Abstract
Bovine viral diarrhea virus (BVDV) is a pestivirus that can establish a persistent infection in the developing fetus and has the ability to disable the production of type I interferon. In this report, we extend our previous observations that BVDV encodes a protein able to specifically block the activity of interferon regulatory factor 3 (IRF-3), a transcription factor essential for interferon promoter activation, by demonstrating that this is a property of the N-terminal protease fragment (NPro) of the BVDV polyprotein. Although BVDV infections cause relocalization of cellular IRF-3 from the cytoplasm to the nucleus early in infection, NPro blocks IRF-3 from binding to DNA. NPro has the additional property of targeting IRF-3 for polyubiquitination and subsequent destruction by cellular multicatalytic proteasomes. The autoprotease activity of NPro is not required for the inhibition of type I interferon induction or the targeting of IRF-3 for degradation.
Bovine viral diarrhea virus (BVDV) has a global spread and is a major reproductive pathogen of cattle (12). BVDV, along with classical Swine fever virus (CSFV), Border disease virus of sheep, and a small number of isolates from undomesticated species, make up the genus Pestivirus in the family Flaviviridae. The viruses are positive-strand RNA viruses with a genome on the order of 12.5 kb in length. The RNA is translated into a single viral polyprotein that is processed by both viral and host proteases to either 11 or 12 polypeptides, depending on the virus biotype. BVDV is generally noncytopathogenic (ncp), although the virus associated with the development of mucosal disease in persistently infected animals is a cytopathogenic (cp) biotype. The result of infection of a susceptible host with BVDV is unusual; BVDV can cause an acute infection like other viruses, but infection of a pregnant animal can result in transmission of virus from the dam to the developing fetus, where the virus can replicate. Infection of the fetus prior to immune competence results in a failure of the fetus to control virus infection and can result in the birth of persistently infected offspring that fail to mount an acquired immune response to BVDV. These offspring then serve as a reservoir for further acute virus infection and are held to be critical for BVDV transmission in areas of endemicity.
Although infection during fetal development takes place in the absence of a functional acquired immune response, viruses still have to evade innate immunity in order to establish a persistent infection; the ability of ncpBVDV to avoid the induction of interferon (IFN) seems critical in this regard (5). We and others have shown that ncpBVDV encodes an active block to type I IFN induction (3, 46), and there is evidence that ncpBVDV employs more than one mechanism to bring about this block. BVDV-infected cells are refractory to the addition of double-stranded RNA (dsRNA) to the medium. At least in part, the failure of this extracellular dsRNA to induce type I IFN can be explained by the action of the secreted viral Erns glycoprotein, which can bind to dsRNA and degrade it, probably by internalization and activation of the Erns dsRNase in endosomes (18). In addition to the inhibition of activation of IFN induction by Erns, BVDV blocks the activation of beta interferon (IFN-β) in an Erns-independent manner when either Semliki Forest virus (SFV) (18) or transfected dsRNA (unpublished data) is used as an inducer; we presume this mechanism is required to block the actions of an IFN inducer generated inside the cell during the BVDV life cycle.
The induction of type I IFN has been studied in great detail. Induction of IFN-β in response to dsRNA or virus infection is a complex process requiring the activation of NF-κB and interferon regulatory factor 3 (IRF-3). These proteins are then assembled on the IFN-β promoter in association with the accessory factor HMG-I/Y (reviewed in reference 32). There appear to be two pathways of activation in response to dsRNA (reviewed in reference 21). One of these is initiated by Toll-like receptor 3 (TLR3) in response to exogenous dsRNA, presumably released as a result of lytic infection. TLR3 engagement recruits the adaptor protein TRIF (45), which coordinates the bifurcation of the signaling response, leading to the activation of NF-κB and IRF-3. The activation of NF-κB is brought about by the recruitment of the IKK complex to TRIF via intermediate interactions with components in either the TRAF6/TAB2/TAB3/TAK-1 pathway (15, 19, 45) or the RIP/FADD/caspase 8 pathway (6, 33), while IRF-3 is directly phosphorylated by the related kinases TBK1 and IKKɛ (11, 48), which are recruited to TRIF by either a TRAF3-dependent (36) or a NAP1-dependent (43) mechanism. The other dsRNA response pathway is TLR3 independent and is initiated by the RNA helicases RIG-I and Mda-5 in response to intracellular dsRNA (1, 55, 56). These molecules are activated in an as-yet-uncharacterized manner but utilize their N-terminal CARD homology domains to engage an adaptor molecule (variously called IPS-1, MAVS, VISA, or Cardif and referred to below as Cardif), which in turn can signal to IRF-3 and NF-κB, probably by recruitment of key signaling molecules (22, 34, 47, 54; reviewed in references 17 and 20).
We previously demonstrated that ncpBVDV infections are capable of mobilizing IRF-3 from the cytoplasm to the nucleus but that IRF-3 is unable to bind to DNA; indeed, ncpBVDV infections are also able to block IRF-3-DNA complex formation induced by a coinfecting virus (3). In contrast, ncpBVDV infections fail to induce NF-κB or IRF-7 (2, 3), demonstrating that BVDV encodes a mechanism for blocking the function of the one factor it activates. Studies of other flaviviruses have indicated several potential viral polypeptides that could play a role in blocking IFN-β induction. The NS3/4a protease of Hepatitis C virus (HCV) is able to cleave TRIF so that the response to exogenous dsRNA is inhibited (10), and this protein is also able to inhibit the intracellular dsRNA response by cleaving the RNA helicase adaptor Cardif (28, 34). The NS5a product of HCV has been demonstrated to inhibit the function of the dsRNA-dependent protein kinase PKR (reviewed in reference 16), a molecule that is frequently implicated in cell signaling in response to dsRNA. It has been reported recently that the NPro polypeptide of CSFV may be a critical factor for the control of IFN synthesis, since CSFV engineered with a deletion in NPro induces IFN and does not inhibit the induction of IFN by a heterologous virus (40), while exogenous expression of the CSFV NPro protein down-regulates IFN induction through a block to IRF-3 transcription (26, 39). Expression of BVDV NPro has also been recently shown to antagonize IFN production in an uncharacterized manner (13). Here, we show that the NPro product of ncpBVDV is necessary and sufficient for the inhibition of IFN-β induction. Like CSFV, the BVDV NPro targets the activity of IRF-3, but in contrast to the suggested property of CSFV (26), the BVDV NPro product does not appear to target the transcription of the IRF-3 gene but rather both prevents IRF-3 from binding to DNA and targets IRF-3 for degradation using the host cell's proteasome machinery.
MATERIALS AND METHODS
Viruses and cells.
The ncpBVDV strain pe515-ncp was obtained from M. C. Clarke (Institute for Animal Health, Compton, United Kingdom) and cp7 from a cDNA clone, a gift from Gregor Meyers, BFAV, Tubingen, Germany. Stocks were prepared in calf testis cell cultures maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 2% fetal bovine serum free of both BVDV and anti-BVDV antibody or in 2% chick serum. Calf testis cells were used between passages 4 and 7. For infection with ncpBVDV, calf testis cells were inoculated at a multiplicity of infection (MOI) of approximately 2 PFU per cell. After 1 h at 37°C, the inoculum was removed and replaced with warm DMEM containing 2% fetal bovine serum or 2% chick serum. Mock infection was carried out with DMEM and fetal bovine serum or chick serum.
HEp2 (human epithelial) and Vero (African green monkey kidney) cells were obtained from ATCC and were routinely passaged in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. To construct Hep2 cell lines stably expressing NPro or the C69S variant of NPro, the NPro gene of BVDV, amplified from pEF.NPro(pe515) or pEF.NPro:C69S(pe515) (see below) by PCR, was cloned into a bicistronic expression vector derived from the self-inactivating lentivirus vector pHR-SIN-CSGW (7). The resulting construct, pdl.NPro(pe515) or pdl.NPro:C69S(pe515), encodes both BVDV NPro (fused to a Vs epitope tag [38] at the carboxyl terminus) and puromycin resistance. To generate recombinant lentiviral particles, 293FT cells (InVitrogen) were transfected with pdl.NPro(pe515) or pdl.NPro:C69S(pe515), pCMVR8.91 (expressing gal/pol, tat, and rev genes of human immunodeficiency virus type 1), and pMD-G (expressing the vesicular stomatitis virus G gene). Two days after transfection, viruses were harvested from the culture medium and filtered through 0.45-μm Tuffryn Membrane filters (InVitrogen), and then HEp2 cells were transduced with the harvested recombinant lentiviral particles at a multiplicity of infection of approximately 1 in the presence of Polybrene (Sigma; 8 μg/ml). Two days posttransduction, pools of transformed cells were selected by resistance to puromycin (2 μg/ml). Expression of the NProC69S transgene was confirmed by Western blotting using a monoclonal antibody against the Vs epitope tag.
Plasmids.
The reporter plasmids with the firefly luciferase gene under the control of the human IFN-β promoter [pIFΔ(−116)lucter] and the synthetic PRD II multimer reporter [p(PRDII)5tkΔ(−39)lucter] have been described previously (23, 51), as has the constitutive β-galactosidase transfection control reporter plasmid pJATlac (31). A luciferase reporter [p(ISG15ISRE)4tkΔ(−39)lucter] under the control of a multimer of the IRF-3-responsive ISG15 interferon-stimulated response element (ISRE) was constructed in the same way as p(PRDII)5tkΔ(−39)lucter. pISG54lucter contains the luciferase gene under the control of the human ISG54 promoter.
Expression in mammalian cells was achieved by cloning cDNAs between the NcoI and XbaI sites of the EF1α promoter vector, pEF.plink2 (30). Clones expressing fragments of the pe515 strain of ncpBVDV polyprotein were generated from existing DNA templates by PCR using primers that incorporated NcoI and XbaI sites at the 5′ and 3′ ends, respectively. The following fragments were generated: pEF.NPro(pe515), polyprotein amino acids 1 to 168; pEF.NS2/3(pe515), polyprotein amino acids 1132 to 2272; pEF.NS3(pe515), polyprotein amino acids 1589 to 2272; pEF.NS2/3/4(pe515), polyprotein amino acids 1132 to 2336; and pEF.NS5a(pe515), polyprotein amino acids 2683 to 3178. To construct equivalent clones from the cp7 strain of BVDV, the 27-base-pair duplication in the NS2 coding region associated with the cytopathogenic conversion (50) was deleted by PCR-mediated mutagenesis, and the resultant template was used in subsequent manipulations [plasmids pEF.NPro(cp7), pEF.NS2/3(cp7), pEF.NS3(cp7), and pEF.NS2/3/4(cp7) thus have the same polyprotein amino acid coordinates as their pe515 counterparts]. We also generated a variant of pEF.NPro(pe515) containing a cysteine-to-serine change at amino acid 69 in NPro [pEF.NPro:C69S(pe515)], a variant of pEF.NPro(pe515) in which the NPro fragment included the C-terminal protein normally cleaved by NPro autoprotease activity [designated pEF.NProC(1-201) to indicate that the NPro fragment is 201 amino acids] and a series of N- and C-terminal deletions of pEF.NProC(1-201). Each of the clones discussed in this section was generated using PCR-mediated site mutagenesis, and specific details of the primers used in PCR steps are available on request. The products were routinely sequenced on an ABI Prism 3100 Genetic Analyzer. The expression vectors for the V protein of simian virus 5 (SV5) (pEF.SV5/V) and Mda-5 (pEF.mda-5) have been described previously (1, 8). Expression vectors for full-length RIG-I, TBK1, and TRIF were made by reverse transcription-PCR from human placental RNA and cloned into pEF.plink2 (K. Childs and S. Goodbourn, unpublished data).
Transfection and analysis of gene expression.
Transfections were carried out using either Lipofectamine (InVitrogen) or Polyfect (QIAGEN). For induction of cells by synthetic dsRNA, poly(I) · poly(C) (Amersham Biosciences) was transfected into cells using Lipofectamine under conditions specified by the manufacturer. For reporter gene assays, lysates were prepared and analyzed for luciferase and β-galactosidase activities as previously described (23). Luciferase activity was corrected to the β-galactosidase activity to normalize for variations in the transfection efficiency. RNA was prepared from 9-cm dishes of confluent cultures of HEp2 or HEp2:NPro cells by using the acid phenol method and was analyzed by RNase protection using probes for IFN-β (58) and γ-actin (9). To generate a probe for human IRF-3 mRNA, a 191-bp fragment of cDNA was amplified by PCR using the primers 5′…TAATACGACTCACTATAGCTGGCTTATCCCTCCCGGGAA…3′ and 5′…GCCACGGATCCTGCCCTGGCT…3′. The underlined region of the first primer contains the T7 promoter, and thus, the PCR product can be used as a template to generate an RNA probe by transcription with T7 RNA polymerase. To generate a probe for bovine IRF-3, a full-length cDNA was generated from calf testis cells by reverse transcription-PCR and cloned into pCRBluntII.TOPO (InVitrogen); this plasmid was then linearized with NcoI and used as a template for SP6 RNA polymerase to generate a 285-base-pair probe complementary to the region between the NcoI site and the stop codon.
Electrophoretic mobility shift assays (EMSAs).
Nuclear extracts on calf testis or HEp2 cells were prepared and analyzed for IRF-3 or NF-κB binding activity as previously described (3).
Proteasomal inhibitors and polyubiquitination assay.
Epoxomicin (Alexis Biochemicals) and benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone (ZVAD; Bachem) were dissolved in dimethyl sulfoxide at 100 μM and 10 mM, respectively, and used at 100 nM (epoxomicin) and 100 μM (ZVAD). Equivalent volumes of dimethyl sulfoxide were added to control samples. For polyubiquitination assays, cell samples were treated as indicated below (see the legend to Fig. 6), and whole-cell extracts were prepared from 9-cm cell culture dishes using M-PER mammalian protein extraction reagent (Pierce) containing protease inhibitors. The protein concentration was determined by the Bradford assay, and 150-μg aliquots of each sample were incubated with 20 μl of Polyubiquitin Affinity Resin (Pierce) at 4°C for 16 h. The slurry was washed, and polyubiquitinated protein was recovered as described by the manufacturer. The polyubiquitin control sample was purchased from Pierce.
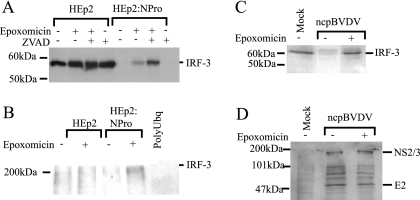
FIG. 6.
The BVDV NPro polypeptide targets IRF-3 for proteasomal degradation. (A) HEp2 and HEp2:NPro cells were either left untreated or treated with epoxomicin and/or ZVAD for 48 h, and then whole-cell extracts were prepared and analyzed for IRF-3 by Western blotting. (B) HEp2 and HEp2:NPro cells were either left untreated or treated with epoxomicin for 10 h, and then whole-cell extracts were prepared and incubated with a polyubiquitin-binding resin as described in Materials and Methods. The resin was washed, and the bound protein was eluted and separated by polyacrylamide gel electrophoresis. IRF-3-containing material was determined by Western blotting. (C) Whole-cell extracts were prepared from calf testis cells either mock infected or infected for 24 h by the pe515 strain of ncpBVDV at an MOI of 2. Infected cells were also treated with epoxomicin where indicated (inhibitors were present throughout the infection). The extracts were examined by Western blotting for the presence of IRF-3. (D) Whole-cell extracts were prepared from calf testis cells either mock infected or infected for 24 h by the pe515 strain of ncpBVDV at an MOI of 2 in the presence or absence of epoxomicin where indicated. The extracts were examined by Western blotting for the presence of BVDV polypeptides. The mobilities of two of these proteins (NS2/3 and E2) are indicated to the right of the panel.
Cell fractionation, antisera, and Western blots.
Cells were harvested by scraping them with a rubber policeman, recovered by centrifugation, and washed with phosphate-buffered saline, and the pellets were frozen until further use. The cell pellets were extracted by hypotonic lysis in the presence of 0.1% NP-40 and separated into cytoplasmic and nuclear fractions as previously described (3). Proteins were fractionated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels by standard procedures and were transferred to polyvinylidene difluoride membranes using semidry blotting. The filters were blocked and incubated with primary antibody overnight at 4°C. IRF-3 was detected using a rabbit polyclonal antibody raised against full-length recombinant human IRF-3 (AbCam AB11978). BVDV proteins were detected using polyclonal serum (39/97) from a gnotobiotic calf that had been challenged with pe515 ncpBVDV. Both antibodies were used at a dilution of 1:5,000. Rabbit immunoglobulin (Ig) was detected using anti-rabbit Ig-horseradish peroxidase (HRP) conjugate (Amersham) diluted 1:10,000. Bovine Ig was detected using an anti-bovine-HRP conjugate (Sigma-Aldrich) diluted at 1:5,000. The cytoplasmic marker protein GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and the nuclear marker protein TATA-binding protein (TBP) were detected using mouse monoclonal antibodies (mAbcam 9484 and ab818; AbCam) at dilutions of 1:10,000 and 1:2,000, respectively, and detected using anti-mouse Ig-HRP conjugate (Amersham) diluted 1:2,000. Bound secondary-antibody conjugates were detected using LumiGLO (Cell Signaling).
Coupled in vitro transcription/translation.
For coupled in vitro transcription/translation reactions, the NPro(cp7), NPro(pe515), and NProC(pe515) fragments and IRF-3 were cloned into pGBKT7 (Clontech). [35S]methionine-labeled proteins were generated from supercoiled plasmid templates using a T7 Quick coupled transcription/translation lysate according to the manufacturer's instructions (Promega).
RESULTS
The BVDV NPro polypeptide is necessary and sufficient to block IFN-β induction by dsRNA.
We previously reported that the ncp biotype of BVDV strain pe515 not only fails to induce the production of type I IFN, but also actively blocks the induction of IFN-β by a coinfecting virus (3), indicating that BVDV encodes a trans-acting inhibitor of IFN activation. To test the abilities of individual BVDV polypeptides to block IFN-β induction, we cloned cDNAs encoding discrete fragments of the polyproteins of the pe515 and cp7 strains into a mammalian expression vector and examined their abilities to block the activation of an IFN-β reporter construct in calf testis cells in response to synthetic dsRNA. Figure 1A shows that, in contrast to the NS3, NS2/3, NS2/3/4, and NS5a (NS5a data not shown) proteins from the pe515 or the cp7 strain of BVDV, the NPro fragment of either strain was able to block IFN-β induction by dsRNA and could do so to an extent similar to that of the V protein of SV5, a protein that functions by inhibiting the RNA helicase Mda-5 (1).
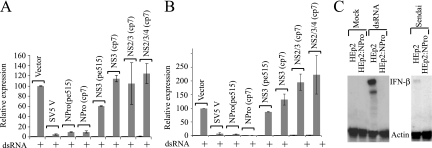
FIG. 1.
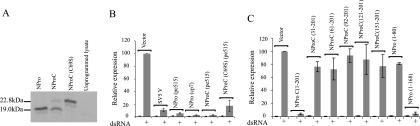
The BVDV NPro polypeptide is necessary and sufficient to block IFN-β induction by dsRNA. (A) Calf testis cells were transfected with a reporter for IFN-β promoter activity [pIFΔ(−116)lucter], the β-galactosidase expression vector pJATlac, and either a mammalian expression plasmid driving the overexpression of SV5 V, fragments of the cp7 or pe515 BVDV polyprotein as indicated, or the control “empty vector” pEF.plink2. Transfected cells were either mock treated or lipofected with the synthetic dsRNA poly(I) · poly(C), and cell extracts were prepared. (B) Experiments were performed identically to those for panel A, except that Vero cells were used. For both panels A and B, luciferase and β-galactosidase activities were determined from cellular extracts, and relative expression values were calculated accordingly. A reference value of 100 was assigned to the level of expression seen with dsRNA-treated control cells. The values shown represent data from three independent experiments, with the standard error of the mean indicated. (C) HEp2 cells, or a pool of HEp2 cells expressing the NPro product of the pe515 strain of BVDV (HEp2:NPro), were treated with dsRNA or infected for 18 h with Sendai virus, as indicated. RNA was harvested, and the expression of IFN-β was determined by RNase protection. The level of control γ-actin mRNA is also shown.
We next tested whether the block to IFN-β induction was restricted to cells of bovine origin. Figure 1B demonstrates that the NPro fragment was equally effective at blocking induction in Vero cells. We have seen equivalent results in human epithelial cells (see below) and chicken fibroblasts (unpublished data), indicating that the NPro-mediated effect on IFN production is not responsible for the restricted host range of BVDV. We decided to investigate whether the block to IFN-β induction was also seen at the level of the endogenous IFN-β gene. To do this, we engineered a human epithelial cell line (HEp2) to constitutively express the NPro protein of the pe515 strain of ncpBVDV (HEp2:NPro). When challenged with dsRNA or Sendai virus, these cells failed to produce detectable IFN-β mRNA, in contrast to the parental cell line (HEp2) (Fig. 1C). We also determined that an NPro-deficient strain of ncpBVDV was an efficient inducer of IFN-β, in contrast to the wild-type strain (unpublished data). These data indicate that the NPro region of BVDV is necessary and sufficient to actively block the production of type I IFN. These results are similar to those observed for the related pestivirus CSFV, in that an NPro-defective virus can induce type I IFN (40) and the isolated NPro protein can block IFN-β induction by a coinfecting virus (26, 39).
The BVDV NPro polypeptide blocks activation of IRF-3, but not NF-κB.
Induction of IFN-β in response to dsRNA by either TLR3-dependent or TLR3-independent pathways requires the activation of both NF-κB and IRF-3 (reviewed in reference 21). We previously showed that infection by ncpBVDV was able to block the activation of IRF-3, but not NF-κB (3). We therefore tested whether this distinction was retained when only the NPro protein was expressed. Figure 2A shows that expression of NPro from either the BVDV strain pe515 or cp7 was unable to block the activation of an NF-κB-dependent reporter gene construct in response to dsRNA; in agreement with this, stable expression of NPro had no effect on the activation of NF-κB as determined by EMSAs (Fig. 2B). In contrast, expression of NPro was able to block the induction of two IRF-3-dependent reporter constructs (a multimeric ISRE from the ISG15 promoter [Fig. 2C] and ISG54 [data not shown]) and was able to block the production of detectable DNA binding to IRF-3 in EMSAs (Fig. 2D). In the same experiments, the V protein of SV5 was able to block the activation of both IRF-3 and NF-κB (Fig. 2A and C), consistent with its known function as an inhibitor of Mda-5, a molecule that acts upstream of both NF-κB and IRF-3. These results indicate that NPro acts at a point that is downstream of a common activator in the dsRNA signaling pathway and is restricted to the IRF-3 response.
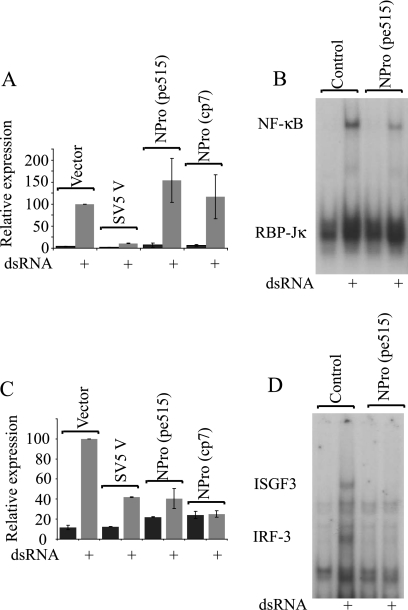
FIG. 2.
The BVDV NPro polypeptide blocks activation of IRF-3, but not NF-κB. Vero cells were transfected with a reporter for (A) NF-κB activity [p(PRDII)5tkΔ(−39)lucter] or (C) IRF-3 activity [p(ISG15ISRE)4tkΔ(−39)lucter], the constitutive β-galactosidase transfection control reporter plasmid pJATlac, and either a mammalian expression plasmid driving the overexpression of SV5 V, the NPro fragment of the pe515 or cp7 BVDV polyprotein, or the control “empty vector” pEF.plink2. Transfected cells were either mock treated or lipofected with the synthetic dsRNA poly(I) · poly(C), and cell extracts were prepared and analyzed for luciferase and β-galactosidase levels. Relative expression values were calculated accordingly. A reference value of 100 was assigned to the level of expression seen with dsRNA-treated control cells. The values shown represent data from three independent experiments, with the standard error of the mean indicated. (B) HEp2 cells, or a pool of HEp2 cells expressing the NPro product of the pe515 strain of BVDV (HEp2:NPro), were treated with dsRNA (+), and nuclear extracts were analyzed for NF-κB DNA binding activity using the PRD II region of the IFN-β promoter as a probe; this probe also identifies a complex containing the nuclear protein RBP-Jκ. (D) The nuclear extracts described for panel B were also analyzed for IRF-3 and ISGF3 binding using the ISRE from the ISG15 promoter as a probe; the identities of the indicated complexes in each case were previously established using specific antisera (3).
Since the induction of IFN-β by dsRNA in the above-mentioned experiments was sensitive to inhibition by the V protein of SV5, it appeared to be proceeding through the activation of the RNA helicase Mda-5 (this protein is unable to block TLR3- or RIG-I-dependent pathways [reference 1 and unpublished data]). Given that IRF-3 can be activated by alternative pathways, including the TLR3/TRIF pathway and the RNA helicase RIG-I, it was of interest to determine whether NPro could target IRF-3 activation by pathways other than Mda-5. We therefore examined the effect of NPro on signaling induced by overexpression of key signaling molecules. Figure 3A shows that NPro is able to block activation of IRF-3-dependent transcription by TRIF (a component of the exogenous dsRNA signaling pathway), by the kinase TBK1, and by the RNA helicases Mda-5 and RIG-I. By comparison, although overexpression of each of these signaling molecules was able to activate NF-κB, none of them was blocked by NPro (Fig. 3B); indeed, NPro expression was able to enhance the activation of the NF-κB-dependent reporter in each case, although we have not investigated this further. These results are consistent with NPro targeting IRF-3 as the final common point in several activation pathways rather than an upstream activation step.
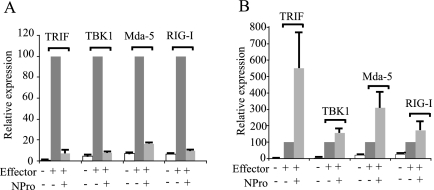
FIG. 3.
The BVDV NPro polypeptide blocks activation of IRF-3 by multiple signaling pathways. (A) Vero cells were transfected with the IFN-β reporter pIFΔ(−116)lucter, the β-galactosidase expression vector pJATlac, either the empty vector pEF.plink2 or the NPro-expressing plasmid pEF.NPro, and a mammalian expression plasmid driving the overexpression of TRIF, TBK1, Mda-5, or RIG-I as indicated above the bars. Cell extracts were prepared from transfected cells and analyzed for luciferase and β-galactosidase levels. Relative expression values were calculated by dividing the background-corrected luciferase values by the background-corrected β-galactosidase levels. A reference value of 100 was assigned to the level of expression seen with each of the effectors (TRIF, TBK1, Mda-5, or RIG-I) in the absence of NPro. (B) The experiment was the same as for panel A, except the reporter was replaced with the NF-κB reporter [p(PRDII)5tkΔ(−39)lucter]).
The catalytic activity of the NPro polypeptide is not required to inhibit IRF-3-dependent activation of transcription.
NPro is able to specifically cleave itself from the developing polyprotein by an autoprotease activity (53). It is therefore possible that it inactivates IRF-3 by site-specific cleavage. To examine this possibility, we generated recombinant forms of both NPro and IRF-3 but failed to observe cleavage of IRF-3 under a variety of conditions (data not shown). To eliminate a role for the autoprotease function, we constructed a mutant form of NPro (C69S) that has been reported to lack activity (41). Figure 4A shows that, as expected, this alteration eliminated autoprotease activity, and Fig. 4B shows that in reporter gene assays this protein is able to block IFN-β induction by dsRNA to almost the same extent as the nonmutated form of the protein.
FIG. 4.
The catalytic activity of the NPro polypeptide is not required to inhibit IRF-3-dependent activation of transcription. (A) The NPro protein from BVDV strain pe515 was expressed as a [35S]methionine-labeled fusion with the C-terminal peptide (NProC) by translation in reticulocyte lysates. The resulting product of 201 amino acids has a predicted molecular mass of 22.8 kDa, but as a result of the autoproteolytic activity of NPro, this is cleaved to give the observed fragment of 19.0 kDa. Changing amino acid 69 from a cysteine to a serine (NProC[C69S]) abolishes the autoprotease activity, as seen by the failure to generate the 19.0-kDa protein from the NProC fusion. The mobility of the 168-amino-acid NPro expressed without fusion to the C-terminal peptide is also shown (NPro). (B) Vero cells were transfected with the IFN-β reporter pIFΔ(−116)lucter, the β-galactosidase expression vector pJATlac, and either the empty vector pEF.plink2 or the indicated NPro-expressing plasmids. Transfected cells were either mock treated or lipofected with the synthetic dsRNA poly(I) · poly(C), and cell extracts were prepared and analyzed for luciferase and β-galactosidase levels. (C) Vero cells were transfected with pIFΔ(−116)lucter, pJATlac, and either the empty vector pEF.plink2 or the indicated NPro-expressing plasmids. NPro fragments are numbered relative to the ncpBVDV pe515 polyprotein. Thus, the full-length 168-amino-acid cleavage product of NPro is numbered 1-168, and C-terminal truncations are numbered 1-x [e.g., NPro(1-80) encodes a protein with the first 80 amino acids of NPro and lacking amino acids 81 to 168]. NPro protein fused with the C-terminal peptide (NProC) encodes an extended protein of 201 amino acids and is therefore numbered 1-201. The N-terminal truncations were constructed using this backbone and are numbered x-201 [e.g., NPro(31-201) encodes a protein lacking the first 30 amino acids of the NProC fusion protein]. Transfected cells were either mock treated or lipofected with the synthetic dsRNA poly(I) · poly(C), and cell extracts were prepared and analyzed for luciferase and β-galactosidase levels. For both panels B and C, relative expression values were calculated accordingly. A reference value of 100 was assigned to the level of expression seen with dsRNA-treated control cells. The values shown represent data from three independent experiments, with the standard error of the mean indicated.
We also constructed a series of NPro variants with deletions of amino acids from the N terminus or the C terminus. Each of these forms of NPro was expressed in mammalian cells, and their abilities to block the induction of IFN-β by dsRNA were compared to that of intact NPro. Figure 4C demonstrates that removal of 30 amino acids from the N terminus or 88 amino acids from the C terminus of the native NPro protein completely abolished the ability to inhibit IFN-β production.
BVDV infection leads to loss of cellular IRF-3 as a function of NPro expression.
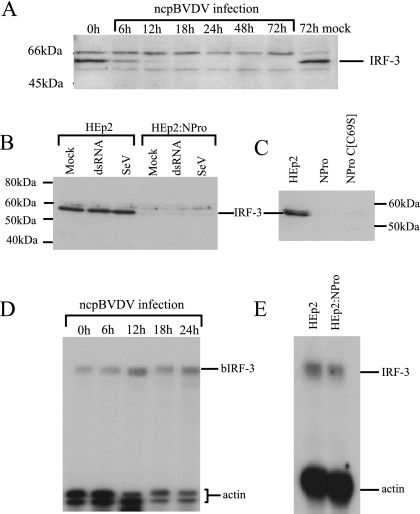
During the course of this work, it was reported that infection by CSFV led to a slow decrease in IRF-3 levels, while expression of NPro in cell lines was associated with a loss of IRF-3 expression (26). An equivalent effect of BVDV NPro on IRF-3 would explain the inhibitory effects described above. We therefore determined whether a loss of IRF-3 expression is seen in ncpBVDV infections. Figure 5A shows that IRF-3 levels are reduced following ncpBVDV infection, declining progressively from within 6 h of infection.
FIG. 5.
BVDV infection leads to loss of cellular IRF-3 as a function of NPro expression. (A) Calf testis cells were infected with the pe515 strain of ncpBVDV at an MOI of 2, and whole-cell extracts were prepared at the indicated times postinfection. The extracts were examined by Western blotting for the presence of IRF-3. (B) Whole-cell extracts were prepared from HEp2 and HEp2:NPro cells either mock infected, treated with dsRNA for 8 h, or infected with Sendai virus (SeV) for 16 h. The extracts were examined by Western blotting for the presence of IRF-3. (C) Whole-cell extracts were prepared from HEp2, HEp2:NPro, or HEp2:NProC[C69S] cells and were examined by Western blotting for the presence of IRF-3. (D) Total RNA was prepared from ncpBVDV-infected calf testis cells (MOI = 2) at the indicated times postinfection and analyzed for the presence of bovine IRF-3 and actin by RNase protection. The indicated sizes of the bovine IRF-3 (bIRF-3) and reference (γ-actin) transcripts are indicated. (E) Total RNA was prepared from HEp2 and HEp2:NPro cells and analyzed for the presence of IRF-3 transcripts by RNase protection. The sizes of the IRF-3 and reference (γ-actin) transcripts are indicated.
We next determined whether the loss of cellular IRF-3 observed in association with ncpBVDV infection was also seen when the NPro product was expressed in the absence of other BVDV proteins. We therefore examined IRF-3 protein levels in HEp2 cell lines stably expressing the NPro product or the autoprotease-negative form of NPro (NProC[C69S]). Figure 5B and C shows that, in contrast to the parental HEp2 cell line, IRF-3 was not detectable when either wild-type NPro or NProC[C69S] was expressed. Thus, long-term expression of NPro, such as that found at later times of infection or in the stable cell lines, clearly leads to a loss of IRF-3 at the level of protein expression in a manner that does not depend upon the autoprotease activity.
It has recently been demonstrated that CSFV infection of porcine cells leads to a down-regulation of the IRF-3 promoter, although this has not been directly linked to the NPro protein (26). We therefore investigated whether a similar phenomenon is observed when calf testis cells are infected with BVDV. Figure 5D shows that IRF-3 mRNA levels remain stable during BVDV infection, and furthermore, the IRF-3 mRNA levels in human HEp2 cells were only slightly lowered in cells expressing BVDV NPro (Fig. 5E). Thus, the decline in IRF-3 protein levels seen within 6 h of infection is not preceded or even mirrored by a decline in IRF-3 transcript, suggesting that the NPro effect is predominantly posttranscriptional.
The BVDV NPro polypeptide targets IRF-3 for proteasomal degradation.
Since long-term stable expression of NPro clearly leads to a loss of IRF-3 at the level of protein expression, rather than of transcriptional inhibition or loss of mRNA stability, yet does not require the autoprotease activity of NPro, we considered the possibility that NPro might be working via an indirect form of proteolysis. We therefore examined the effects of a variety of protease inhibitors on IRF-3 levels and determined that treatment with either of the proteasome inhibitors epoxomicin or MG115 (data not shown) led to a partial recovery in the IRF-3 status of the NPro-expressing HEp2 cell line (Fig. 6A). The degree of recovery was markedly stimulated in cells that were also treated with the caspase inhibitor ZVAD, the addition of which limited the apoptotic side effects of long-term treatment with proteasome inhibitors (Fig. 6A). In contrast, IRF-3 levels were unaffected by proteasome inhibitors in the parental HEp2 cells (Fig. 6A). Consistent with the involvement of proteasomes in the degradation of IRF-3, treatment with epoxomicin was also associated with an increase in the level of IRF-3 in the polyubiquitin-associated protein fraction; this epoxomicin-stimulated increase was seen only in cells expressing NPro (Fig. 6B). Treatment with epoxomicin prevented any loss in cellular IRF-3 levels in ncpBVDV-infected calf testis cells (Fig. 6C). These inhibitors do not affect the synthesis of ncpBVDV proteins (Fig. 6D) and hence do not act to block some aspect of the viral life cycle, such as virus entry. These results are consistent with a requirement for proteasome activity to lower IRF-3 levels in response to NPro expression.
Nuclear IRF-3 is resistant to degradation but is prevented from binding to DNA by NPro.
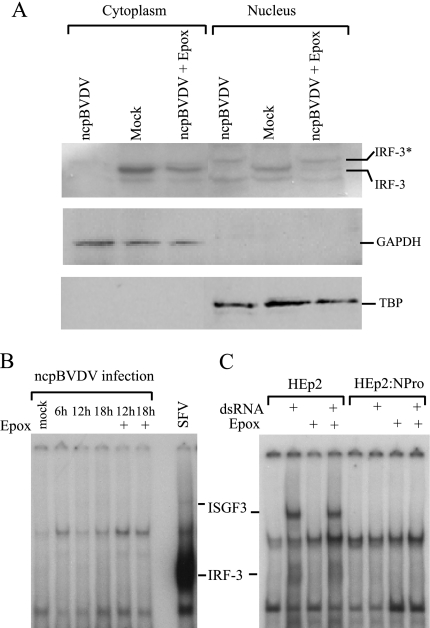
Despite the NPro-dependent decrease in IRF-3 levels, ncpBVDV and cpBVDV infections mobilize IRF-3 to the nucleus (3), suggesting that the degradation must lag behind an initial infection-dependent mobilization event. We repeated our immunofluorescence analysis of IRF-3 activation in response to BVDV (3), concentrating on relatively early time points after infection, and observed that IRF-3 had relocalized from the cytoplasm to the nucleus within 12 h of infection (data not shown). This was at a time postinfection when total IRF-3 levels were low but remained detectable (Fig. 5A). These data suggest that a fraction of cellular IRF-3 is mobilized to the nucleus as a result of infection but the bulk of the IRF-3 is degraded. To investigate this further, we investigated whether IRF-3 could be detected in nuclear extracts of BVDV-infected calf testis cells. Figure 7A shows that IRF-3 could be detected in both the cytoplasm and the nucleus of mock-infected cells. The latter observation is consistent with the constitutive shuttling of nonactivated IRF-3 between compartments (25). Infection of cells with ncpBVDV caused a complete loss of IRF-3 from the cytoplasm. However, in nuclear extracts of ncpBVDV-infected cells (12 h postinfection), an IRF-3-immunoreactive polypeptide was detected (IRF-3*) (Fig. 7A), but strikingly, it showed a decrease in electrophoretic mobility relative to the IRF-3-immunoreactive polypeptide seen in mock-infected cells; this pattern of mobility shift has been frequently reported to be associated with infection-induced phosphorylation of IRF-3 (29, 44, 52, 57). When infection was repeated in the presence of epoxomicin, cytoplasmic IRF-3 remained intact (although the levels detected were lower than in mock-infected cells because some of the IRF-3 was mobilized to the nucleus as a result of infection), but both electrophoretically distinct forms of IRF-3 were detected in the nucleus. We presume that this reflects the stable nuclear localization of the modified form of IRF-3 caused by the BVDV infection and the presence of undegraded, unmodified IRF-3 being constitutively shuttled between the cytoplasm and the nucleus.
FIG. 7.
Activated IRF-3 is unable to bind to DNA in ncpBVDV-infected cells or in the presence of NPro. (A) Calf testis cells were mock infected or infected with the pe515 strain of ncpBVDV at an MOI of 2 in the presence or absence of epoxomicin as indicated, and extracts were prepared from both the cytoplasm and nucleus. The extracts were examined by Western blotting for the presence of IRF-3 (top) or for cytoplasmic (GAPDH) (middle) or nuclear (TBP) (bottom) proteins. The mobility of IRF-3 is indicated; also indicated is a presumptive modified form of IRF-3 (IRF-3*) present in nuclear extracts of infected cells. (B) Calf testis cells were infected with the pe515 strain of ncpBVDV at an MOI of 2 in the presence or absence of epoxomicin (Epox) where indicated. Nuclear extracts were prepared and analyzed for the presence of ISGF3 and IRF-3 in DNA binding form by EMSA using the ISRE from the ISG15 promoter as a probe. The lane marked SFV is an extract from SFV-infected calf testis cells and clearly shows the activation of IRF-3. (C) HEp2 and HEp2:NPro cells were either left untreated or treated with dsRNA in the presence or absence of epoxomicin as indicated. Nuclear extracts were prepared and analyzed for the presence of ISGF3 and IRF-3 in DNA binding form by EMSA using the ISRE from the ISG15 promoter as a probe. Although IRF-3 levels in the Hep2:NPro cells were only partially restored by blocking proteasome function (Fig. 6A), it should be emphasized that even on very prolonged autoradiographic exposure, IRF-3 and ISGF3 complexes were not detected; we also estimate from mixing experiments that under the same EMSA conditions we would easily detect the activation of IRF-3 in the HEp2:NPro cells to 10% of the levels seen in the HEp2 cells. The identities of the indicated complexes in panels B and C were previously established using specific antisera (3).
These observations show that the relocalization of IRF-3 to the nucleus caused by BVDV infections is likely associated with modification of the protein, and significantly, this material is resistant to proteasomal degradation, which presumably is restricted to the cytoplasm. It is thus pertinent to ask whether the nuclear IRF-3 can bind to DNA. Figure 7B shows that, in contrast to infection by SFV, which generates an abundant IRF-3-containing complex in EMSA experiments, infection of calf testis cells by ncpBVDV generated no detectable IRF-3-containing complexes at any time point tested. We also investigated whether extracts from infected cells treated with epoxomicin could generate DNA binding complexes containing IRF-3. Figure 7B shows that they cannot, and we conclude that even when IRF-3 is not degraded and is apparently able to enter the nucleus and be modified (Fig. 7A), it is unable to bind to DNA. The failure to observe DNA binding forms of IRF-3 even when degradation is inhibited indicates either that ncpBVDV encodes an inhibitor of DNA binding or that the ncpBVDV-activated nuclear IRF-3 is somehow not fully competent to bind to DNA.
Finally, we assessed whether the increased level of cellular IRF-3 observed in the presence of proteasome inhibitors was associated with the detection of a DNA binding form of IRF-3 in the cell line expressing NPro. HEp2 cells and the HEp2:NPro stable line were treated with epoxomicin and then treated with dsRNA. Figure 7C shows that, in contrast to the parental line, in which an IRF-3-containing complex can be detected by EMSAs using extracts from cells treated with dsRNA, an equivalent complex was not seen in cells expressing NPro. Thus, the presence of NPro is sufficient to block the generation of a DNA binding form of IRF-3 even when the proteasomal degradation of IRF-3 is blocked.
DISCUSSION
In this paper, we have extended our previous observations that BVDV encodes a protein able to specifically block the activity of IRF-3 (2, 3). We have demonstrated that this is a property of the NPro fragment of the BVDV polyprotein and that NPro expression inhibits IRF-3 from binding to DNA and leads to a long-term down-regulation of IRF-3 protein levels by targeting IRF-3 for polyubiquitination and subsequent proteasomal degradation. Our results are consistent with those recently published by Gil et al., who have shown that the BVDV NPro protein is necessary in an uncharacterized manner for the antagonism of type I IFN production (13). Our results are also consistent with those observed for CSFV, where it has been reported that CSFV engineered with a deletion in NPro induces IFN (40) while overexpression of the CSFV NPro protein down-regulates IFN induction (26, 39). However, there appear to be major differences between the IFN evasion mechanisms employed by CSFV and BVDV. First, unlike BVDV, CSFV infection fails to cause IRF-3 mobilization to the nucleus (26), and second, the decrease in IRF-3 protein levels was suggested to be a consequence of an NPro-dependent inhibition of IRF-3 transcription (26). It is surprising that two such closely related viruses (whose NPro proteins are 70% identical) should differ so markedly in their mechanisms of action, although it should be noted that many paramyxoviruses evade IFN signaling using their accessory V proteins yet do so by a variety of distinct means (reviewed in reference 49). It will be of interest to examine the differences between BVDV and CSFV NPro proteins in more detail.
We note that it has been reported that cellular IRF-3 can be targeted for proteasomal degradation following virus-induced phosphorylation (29), and during the preparation of the manuscript, it was also reported that the stability of IRF-3 could be decreased in response to dsRNA treatment in a manner that depended on phosphorylation, polyubiquitination, and eventual degradation, targeted by the prolyl isomerase Pin1 (42). This is in marked contrast to the stability of unmodified IRF-3, which has a half-life of over 24 h in uninfected cells (reference 37 and our unpublished data). In the absence of conflicting data, it might be concluded that BVDV infection triggers phosphorylation of IRF-3, which is thus degraded, and the sole function of NPro in this scenario would be to prevent IRF-3 from binding to DNA. However, our observations that modified IRF-3 is more stable than the unmodified form and that IRF-3 is polyubiquitinated and degraded by proteasomes in uninfected cells that express NPro clearly indicate that NPro actively targets unmodified IRF-3 and dramatically decreases its stability. A similar situation appears to occur during rotavirus infections, where IRF-3 is targeted for proteasomal degradation (4); in this case, the viral NSP1 protein that mediates this response interacts directly with IRF-3. To date, we have not been able to demonstrate a direct interaction between the BVDV NPro protein and IRF-3 (unpublished results). Since the prolyl isomerase Pin1 can play a role in targeting phosphorylated IRF-3 for ubiquitination and degradation (42), it is possible that NPro utilizes this factor to enhance the normal pathways for the degradation of unmodified IRF-3.
Although the autoprotease activity of NPro is not essential for blocking IRF-3 function, the structural integrity of the BVDV NPro protein appears important, since deletions from either end of the protein impair its function in transient-transfection experiments. Consistent with this, Gil et al. also determined that alterations in the N-terminal domain of NPro disrupted its ability to act as an IFN antagonist (13). It remains to be determined whether the apparently separate functions of NPro map to distinct domains of the protein.
Many viruses encode proteins that act to limit the production of IFN, although most of these act at points upstream of the activation of IRF-3 or NF-κB. By targeting a component that is at the end of several pathways for signaling in response to viral infection (IRF-3 can be activated by pathways initiated by TLR, Mda-5, and RIG-I), the actions of NPro ensure that IFN production is completely blocked. In this context, it is interesting to speculate on the role of the BVDV Erns protein, which we have demonstrated has the property of blocking signaling in response to extracellular dsRNA by binding and internalizing the dsRNA to endosomes, where the low pH activates the dsRNase function of Erns (18). Since extracellular dsRNA signals through the TLR3/TRIF pathway to activate IRF-3, it would be expected that NPro would inactivate signaling through this pathway (and indeed, this is the case [Fig. 3A]), and hence, a role for Erns would be redundant. However, Erns has the unusual property of being a glycoprotein that is secreted from infected cells and thus can act on neighboring uninfected cells. In this capacity, it could act to prevent any activation of IFN production in an uninfected cell caused by release of viral dsRNA following lysis of an infected cell. Such a mechanism would aid infection, since BVDV has no apparent means to protect itself from the antiviral actions of IFN (3).
In the case of the related flavivirus HCV, induction of type I IFN is prevented by the proteolytic cleavage by the NS3/4a protease of the RNA helicase adaptor Cardif (28, 34). Although this adaptor appears to be a downstream component that is common to activation by Mda-5 or RIG-I (22, 34, 47, 54), this would leave the virus open to IFN production through the TLR3/TRIF pathway. The finding that the NS3/4a protease also cleaves and inactivates TRIF (10) indicates that HCV protects itself against both internal and external dsRNA responses. In this context, it is interesting that the highly attenuated phenotype of NPro-deficient BVDV can be rescued by the NS3 protease of HCV (27).
It is notable that in contrast to ncpBVDV, cpBVDV infections are associated with the production of type I IFN (3, 5, 46). However, cpBVDV infections retain the ability to prevent mobilized IRF-3 from binding to DNA (2), consistent with their expression of NPro. Since the activated IRF-3 associated with cpBVDV is unable to bind to DNA, it is interesting to speculate on the molecular basis of the IFN-β induction. In contrast to ncpBVDV infections, cpBVDV infections cause the activation of NF-κB (3, 14), possibly due to an increase in the amount of dsRNA produced relative to that seen with ncpBVDV (14). The activation of NF-κB by cpBVDV is insensitive to the presence of the Erns glycoprotein (18) because it is produced during viral replication and thus is not extracellular. We presume that the elevated levels of NF-κB play an important role in IFN-β induction by cpBVDV. However, although NF-κB is an essential component of the IFN-β enhanceosome, it is not able to activate IFN-β transcription in the absence of a member of the IRF family (reviewed in reference 32), and hence, some factor other than IRF-3 must be able to substitute. This is unlikely to be IRF-7, because BVDV infections do not cause IRF-7 activation (2), and we hypothesize that the substituting factor is IRF-1 on the grounds that IRF-1 is inducible by NF-κB (24, 35) and that BVDV infections are associated with an increase in the levels of IRF-1 (unpublished observations) and do not block its ability to bind to DNA (3). It is important to stress that ncpBVDV infections are unable to induce IFN-β, not because they block this putative alternative pathway, but because they do not induce the activation of NF-κB.
Acknowledgments
This work was supported by the BBSRC Controlling Viral Diseases in Livestock Initiative.
We thank Phil Dash for advice on immunofluorescence and confocal microscopy, Kay Childs for pEF.RIG-I, Dushen Murugiah for assistance during the construction of the NPro deletion plasmids, and Kay Childs and Craig Ross for comments on the manuscript.
Footnotes
Published ahead of print on 13 September 2006.
REFERENCES
- 1.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baigent, S. J., S. Goodbourn, and J. W. McCauley. 2004. Differential activation of interferon regulatory factors-3 and -7 by non-cytopathogenic and cytopathogenic bovine viral diarrhoea virus. Vet. Immunol. Immunopathol. 100:135-144. [DOI] [PubMed] [Google Scholar]
- 3.Baigent, S. S., G. Zhang, M. D. Fray, H. Flick-Smith, S. Goodbourn, and J. W. McCauley. 2002. Inhibition of interferon-beta transcription by noncytopathic bovine viral diarrhoea virus is through an interferon regulatory factor 3 (IRF-3)-dependent mechanism. J. Virol. 76:8979-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barro, M., and J. T. Patton. 2005. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc. Natl. Acad. Sci. USA 102:4114-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charleston, B., M. D. Fray, S. Baigent, B. V. Carr, and W. I. Morrison. 2001. Establishment of persistent infection with non-cytopathic bovine viral diarrhoea virus in cattle is associated with a failure to induce type I interferon. J. Gen. Virol. 82:1893-1897. [DOI] [PubMed] [Google Scholar]
- 6.Cusson-Hermance, N., S. Khurana, T. H. Lee, K. A. Fitzgerald, and A. M. Kelliher. 2005. Rip1 mediates the Trif-dependent Toll-like receptor 3- and 4-induced NF-κB activation but does not contribute to interferon regulatory factor 3 activation. J. Biol. Chem. 280:36560-36566. [DOI] [PubMed] [Google Scholar]
- 7.Demaison, C., K. Parsley, G. Brouns, M. Scherr, K. Battmer, C. Kinnon, M. Grez, and A. J. Thrasher. 2002. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum. Gene Ther. 13:803-813. [DOI] [PubMed] [Google Scholar]
- 8.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enoch, T., K. Zinn, and T. Maniatis. 1986. Activation of the human β-interferon gene requires an interferon inducible factor. Mol. Cell. Biol. 6:801-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferreon, J. C., A. C. Ferreon, K. Li, and S. M. Lemon. 2005. Molecular determinants of TRIF proteolysis mediated by the hepatitis C virus NS3/4A protease. J. Biol. Chem. 280:20483-20492. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKɛ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 12.Fray, M. D., D. J. Paton, and S. Alenius. 2000. The effects of bovine viral diarrhoea virus on cattle reproduction in relation to disease control. Anim. Reprod. Sci. 60:615-627. [DOI] [PubMed] [Google Scholar]
- 13.Gil, L. H., I. H. Ansari, V. Vassilev, D. Liang, V. C. Lai, W. Zhong, Z. Hong, E. J. Dubovi, and R. O. Donis. 2006. The amino-terminal domain of bovine viral diarrhea virus Npro protein is necessary for alpha/beta interferon antagonism. J. Virol. 80:900-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gil, L. H., A. L. van Olphen, S. K. Mittal, and R. O. Donis. 2006. Modulation of PKR activity in cells infected by bovine viral diarrhea virus. Virus Res. 116:69-77. [DOI] [PubMed] [Google Scholar]
- 15.Han, K. J., X. Su, L. G. Xu, L. H. Bin, J. Zhang, and H. B. Shu. 2004. Mechanisms of the TRIF-induced interferon-stimulated response element and NF-κB activation and apoptosis pathways. J. Biol. Chem. 279:15652-15661. [DOI] [PubMed] [Google Scholar]
- 16.He, Y., and M. G. Katze. 2002. To interfere and to anti-interfere: the interplay between hepatitis C virus and interferon. Viral Immunol. 15:95-119. [DOI] [PubMed] [Google Scholar]
- 17.Hiscott, J., R. Lin, P. Nakhaei, and S. Paz. 2006. MasterCARD: a priceless link to innate immunity. Trends Mol. Med. 12:53-56. [DOI] [PubMed] [Google Scholar]
- 18.Iqbal, M., E. Poole, S. Goodbourn, and J. W. McCauley. 2004. Role for bovine viral diarrhea virus Erns glycoprotein in the control of activation of beta interferon by double-stranded RNA. J. Virol. 78:136-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang, Z., T. W. Mak, G. Sen, and X. Li. 2004. Toll-like receptor 3-mediated activation of NF-κB and IRF3 diverges at Toll-IL-1 receptor domain-containing adapter inducing IFN-beta. Proc. Natl. Acad. Sci. USA 101:3533-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, C. L., and M. Gale, Jr. 2006. CARD games between virus and host get a new player. Trends Immunol. 27:1-4. [DOI] [PubMed] [Google Scholar]
- 21.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 22.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 23.King, P., and S. Goodbourn. 1994. The β-interferon promoter responds to priming through multiple independent regulatory elements. J. Biol. Chem. 269:30609-30615. [PubMed] [Google Scholar]
- 24.Kumar, A., Y. L. Yang, V. Flati, S. Der, S. Kadereit, A. Deb, J. Haque, L. Reis, C. Weissmann, and B. R. Williams. 1997. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J. 16:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar, K. P., K. M. McBride, B. K. Weaver, C. Dingwall, and N. C. Reich. 2000. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol. 20:4159-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.La Rocca, S. A., R. J. Herbert, H. Crooke, T. W. Drew, T. E. Wileman, and P. P. Powell. 2005. Loss of interferon regulatory factor 3 in cells infected with classical swine fever virus involves the N-terminal protease, Npro. J. Virol. 79:7239-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai, V. C., W. Zhong, A. Skelton, P. Ingravallo, V. Vassilev, R. O. Donis, Z. Hong, and J. Y. Lau. 2000. Generation and characterization of a hepatitis C virus NS3 protease-dependent bovine viral diarrhea virus. J. Virol. 74:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, X. D., L. Sun, R. B. Seth, G. Pineda, and Z. J. Chen. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. USA 102:17717-17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marais, R., Y. Light, H. F. Paterson, and C. J. Marshall. 1995. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 14:3136-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masson, N., M. Ellis, S. Goodbourn, and K. A. W. Lee. 1992. Cyclic-AMP response element-binding protein and the catalytic subunit of protein kinase A are present in F9 embryonal carcinoma cells but are unable to activate the somatostatin promoter. Mol. Cell. Biol. 12:1096-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merika, M., and D. Thanos. 2001. Enhanceosomes. Curr. Opin. Genet. Dev. 11:205-208. [DOI] [PubMed] [Google Scholar]
- 33.Meylan, E., K. Burns, K. Hofmann, V. Blancheteau, F. Martinon, M. Kelliher, and J. Tschopp. 2004. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat. Immunol. 5:503-507. [DOI] [PubMed] [Google Scholar]
- 34.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 35.Mori, K., K. Yoshida, J. Tani, Y. Nakagawa, S. Hoshikawa, and S. Ito. 2001. Double-stranded RNA-induced interferon regulatory factor-1 gene expression in FRTL-5 rat thyroid cells. Mol. Cell Endocrinol. 184:77-86. [DOI] [PubMed] [Google Scholar]
- 36.Oganesyan, G., S. K. Saha, B. Guo, J. Q. He, A. Shahangian, B. Zarnegar, A. Perry, and G. Cheng. 2006. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439:208-211. [DOI] [PubMed] [Google Scholar]
- 37.Prakash, A., and D. E. Levy. 2006. Regulation of IRF7 through cell type-specific protein stability. Biochem. Biophys. Res. Commun. 342:50-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Randall, R. E., D. F. Young, K. K. Goswami, and W. C. Russell. 1987. Isolation and characterization of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J. Gen. Virol. 68:2769-2780. [DOI] [PubMed] [Google Scholar]
- 39.Ruggli, N., B. H. Bird, L. Liu, O. Bauhofer, J. D. Tratschin, and M. A. Hofmann. 2005. N(pro) of classical swine fever virus is an antagonist of double-stranded RNA-mediated apoptosis and IFN-alpha/beta induction. Virology 340:265-276. [DOI] [PubMed] [Google Scholar]
- 40.Ruggli, N., J. D. Tratschin, M. Schweizer, K. C. McCullough, M. A. Hofmann, and A. Summerfield. 2003. Classical swine fever virus interferes with cellular antiviral defense: evidence for a novel function of N(pro). J. Virol 77:7645-7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rumenapf, T., R. Stark, M. Heimann, and H. J. Thiel. 1998. N-terminal protease of pestiviruses: identification of putative catalytic residues by site-directed mutagenesis. J. Virol. 72:2544-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saitoh, T., A. Tun-Kyi, A. Ryo, M. Yamamoto, G. Finn, T. Fujita, S. Akira, N. Yamamoto, K. P. Lu, and S. Yamaoka. 2006. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 7:598-605. [DOI] [PubMed] [Google Scholar]
- 43.Sasai, M., H. Osiumi, M. Matsumoto, N. Inoue, F. Fujita, M. Nainishi, and T. Seya. 2005. NF-κB-activating kinase-associated protein 1 participates in TLR3/Toll-IL-1 homology domain-containing adapter molecule-1-mediated IFN regulatory factor 3 activation. J. Immunol. 174:27-30. [DOI] [PubMed] [Google Scholar]
- 44.Sato, M., N. Tanaka, N. Hata, E. Oda, and T. Taniguchi. 1998. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-beta gene. FEBS Lett. 425:112-116. [DOI] [PubMed] [Google Scholar]
- 45.Sato, S., M. Sugiyama, M. Yamamoto, Y. Watanabe, T. Kawai, K. Takeda, and S. Akira. 2003. Toll-IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase I, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 171:4304-4310. [DOI] [PubMed] [Google Scholar]
- 46.Schweizer, M., and E. Peterhans. 2001. Noncytopathic bovine viral diarrhea virus inhibits double-stranded RNA-induced apoptosis and interferon synthesis. J. Virol. 75:4692-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF-3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 48.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 49.Stock, N., S. Goodbourn, and R. E. Randall. 2004. The anti-interferon mechanisms of paramyxoviruses, p. 115-140. In S. P. Changeux and P. Palese (ed.), Modulation of host gene expression and innate immunity by viruses. Springer, Heidelberg, Germany.
- 50.Tautz, N., G. Meyers, R. Stark, E. J. Dubovi, and H. J. Thiel. 1996. Cytopathogenicity of a pestivirus correlates with a 27-nucleotide insertion. J. Virol. 70:7851-7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Visvanathan, K. V., and S. Goodbourn. 1989. Double-stranded RNA activates binding of NF-κB to an inducible element in the human β-interferon promoter. EMBO J. 8:1129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiskerchen, M., S. K. Belzer, and M. S. Collett. 1991. Pestivirus gene expression: the first protein product of the bovine viral diarrhea virus large open reading frame, p20, possesses proteolytic activity. J. Virol. 65:4508-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 55.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., and S. Akira. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851-2858. [DOI] [PubMed] [Google Scholar]
- 56.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 57.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zinn, K., D. DiMaio, and T. Maniatis. 1983. Identification of two distinct regulatory regions adjacent to the human β-interferon gene. Cell 34:865-879. [DOI] [PubMed] [Google Scholar]