effect, an effect that LMP-1's negative regulation of IFN signaling would effectively curtail to the advantage of viral persistence. LMP-1's potential contribution to regulation of IFN signaling during EBV's life cycle in vivo will be restricted to infected cells expressing LMP-1 (latency II/III) and not to memory cells in which EBV is maintained persistently for the life of the host. In this context, LMP-1-positive infected cells include newly infected naive B cells and germinal center B cells in the immunocompetent host and tumors in which LMP-1 is expressed (i.e., posttransplant lymphoproliferative disease, Hodgkin's disease, and immunoblastic lymphoma) (45). Accumulating evidence suggests that EBV infects epithelial cells of the oropharynx (47) and that LMP-1, in addition to being expressed in a fraction of nasopharyngeal carcinoma tumors, may also be expressed in infected epithelial cells in vivo (33). Thus, there are a variety of infected cell types and stages of EBV infection in vivo in which LMP-1's regulation of IFN signaling could have an impact on the establishment of EBV persistence. Regulation of IFN-α signaling may represent an evolved strategy balancing the needs of the virus to persist and of the host cell to inactivate the virus. Thus, analysis of EBV-targeted signaling events, including Tyk2-dependent pathways, provides opportunities to identify and understand complexities in cellular signaling that play a role in EBV persistence in vivo.
.
Acknowledgments
We thank members of the Martin lab and Brad Olwin, Tin Tin Su, Xuedong Liu, and Kelly Geiger for critical reading of the manuscript. We thank John Krolewski, Curt Horvath, and Luwen Zhang for providing plasmids.
This work was supported in part by NIH grants CA095043 and AI-01537 to J. M. Martin.
Footnotes
Published ahead of print on 20 September 2006.
REFERENCES
- 1.Bacon, C. M., D. W. McVicar, J. R. Ortaldo, R. C. Rees, J. J. O'Shea, and J. A. Johnston. 1995. Interleukin 12 (IL-12) induces tyrosine phosphorylation of JAK2 and TYK2: differential use of Janus family tyrosine kinases by IL-2 and IL-12. J. Exp. Med. 181:399-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baichwal, V. R., and B. Sugden. 1988. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus. Oncogene 2:461-467. [PubMed] [Google Scholar]
- 3.Coffin, W. F., III, K. D. Erickson, M. Hoedt-Miller, and J. M. Martin. 2001. The cytoplasmic amino-terminus of the latent membrane protein-1 of Epstein-Barr virus: relationship Between transmembrane orientation and effector functions of the carboxy-terminus. Oncogene 20:5313-5330. [DOI] [PubMed] [Google Scholar]
- 4.Coffin, W. F., III, T. Geiger, and J. M. Martin. 2003. Transmembrane domains 1 and 2 of the latent membrane protein 1 (LMP-1) of Epstein-Barr virus contain a lipid raft targeting signal and play a critical role in cytostasis. J. Virol. 77:3749-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devergne, O., E. Hatzivassiliou, K. Izumi, K. Kaye, M. Kleijnen, E. Kieff, and G. Mosialos. 1996. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-kB activation. Mol. Cell. Biol. 16:7098-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eilers, A., K. Kanda, B. Klose, J. Krolewski, and T. Decker. 1996. Constitutive STAT1 tyrosine phosphorylation in U937 monocytes overexpressing the TYK2 protein tyrosine kinase does not induce gene transcription. Cell Growth Differ. 7:833-840. [PubMed] [Google Scholar]
- 7.Erickson, K. D., and J. M. Martin. 1997. Early detection of the lytic LMP-1 protein in EBV-infected B-cells suggests its presence in the virion. Virology 234:1-13. [DOI] [PubMed] [Google Scholar]
- 8.Garner, J. G., M. S. Hirsch, and R. T. Schooley. 1984. Prevention of Epstein-Barr virus-induced B-cell outgrowth by interferon alpha. Infect. Immun. 43:920-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gires, O., F. Kohlhuber, E. Kilger, M. Baumann, A. Kieser, C. Kaiser, R. Zeidler, B. Scheffer, M. Ueffing, and W. Hammerschmidt. 1999. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 18:3064-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gires, O., U. Zimber-Strobl, R. Gonnella, M. Ueffing, G. Marschall, R. Zeidler, D. Pich, and W. Hammerschmidt. 1997. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 16:6131-6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammerschmidt, W., B. Sugden, and V. R. Baichwal. 1989. The transforming domain alone of the latent membrane protein of Epstein-Barr virus is toxic to cells when expressed at high levels. J. Virol. 63:2469-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanda, K., T. Decker, P. Aman, M. Wahlstrom, A. von Gabain, and B. Kallin. 1992. The EBNA2-related resistance towards alpha interferon (IFN-alpha) in Burkitt's lymphoma cells effects induction of IFN-induced genes but not the activation of transcription factor ISGF-3. Mol. Cell. Biol. 12:4930-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanda, K., B. Kempkes, G. W. Bornkamm, A. von Gabain, and T. Decker. 1999. The Epstein-Barr virus nuclear antigen 2 (EBNA2), a protein required for B lymphocyte immortalization, induces the synthesis of type I interferon in Burkitt's lymphoma cell lines. Biol Chem. 380:213-221. [DOI] [PubMed] [Google Scholar]
- 14.Karaghiosoff, M., H. Neubauer, C. Lassnig, P. Kovarik, H. Schindler, H. Pircher, B. McCoy, C. Bogdan, T. Decker, G. Brem, K. Pfeffer, and M. Muller. 2000. Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity 13:549-560. [DOI] [PubMed] [Google Scholar]
- 15.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte transformation. Proc. Natl. Acad. Sci. USA 90:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kieff, E. 1996. Epstein-Barr virus and its replication, 3rd ed. Raven Press, Ltd., Philadelphia, Pa.
- 17.Kilger, E., A. Kieser, M. Baumann, and W. Hammerschmidt. 1998. Epstein-Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J. 17:1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraus, T. A., J. F. Lau, J. P. Parisien, and C. M. Horvath. 2003. A hybrid IRF9-STAT2 protein recapitulates interferon-stimulated gene expression and antiviral response. J. Biol. Chem. 278:13033-13038. [DOI] [PubMed] [Google Scholar]
- 19.Lam, N., M. L. Sandberg, and B. Sugden. 2004. High physiological levels of LMP1 result in phosphorylation of eIF2 alpha in Epstein-Barr virus-infected cells. J. Virol. 78:1657-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam, N., and B. Sugden. 2003. CD40 and its viral mimic, LMP1: similar means to different ends. Cell. Signal. 15:9-16. [DOI] [PubMed] [Google Scholar]
- 21.Li, H. P., and Y. S. Chang. 2003. Epstein-Barr virus latent membrane protein 1: structure and functions. J. Biomed. Sci. 10:490-504. [DOI] [PubMed] [Google Scholar]
- 22.Li, S., S. Labrecque, M. C. Gauzzi, A. R. Cuddihy, A. H. Wong, S. Pellegrini, G. J. Matlashewski, and A. E. Koromilas. 1999. The human papilloma virus (HPV)-18 E6 oncoprotein physically associates with Tyk2 and impairs Jak-STAT activation by interferon-alpha. Oncogene 18:5727-5737. [DOI] [PubMed] [Google Scholar]
- 23.Liebowitz, D., R. Kopan, E. Fuchs, J. Sample, and E. Kieff. 1987. An Epstein-Barr virus transforming protein associates with vimentin in lymphocytes. Mol. Cell. Biol. 7:2299-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin, R. J., C. L. Liao, E. Lin, and Y. L. Lin. 2004. Blocking of the alpha interferon-induced Jak-Stat signaling pathway by Japanese encephalitis virus infection. J. Virol. 78:9285-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lotz, M., C. D. Tsoukas, S. Fong, D. A. Carson, and J. H. Vaughan. 1985. Regulation of Epstein-Barr virus infection by recombinant interferons. Selected sensitivity to interferon-gamma. Eur. J. Immunol. 15:520-525. [DOI] [PubMed] [Google Scholar]
- 26.Mann, K. P., and D. Thorley-Lawson. 1987. Posttranslational processing of the Epstein-Barr virus-encoded p63/LMP protein. J. Virol. 61:2100-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, J. M., and B. Sugden. 1991. The LMP onco-protein resembles activated receptors in its properties of turnover. Cell Growth Differ. 2:653-660. [PubMed] [Google Scholar]
- 28.Martin, J. M., and B. Sugden. 1991. Transformation by the oncogenic latent membrane protein correlates with its rapid turnover, membrane localization, and cytoskeletal association. J. Virol. 65:3246-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, G. (ed.). 1985. Epstein-Barr Virus, 1st ed. Raven Press, New York, N.Y.
- 30.Najjar, I., F. Baran-Marszak, C. Le Clorennec, C. Laguillier, O. Schischmanoff, I. Youlyouz-Marfak, M. Schlee, G. W. Bornkamm, M. Raphael, J. Feuillard, and R. Fagard. 2005. Latent membrane protein 1 regulates STAT1 through NF-kB-dependent interferon secretion in Epstein-Barr virus-immortalized B cells. J. Virol. 79:4936-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nanbo, A., K. Inoue, K. Adachi-Takasawa, and K. Takada. 2002. Epstein-Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt's lymphoma. EMBO J. 21:954-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nonkwelo, C., I. K. Ruf, and J. Sample. 1997. Interferon-independent and -induced regulation of Epstein-Barr virus EBNA-1 gene transcription in Burkitt lymphoma. J. Virol. 71:6887-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pegtel, D. M., J. Middeldorp, and D. A. Thorley-Lawson. 2004. Epstein-Barr virus infection in ex vivo tonsil epithelial cell cultures of asymptomatic carriers. J. Virol. 78:12613-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfeffer, L. M., N. Stebbing, and D. B. Donner. 1987. Cytoskeletal association of human alpha-interferon-receptor complexes in interferon-sensitive and -resistant lymphoblastoid cells. Proc. Natl. Acad. Sci. USA 84:3249-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Platanias, L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5:375-386. [DOI] [PubMed] [Google Scholar]
- 36.Rabson, M., L. Heston, and G. Miller. 1983. Identification of a rare Epstein-Barr virus variant that enhances early antigen expression in Raji cells. Proc. Natl. Acad. Sci. USA 80:2762-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandberg, M. L., A. Kaykas, and B. Sugden. 2000. Latent membrane protein 1 of Epstein-Barr virus inhibits as well as stimulates gene expression. J. Virol. 74:9755-9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaefer, B. C., E. Paulson, J. L. Strominger, and S. H. Speck. 1997. Constitutive activation of Epstein-Barr virus (EBV) nuclear antigen 1 gene transcription by IRF1 and IRF2 during restricted EBV latency. Mol. Cell. Biol. 17:873-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sehgal, P. B., G. G. Guo, M. Shah, V. Kumar, and K. Patel. 2002. Cytokine signaling: STATS in plasma membrane rafts. J. Biol. Chem. 277:12067-12074. [DOI] [PubMed] [Google Scholar]
- 40.Shimoda, K., K. Kamesaki, A. Numata, K. Aoki, T. Matsuda, K. Oritani, S. Tamiya, K. Kato, K. Takase, R. Imamura, T. Yamamoto, T. Miyamoto, K. Nagafuji, H. Gondo, S. Nagafuchi, K. Nakayama, and M. Harada. 2002. Cutting edge: tyk2 is required for the induction and nuclear translocation of Daxx which regulates IFN-alpha-induced suppression of B lymphocyte formation. J. Immunol. 169:4707-4711. [DOI] [PubMed] [Google Scholar]
- 41.Shimoda, K., K. Kato, K. Aoki, T. Matsuda, A. Miyamoto, M. Shibamori, M. Yamashita, A. Numata, K. Takase, S. Kobayashi, S. Shibata, Y. Asano, H. Gondo, K. Sekiguchi, K. Nakayama, T. Nakayama, T. Okamura, S. Okamura, and Y. Niho. 2000. Tyk2 plays a restricted role in IFN alpha signaling, although it is required for IL-12-mediated T cell function. Immunity 13:561-571. [DOI] [PubMed] [Google Scholar]
- 42.Sinclair, A. J., I. Palmero, G. Peters, and P. J. Farrell. 1994. EBNA-2 and EBNA-LP cooperate to cause G0 to G1 transition during immortalization of resting human B lymphocytes by Epstein-Barr virus. EMBO J. 13:3321-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su, T. T., G. Feger, and P. H. O'Farrell. 1996. Drosophila MCM protein complexes. Mol. Biol. Cell 7:319-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swaminathan, S., B. S. Huneycutt, C. S. Reiss, and E. Kieff. 1992. Epstein-Barr virus-encoded small RNAs (EBERs) do not modulate interferon effects in infected lymphocytes. J. Virol. 66:5133-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorley-Lawson, D. A. 2005. EBV the prototypical human tumor virus—just how bad is it? J. Allergy Clin. Immunol. 116:251-262. [DOI] [PubMed] [Google Scholar]
- 46.Thorley-Lawson, D. A. 1981. The transformation of adult but not newborn human lymphocytes by Epstein Barr virus and phytohemagglutinin is inhibited by interferon: the early suppression by T cells of Epstein Barr infection is mediated by interferon. J. Immunol. 126:829-833. [PubMed] [Google Scholar]
- 47.Tugizov, S. M., J. W. Berline, and J. M. Palefsky. 2003. Epstein-Barr virus infection of polarized tongue and nasopharyngeal epithelial cells. Nat. Med. 9:307-314. [DOI] [PubMed] [Google Scholar]
- 48.Verma, A., S. Kambhampati, S. Parmar, and L. C. Platanias. 2003. Jak family of kinases in cancer. Cancer Metastasis Rev. 22:423-434. [DOI] [PubMed] [Google Scholar]
- 49.Xu, D., K. Brumm, and L. Zhang. 2006. The latent membrane protein 1 of EBV primes EBV latency cells for type 1 interferon production. J. Biol. Chem. 281:9163-9169. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, L., and J. S. Pagano. 1999. Interferon regulatory factor 2 represses the Epstein-Barr virus BamHI Q latency promoter in type III latency. Mol. Cell. Biol. 19:3216-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, L., and J. S. Pagano. 2000. Interferon regulatory factor 7 is induced by Epstein-Barr virus latent membrane protein 1. J. Virol. 74:1061-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, L., and J. S. Pagano. 2001. Interferon regulatory factor 7 mediates activation of Tap-2 by Epstein-Barr virus latent membrane protein 1. J. Virol. 75:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, L., and J. S. Pagano. 1997. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol. Cell. Biol. 17:5748-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, L., L. Wu, K. Hong, and J. S. Pagano. 2001. Intracellular signaling molecules activated by Epstein-Barr virus for induction of interferon regulatory factor 7. J. Virol. 75:12393-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]