Abstract
The unfolded protein response (UPR) is a coordinated change in gene expression triggered by perturbations in functions of the endoplasmic reticulum (ER). XBP1, a key transcription factor of the UPR, is activated by an IRE1-mediated splicing event, which results in a frameshift and encodes a protein with transcriptional activity. Here, we report that XBP1 was activated during flaviviral infection, as evidenced by XBP1 mRNA splicing and protein expression, as well as induction of the downstream genes ERdj4, EDEM1, and p58(IPK) in Japanese encephalitis virus (JEV)- and dengue virus serotype 2 (DEN-2)-infected cells. Reporter systems based on IRE1-mediated XBP1 splicing were established, and several flaviviral proteins associated with the ER, including glycoproteins and small hydrophobic membrane-anchored proteins, were found to trigger the splicing event. Notably, nonstructural protein NS2B-3 of DEN-2, but not of JEV, was a potent inducer of XBP1 splicing through an unclear mechanism(s). Reduction of XBP1 by a small interfering RNA had no effect on cells' susceptibility to the two viruses but exacerbated the flavivirus-induced cytopathic effects. Overall, flaviviruses trigger the XBP1 signaling pathway and take advantage of this cellular response to alleviate virus-induced cytotoxicity.
The flaviviruses are a group of arthropod-borne viruses, members of which, such as yellow fever virus (49, 62), West Nile virus (31), Japanese encephalitis virus (JEV) (55, 56), and dengue viruses (DEN), cause medically important problems worldwide (17). The flaviviral genome is a single-stranded positive-sense RNA encoding a polyprotein precursor that is cleaved into several structural and nonstructural proteins. These are the core (C) protein, the precursor membrane (prM) protein, the envelope (E) protein, and seven nonstructural (NS) proteins. The virion is enveloped with lipid bilayers and viral structural proteins (40). Three flaviviral proteins, prM, E, and NS1, enter the endoplasmic reticulum (ER) and are glycosylated. Four small hydrophobic NS proteins, NS2A, NS2B, NS4A, and NS4B, are anchored to the ER membrane. The cytoplasmic viral proteins NS3 and NS5 are enzymatic components of protease/helicase/NTPase and methyltransferase/RNA-dependent RNA polymerase, respectively. Flaviviruses are thought to replicate in the cytoplasm, and employing the intrinsic secretory pathways, flavivirus buds from the membranes of the ER and the Golgi apparatus to release mature virions (40). One of the major morphological changes in flavivirus-infected cells is proliferation and hypertrophy of the ER membranes, where virus particles accumulate (6, 19).
The ER of eukaryotic cells consists of an extensive membranous network that provides a specialized compartment for posttranslational modification, folding, and oligomerization of newly synthesized proteins. The ER is also considered the main intracellular signal-transducing organelle, and one of its actions is to release calcium from the ER reservoir when the cell responds to environmental cues (11, 28, 46). The ER is extremely sensitive to alterations in homeostasis. In response to a variety of stimuli, certain signals are transduced from the ER to both the cytoplasm and the nucleus, resulting in either cellular adaptation for survival or induction of apoptosis (4). Several endogenous imbalances in cells, such as massive protein production, loss of calcium homeostasis, inhibition of N-linked glycosylation, and accumulation of mutant proteins, often contribute to malfunction of the ER. Such malfunction is termed ER stress (28).
To alleviate ER stress, the cell induces a coordinated adaptive program termed the unfolded protein response (UPR) (51, 54). The UPR is designed to eliminate malfolded ER proteins either by suppressing new protein synthesis or by elevating processes of protein folding and degradation (18). The UPR is characterized by the enhanced expression of several chaperones, such as the glucose-regulated proteins GRP78 (also known as BiP) and GRP94 (29), and other ER-resident proteins. In addition, the UPR triggers a cellular response resulting in inhibition of translation initiation, thereby preventing further accumulation of unfolded protein in the ER. The UPR signaling pathway, as first described in yeast, is a rather simple linear pathway involving transcriptional regulation mediated by Ire1p (13, 14, 44). Higher eukaryotic cells have conserved the essential and unique properties of Ire1p-mediated UPR signaling but have also evolved additional transducers to generate a diversity of responses. So far, three ER stress sensors, IRE1, ATF6 (activating transcription factor 6), and PERK (PKR-like ER protein kinase), have been identified in mammals to achieve the various cellular adaptations against ER dysfunction (4, 66). The PERK pathway results in phosphorylation of eukaryotic translation initiation factor 2 α subunit (eIF2α), leading to translation attenuation (18). Activation of the IRE1 and ATF6 pathways produces two transcriptional factors, XBP1 and ATF6, responsible for induction of many genes involved in the UPR (33).
IRE1 is a bifunctional ER transmembrane protein with both serine/threonine kinase and RNase activities (61, 65). Under nonstressed conditions, the luminal kinase domain of IRE1 binds with BiP (3, 41). As unfolded proteins accumulate, IRE1 is released from BiP and then oligomerizes. The RNase activity of the protein is then activated by autophosphorylation. The transcript from the X box binding protein 1 gene (XBP1) is the only substrate for IRE1 RNase activity identified in mammalian cells to date (7, 34, 53, 69). Upon activation, IRE1 removes a 26-nucleotide (nt) intron from unspliced XBP1 mRNA (XBP1u), resulting in a frameshift. The spliced form of XBP1 (XBP1s) encodes a protein with a novel C terminus that acts as a potent transcriptional activator of many genes involved in the UPR. Expression of XBP1 is necessary for survival, as deletion of mouse XBP1 results in liver hypoplasia, followed by embryonic lethality (47). Overexpression of XBP1s in B cells induces a wide spectrum of secretory-pathway genes and results in expansion of the ER (52). These observations may explain why XBP1 is required for terminal B-cell differentiation and efficient antibody production (23, 24). XBP1 may also link the mammalian UPR to phospholipid biosynthesis and ER biogenesis, as fibroblasts overexpressing XBP1s exhibit enhanced phosphatidylcholine biosynthesis, elevated levels of membrane phospholipids, and an expanded ER (57). Expression of a subset of UPR target genes, including EDEM (ER degradation-enhancing α-mannosidase-like protein), which is critically involved in the ER-associated protein degradation (ERAD) pathway, depends on XBP1 (33, 45, 63, 68). Thus, the IRE1-XBP1 pathway is required for ER biogenesis, as well as for efficient protein folding, maturation, and degradation in the ER.
Flaviviruses use the ER of host cells as the primary site of glycoprotein synthesis, genomic RNA replication, and virus particle maturation. ER stress-mediated signaling has been implicated in apoptosis induced by JEV (58) and bovine viral diarrhea virus, a member of the family Flaviviridae (27). Infection of JEV triggers not only ER stress, but also the UPR, as evidenced by substantial induction of the expression of several chaperones (58). Replication of another member of the family Flaviviridae, hepatitis C virus (HCV) replicon, induces ER stress and the UPR (60). However, XBP1 trans-activating activity, such as EDEM induction, is repressed in cells expressing HCV replicon, even though splicing of XBP1 mRNA occurs and XBP1s expression is elevated (59). The E2 envelope protein of HCV is able to activate the BiP gene promoter, and then BiP binds tightly to E2 (36). Expression of the HCV E1 and/or E2 envelope protein has been found to induce XBP1 splicing (8). The HCV core protein has also been shown to trigger ER stress through a mechanism involving ER calcium depletion (2). It thus appears that extensive interaction of ER networks and signaling events takes place in flavivirus-infected cells; however, little is known about the molecular details of the UPR and the roles played by XBP1 in the flavivirus life cycle.
In this study, we analyzed the IRE1-XBP1 arm of the UPR in a mouse neuroblastoma N18 cell line infected with two flaviviruses, JEV and DEN serotype 2 (DEN-2). The XBP1 pathway was activated in cells infected with either JEV or DEN-2, as shown by the occurrence of XBP1 splicing, XBP1s protein expression, and induction of downstream genes. Impairment of XBP1 by using a small interfering RNA (siRNA) exacerbated cytotoxicity induced by these two viruses. We also designed a reporter system, based on the IRE1-mediated 26-nt splicing of XBP1, to screen for flaviviral proteins potentially acting as ER stress inducers. The possible mechanisms of flavivirus-induced UPR are discussed, as is the biological significance of this phenomenon, in an effort to understand how an intracellular pathogen may achieve cross talk with a process normally controlled by the host cell.
MATERIALS AND METHODS
Viruses and cell lines.
JEV strain RP-9 (12) and DEN-2 strain PL046 (39) were used in this study. The propagation of virus was carried out in C6/36 cells utilizing RPMI 1640 medium containing 10% fetal bovine serum (FBS) (Gibco). Virus titers (in PFU/ml) were determined by a plaque-forming assay on BHK-21 cells as previously described (67). N18, a mouse neuroblastoma cell line (1), was cultured in RPMI 1640 medium supplemented with 5% FBS.
Plasmid construction.
As a component of ER stress reporter plasmids, human XBP1u cDNA was amplified from A549 cells by reverse transcriptase-PCR (RT-PCR) using XBP1-specific primers. The forward primer, 5′-AGCTTAAGCTTGCTATGGTGGTGGTGGCAGC-3′, anneals to nt −3 to 17 of XBP1 (the A of the XBP1 initiation codon [underlined] is 1 in the XBP1 numbering system) and has a HindIII recognition site (italic). The reverse primer, 5′-GAGGTGCTTCCTCGATTTTCACTACC-3′, anneals to nt 885 to 860 of XBP1. The sequences of the cloned human XBP1u were identical to the unspliced form of human XBP1 (GenBank accession number NM005080). The PCR product was cloned to the N terminus of green fluorescent protein (GFP) in pEGFP-N1 (Clontech) and named pXBP1u-GFP, in which the GFP was in frame with the second open reading frame (ORF) of XBP1u. To construct pHA-XBP1u-GFP (see Fig. 2), the XBP1u-GFP fragment was subcloned into pcDNA3 (Invitrogen) to give a construct in which a hemagglutinin (HA) tag was fused in frame with the first ORF of XBP1u. To construct pXBP1u-FLuc (see Fig. 3A), the C-terminal region of pXBP1u-GFP from the EcoRV site (at nt 606 of XBP1u) to the end of the GFP gene was replaced by the firefly luciferase gene of pGL3 (Promega). The reporter genes were under the control of the immediate-early promoter of cytomegalovirus. All constructs were handled using standard molecular cloning techniques and verified by sequencing.
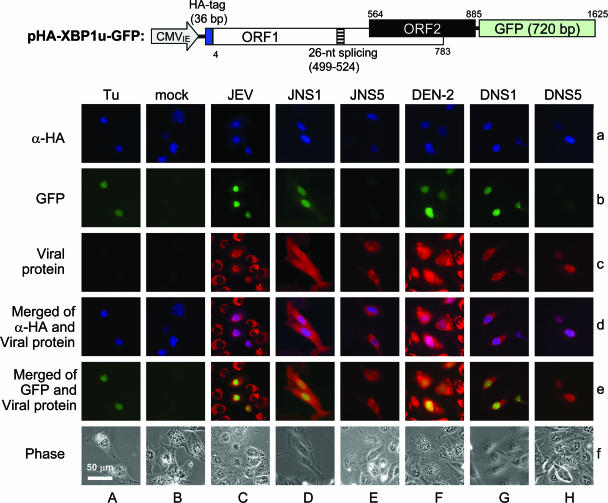
FIG. 2.
Analysis of an XBP1 splicing event using a GFP-based reporter. (C and F) N18 cells with JEV or DEN-2 (MOI, 3) adsorbed for 2 h were transfected with pHA-XBP1u-GFP reporter (1 μg). (D, E, G, and H) For cotransfection, N18 cells were transfected with reporter (1 μg) and plasmids encoding individual viral proteins (1 μg) as indicated. (A) Tunicamycin (Tu) treatment (2 μg/ml for 12 h starting from 8 h post-reporter transfection) served as a positive control. Immunofluorescence assays detecting the HA tag and the viral proteins were performed 20 h after transfection. a, staining with anti-HA antibody plus Cy5-labeled secondary antibody (pseudocolored blue); b, GFP (green); c, viral proteins stained with anti-NS1 or anti-Flag plus rhodamine-labeled secondary antibody (red); d, merged images of a and c; e, merged images of b and c; f, phase-contrast. The fluorescence was observed using an Axiovert 200 M inverted microscope (Zeiss) with a 40× objective (0.6 air). The images were captured using the MetaMorph Imaging System (Molecular Devices). Bar, 50 μm.
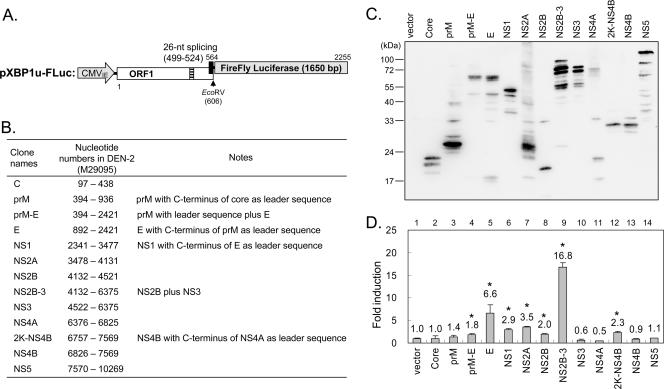
FIG. 3.
Screening of DEN-2 viral proteins as ER stress-inducing agents. (A) Schematic diagram of pXBP1u-FLuc reporter construct. (B) The corresponding nucleotide numbers of DEN-2 (based on the sequences of DEN-2 NGC [GenBank accession number M29095]) contained in each construct. (C) Protein expression from plasmid constructs with DEN-2 cDNA. N18 cells were transfected with plasmids as indicated and immonoblotted with anti-Flag antibody. (D) N18 cells were cotransfected with pXBP1u-FLuc reporter (0.25 μg), pRL-TK (0.15 μg), and plasmids encoding individual viral proteins (0.6 μg) as indicated. Cell lysates were harvested 48 h after transfection and measured for firefly and Renilla luciferase activities using the Dual-Luciferase assay system. Firefly luciferase activity was normalized to Renilla luciferase activity, and multiples of firefly luciferase induction were calculated based on the value of the vector control. The results are means plus standard deviations (n = 3 to 5). *, P < 0.01 by Student's t test.
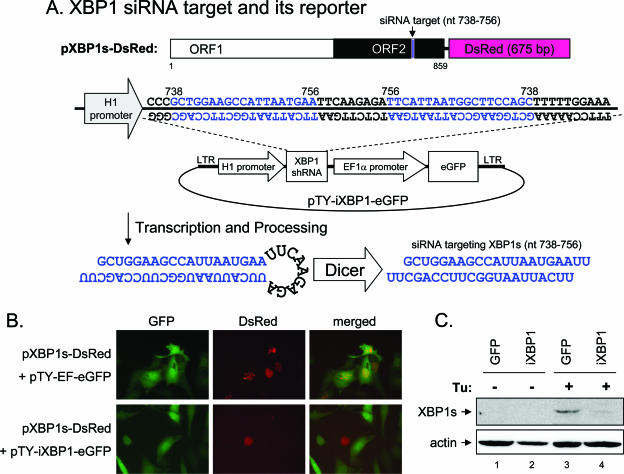
For expression of XBP1 siRNA targeting nt 738 to 756 of human XBP1s (also nt 723 to 741 of mouse XBP1s), two single-stranded DNA oligomers, 5′-GATCCCCGCTGGAAGCCATTAATGAATTCAAGAGATTCATTAATGGCTTCCAGCTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGCTGGAAGCCATTAATGAATCTCTTGAATTCATTAATGGCTTCCAGCGGG-3′, were annealed and cloned into the HindIII and BglII sites of pSUPER (5). The H1 promoter-driven siRNA expression cassette was then subcloned into the ClaI site of pTY-EFeGFP to form pTY-iXBP1-eGFP.
The human XBP1s cDNA was amplified from tunicamycin-treated A549 cells by RT-PCR. The forward primer was as described above for XBP1u, and the reverse primer, 5′-ACCGCGTCGACAGACACTAATCAGCTGGGGAAAGAG-3′, annealed to nt 1131 to 1155 of XBP1s and contained a SalI site (italic). The sequence of the cloned XBP1s was confirmed to be identical to that of human spliced XBP1 (GenBank accession number AB076384). To construct pXBP1s-DsRed, the XBP1s cDNA from nt −3 to 859 was amplified and cloned into pDsRed2-N1 (Clontech).
To express viral proteins, cDNA fragments of JEV RP-9 (GenBank accession number AF014161) and DEN-2 PL046 (based on the sequences of DEN-2 New Guinea-C strain, GenBank accession number M29095) encoding the individual viral proteins were subcloned into Flag-tagged pCR3.1 (Invitrogen). Initiation codons were added to these constructs at their N termini. Detailed information about the JEV constructs has been provided elsewhere (38); see Fig. 3B for the nucleotides included in each of the DEN-2 viral-protein constructs. The S135A and K199A mutants of DEN-2 NS2B-3 were generated by single-primer mutagenesis as previously described (42) using the primers 5′-CTGGACTTTTCTCCTGGAACCGCGGGATCTCCAATCATCGACAAA-3′ and 5′-CCACCCAGGAGCGGGCGCCACGAAGAGATACCTTCCGGC-3′, respectively.
Lentiviral-vector preparation.
Human TE671 cells were cotransfected with the three helper plasmids, pHP-dl-N/A, pHEF-VSV-G, and pCEP4-tat (obtained from Lung-Ji Chang through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH) (9, 15, 25, 71), plus either pTY-EF-eGFP or pTY-iXBP1-eGFP, using GeneJammer transfection reagent (Stratagene). The transfected cells were incubated at 37°C in an atmosphere of 3% CO2 for 4 to 5 h, and then the medium was replaced with fresh medium. Cell supernatants containing the viral vectors were harvested at 36, 48, and 60 h after transfection. The virus was filtered using a 0.45-μm low-protein-binding filter and concentrated by centrifugation at 14,000 rpm at 4°C for 3.5 h using a JA20 (Beckman) rotor. The virus pellets were resuspended with fresh medium and stored at −80°C. The lentiviral vector was then transduced into cells by serial dilution to determine the relative titers by GFP signals.
RNA preparation and RT-PCR analysis.
Total RNAs from cultured cells and mouse brains were prepared with an RNeasy RNA Mini Kit (QIAGEN) and TRIZOL Reagent (Invitrogen), respectively. The cDNA was reverse transcribed from 1 μg of total RNA with oligo(dT) using a ThermoScript RT kit (Invitrogen). PCR was then carried out using the specific primer sets for XBP1 (5′-AAACAGAGTAGCAGCGCAGACTGC-3′ and 5′-GGATCTCTAAAACTAGAGGCTTGGTG-3′, annealing to nt 216 to 239 and 788 to 813 of mouse XBP1) (7), EDEM1 (5′-GCCTCCTTTCTGCTCACAGAATAATAA-3′ and 5′-CTCCTTCTCCTTCATTGCAGGCT-3′, annealing to nt 593 to 619 and 1119 to 1141 of mouse EDEM1), p58(IPK) (p58 inhibitor of PKR) [5′-GAGGTTTGTGTTTGGGATGCAG-3′ and 5′-TGAGCTGAAGGGATTGAACCC-3′, annealing to nt 544 to 565 and 1459 to 1479 of p58(IPK)], ERdj4 (5′-TTAGAAATGGCTACTCCACAGTCA-3′ and 5′-TTGTCCTGAACAATCAGTGTATGTAG-3′, annealing to nt −6 to 18 and 641 to 666), and β-actin (5′-TCCTGTGGCATCCACGAAACT-3′ and 5′-GAAGCATTTGCGGTGGACGAT-3′, annealing to nt 811 to 831 and 1105 to 1125 of actin), respectively. The sizes of the PCR products for XBP1 are listed in Fig. 1A, and those for EDEM1, p58(IPK), ERdj4, and β-actin are 549, 936, 672, and 315 bp, respectively. For detecting JEV genomic RNA, the following primers were used: JEV5678-5639 (5′-GCCCGCATATTCTGTGATCCATTCGTATCCACTGCTCCAC-3′) for reverse transcription, and JEV3098-3123 (5′-CGACACATGGAAACTTGAGAGGGCAG-3′) and JEV4214-4185 (5′-TCTCTTCTTGTTTGGGTTGCAGACCATTAG-3′) for PCR; the expected size of the PCR product was 1,117 bp.
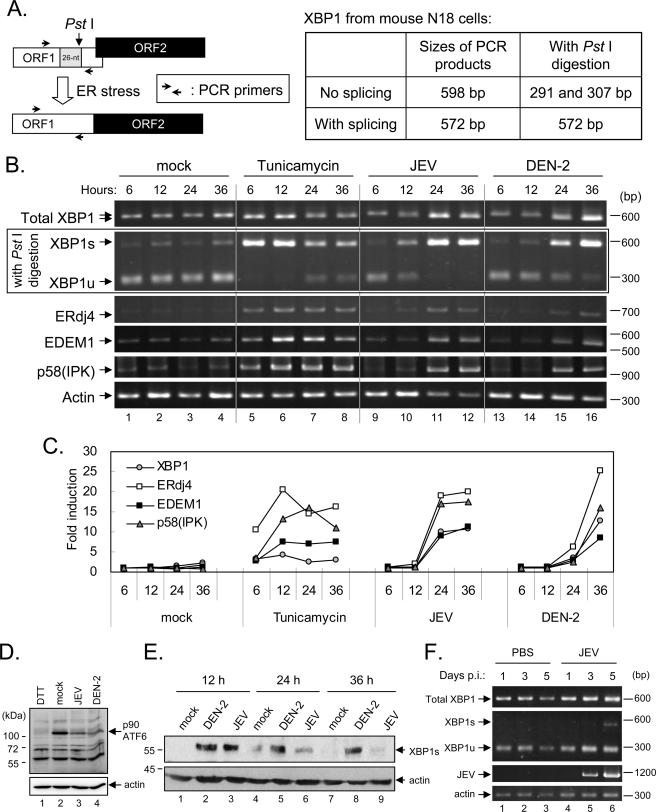
FIG. 1.
Flavivirus induces XBP1 mRNA splicing in infected cultured neuronal cells and mouse brains. (A) The analysis scheme of XBP1 mRNA splicing. The relative locations of the 26-nt intron, the PstI recognition site, and the PCR-amplified region are shown. The sizes of PCR-amplified fragments from spliced and unspliced XBP1 with or without PstI cleavage are also listed. (B) XBP1 mRNA splicing and induction of downstream genes. N18, a mouse neuroblastoma cell line, was mock infected (lanes 1 to 4), treated with tunicamycin (1 μg/ml; lanes 5 to 8), and infected with JEV (lanes 9 to 12) or DEN-2 (lanes 13 to 16). The MOI was 3. Cells were harvested at 6, 12, 24, or 36 h after treatment for RT-PCR analysis using primer sets specific for the genes shown on the left. The PCR product of XBP1 was further analyzed by PstI digestion. DNA molecules were separated by 2 or 1.5% agarose gel electrophoresis, stained with ethidium bromide, and photographed with a Photo-Print photodocumentation system (Vilber Lourmat). (C) The induction (n-fold) of selected UPR downstream genes was quantified by real-time RT-PCR as described in Materials and Methods. (D) Decrease in full-length ATF6 (p90ATF6) in flavivirus-infected cells. N18 cells were treated with 2 mM dithiothreitol (DTT) for 3 h or infected with JEV or DEN-2 (MOI, 5) for 20 h before the cell lysates were harvested (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol) for immunoblotting with anti-ATF6 (IMGENEX) and anti-actin antibodies. (E) The protein expression of XBP1s in N18 cells was analyzed by immunoblotting at several time points after infection with JEV or DEN-2 (MOI, 5) as indicated. The nuclear extracts were separated by 10% SDS-polyacrylamide gel electrophoresis and immunoblotted with the antibody indicated on the right. (F) The XBP1 mRNA status in brain lysates of C57/B6 mice intracranially inoculated with JEV (1 × 104 PFU/mouse) was analyzed by RT-PCR and PstI digestion. The level of JEV replication was also detected by RT-PCR of the NS3 region of the JEV genome. PBS, mice sham inoculated with PBS serving as a negative control. p.i., postinfection.
For quantification, the PCR was performed with the LightCycler FastStart DNA Master PLUS SYBR Green I kit (Roche), according to the manufacturer's recommendations. In brief, 0.5 μM of each primer was mixed with 4 μM of premix FastStart DNA Master PLUS SYBR Green I and 1 μl of cDNA. The following program was used for quantification and melting-curve analysis: 95°C for 10 min; 40 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 20 s; 95°C for 0 s at a ramp rate of 20; 65°C for 15 s at a ramp rate of 20; and 95°C for 0 s at a ramp rate of 0.1. For relative quantification, expression of each target gene was measured with respect to the actin in each sample, and the concentration was calculated from a standard curve. Quantification analysis of target and reference genes was based on the second derivative maximum method (Roche, technical note LC13/2001). Melting curves were used to verify the specificities of products.
Western immunoblot analysis.
The cells were disrupted with RIPA buffer (10 mM Tris, pH 7.5, 5 mM EDTA, 150 mM NaCl, 0.1% sodium dodecyl sulfate [SDS], 1% Triton X-100, 1% sodium deoxycholate) containing a cocktail of protease inhibitors (Roche). For nuclear-extract preparation, we followed the protocol of Heather Harding and Huiqing Zeng of David Ron's laboratory in New York University (http://saturn.med.nyu.edu/research/mp/ronlab/protocols/NucCyto.pdf). Protein samples were separated by SDS-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane (Hybond-C Super; Amersham). The nonspecific antibody binding sites were blocked with skim milk in phospate-buffered saline (PBS) and then reacted with the primary antibody, rabbit-anti-XBP1 antibody (1:5,000), kindly provided by David Ron; anti-actin (1:10,000; Chemicon); anti-ATF6 (1:250; IMG-273; IMGENEX); anti-caspase 3 (1:1,000; no. 9662; Cell Signaling Technology, Inc.); and mouse anti-Flag M2 (1:1,000; Sigma-Aldrich). For XBP1 and ATF6 detection, we then used Biotin-SP-AffiniPure goat anti-rabbit/mouse immunoglobulin G (1:2,000) and peroxidase-streptavidin (1:2,000; Jackson ImmunoResearch). Other blots were treated with horseradish peroxidase-conjugated secondary antibody. The signals were detected using enhanced chemiluminescence (Amersham Biosciences).
Immunofluorescence assay.
Cells were fixed with 4% paraformaldehyde in PBS and permeabilized by 0.5% Triton X-100. After the cells were blocked with skim milk in PBS, primary antibody (1:1,000) was added, and the cells were incubated for 1 h at room temperature. Anti-HA tag antibody (clone DW2) was from Upstate, mouse anti-Flag M2 antibody was obtained from Sigma-Aldrich, and monoclonal antibodies against JEV and DEN-2 were previously described (67). Secondary antibodies conjugated with Alexa Fluor 568 (Molecular Probes), rhodamine (Jackson ImmunoResearch), or Cy-5 (KPL) were used. The cells were examined and photographed under an inverted fluorescence microscope.
Screening of XBP1 splicing-inducing viral proteins.
The reporter and an internal control, pRL-TK (Promega), were cotransfected with the plasmids encoding individual viral proteins by SuperFect (QIAGEN). The transfectants were cultured in 12-well plates with high-glucose Dulbecco's modified Eagle's medium containing 5% FBS, and the medium was replaced frequently to minimize the effect of glucose starvation. Samples were harvested and measured for firefly and Renilla luciferase activities by the Dual-Luciferase assay system (Promega). The firefly luciferase was normalized with the Renilla luciferase activity, and the relative inductions were calculated based on the vector control.
ER expansion analysis.
The cells were pelleted, resuspended in culture medium, and then stained with an ER marker, brefeldin A, and BODIPY 558/568 conjugate (Molecular Probes) (16), by incubating the live cells with the dye at 50 to 100 ng/ml for 10 to 20 min. For flow cytometry, the staining medium was washed off and replaced by PBS, and then analysis was done on a FACSort (Becton Dickinson) using CellQuest software (52).
Cytotoxicity assay.
Virus-induced cytopathic effects (CPE) were assessed by the release of a cytoplasmic enzyme, lactate dehydrogenase (LDH), by using a commercial kit (Cytotoxicity Detection Kit; Roche) and staining the surviving cells with crystal violet. The culture supernatants from cell samples were clarified by centrifugation, mixed with reaction mixture (diaphorase/NADH+, tetrazolium salt INT/sodium lactate), incubated at room temperature for about 30 min, and then read by an enzyme-linked immunosorbent assay reader at 490 nm (Molecular Devices). For crystal violet staining, cells were overlaid with medium containing 1% agarose after virus adsorption. At 2 days postinfection, the agarose was removed and the cells were fixed with 10% formaldehyde and stained with 0.5% crystal violet.
RESULTS
Flavivirus infection activates the XBP1 pathway.
To assess whether flavivirus infection triggers the IRE1-XBP1 pathway, we analyzed XBP1 mRNA splicing in JEV- and DEN-2-infected N18 cells, a mouse neuroblastoma cell line in which JEV has been shown to induce ER stress and the UPR (58). The XBP1 cDNA was amplified by RT-PCR and digested by PstI, which has a recognition site located within the 26-nt region of XBP1 cDNA removed by IRE1-mediated splicing, as previously described (7) (Fig. 1A). Infection of N18 cells with JEV or DEN-2, but not mock infection, caused XBP1 splicing, as evidenced by resistance of the spliced product to PstI digestion (Fig. 1B). Tunicamycin, a well-recognized inducer of ER stress, served as a positive control in these tests. It was also noticed that cells treated with tunicamycin or infected with JEV or DEN-2 seemed to express higher levels of XBP1 than did mock-infected cells (Fig. 1B). As XBP1 is produced after ATF6 activation (34, 69), we suspect that the ATF6 pathway might also be activated by flavivirus infection. In response to ER stress, ATF6 translocates from the ER to the Golgi, and then the 90-kDa full-length ATF6 (p90ATF6) is processed to its active 50-kDa form (20). As evidenced by the much lower p90ATF6 levels in dithiothreitol, an ER stress inducer, and flavivirus-infected cells compared to the mock control (Fig. 1D), cleavage and activation of ATF6 likely occur in JEV- and DEN-2-infected cells. Regarding the other ER stress sensor, PERK, even though translational shutdown of host gene expression was not generally observed in flavivirus-infected cells (48), a potential involvement of the PERK pathway in JEV/DEN-2 infection remains to be studied, as previously discussed (58).
The XBP1 downstream genes ERdj4 (30), EDEM1, and p58(IPK) (33, 64) were elevated in JEV- and DEN-2-infected cells, as seen in tunicamycin-treated N18 cells (Fig. 1B and C), suggesting that unlike the results of HCV infection (59), the XBP1 pathway is activated during flavivirus infection. The time course studies showed that the stable XBP1s, encoding a protein of about 55 kDa, was detected as early as 12 h after infection with JEV or DEN-2 (Fig. 1E). We noticed that at the later stage of viral infection, XBP1s protein levels declined, in spite of an accumulation of XBP1 mRNA (Fig. 1B and C). Whether a negative regulation targeting the XBP1s protein for proteasome-mediated degradation, as reported for XBP1u (70) or certain flaviviral proteins, might be responsible for this observation remains to be studied. Furthermore, XBP1 splicing was seen both in cultured cells and in the animal model, as XBP1 splicing was detected in the mouse brain 5 days post-JEV inoculation but not in the PBS sham-inoculated control (Fig. 1F). As JEV replicated to a peak titer of around 107 PFU at days 4 to 6 postinoculation (12), the levels of JEV replication detected by RT-PCR, shown in Fig. 1F, reflect the fact that XBP1 splicing correlated with JEV production. Thus, our results indicate that XBP1 mRNA is spliced to encode the XBP1s protein with transactivation activity and that the downstream genes of XBP1 are turned on in JEV- and DEN-2-infected N18 cells.
The flaviviral proteins responsible for XBP1 splicing.
To identify the viral components inducing ER stress, we established a reporter system based on the XBP1 splicing event similar to those described previously (26). A reporter plasmid, pHA-XBP1u-GFP (Fig. 2), encodes an N-terminal HA-tagged XBP1u, with the C terminus fused to GFP at the second ORF of XBP1. The GFP is expressed only after XBP1 splicing removes the 26-nt intron. Transfection and expression of pHA-XBP1u-GFP was confirmed by immunofluorescence assay with anti-HA antibody (Fig. 2, row a). GFP signals were noticed in JEV-infected (Fig. 2C), DEN-2-infected (Fig. 2F), and tunicamycin-treated (Fig. 2A) cells, but not in mock-infected cells (Fig. 2B). Cotransfection of the reporter construct with plasmids encoding the JEV or DEN-2 glycoprotein NS1 (Fig. 2D and G) resulted in GFP expression, whereas cotransfection of the reporter construct with plasmids encoding the cytoplasmic NS5 of JEV and DEN-2 did not convert GFP expression (Fig. 2E and H).
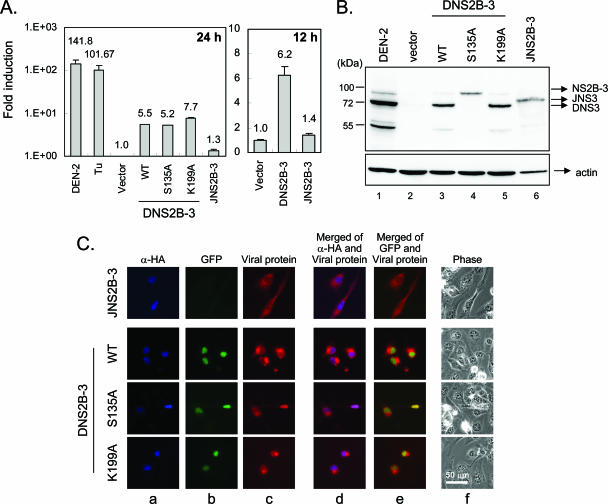
To quantify reporter gene expression, we constructed another reporter plasmid, pXBP1u-FLuc, using the firefly luciferase gene (Fig. 3A). N18 cells cotransfected with pXBP1u-FLuc, pRL-TK (an internal transfection control expressing Renilla luciferase), and plasmids encoding individual DEN-2 viral proteins (Fig. 3B and C) were analyzed for reporter activities, and relative increases in reporter activity (induction compared with that of the vector control) are shown (Fig. 3D). Several ER-associated DEN-2 viral proteins, such as the glycoproteins prM-E, E, and NS1 and the ER membrane-anchored small hydrophobic proteins NS2A, NS2B, and 2,000-molecular-weight protein (2K)-NS4B, increased luciferase reporter activity. Notably, DEN-2 NS2B-3, a fusion of NS2B and NS3, strongly induced XBP1-mediated reporter gene expression. As the protease activity of NS3 depends on the cofactor NS2B (40), the finding that NS2B-3 was much more effective in inducing XBP1 splicing than was NS2B or NS3 alone may suggest that NS2B-3 protease activity is important in triggering the ER stress response. However, a single point mutation changing serine residue 135 of NS3 to alanine (S135A) abolished protease activity, as evidenced by loss of NS2B-3 autocleavage (Fig. 4B, lane 4) (35); this mutation did not hamper the ability of NS2B-3 to trigger XBP1 splicing in both the luciferase and GFP reporter systems (Fig. 4A and C). Furthermore, blocking of the helicase and NTPase activities of NS2B-3 by a K199A mutation (43) also did not affect the ability of DEN-2 NS2B-3 to trigger XBP1 splicing (Fig. 4). We also noticed that, although several viral proteins induced XBP1 splicing in the reporter assay system, none of the individual viral proteins induced XBP1 splicing to the level seen upon viral infection (more than 100-fold higher than the vector control) (Fig. 4A). These results suggest that several viral proteins may cooperate to trigger XBP1 splicing in flavivirus-infected cells or that a role for RNA replication cannot be excluded.
FIG. 4.
DEN-2 NS2B-3 triggers XBP1 splicing. (A) XBP1 splicing by wild-type (WT), protease-dead (S135A), and helicase/NTPase-dead (K199A) constructs of DEN-2 NS2B-3, but not by JEV NS2B-3, was analyzed as described in the legend to Fig. 3, except that data were obtained 24 h or 12 h after transfection. N18 cells infected with DEN-2 (MOI, 3) or treated with tunicamycin (Tu) (1 μg/ml) were also included. The results are means plus standard deviations (n = 3). (B) The expression of proteins from these constructs was detected by immunoblotting with anti-NS3 antibody. (C) The wild-type and mutant forms of DEN-2 NS2B-3 were also tested for their abilities to trigger XBP1 splicing using the GFP-based reporter system, as described in the legend to Fig. 2.
On examination of JEV proteins, we found that the viral glycoproteins prM, E, and NS1, as well as the small hydrophobic viral proteins NS2A, NS2B, and NS4B, also acted as ER stress inducers in our reporter systems (data not shown). However, in marked contrast to the situation with DEN-2 NS2B-3, we were not able to detect significant XBP1 splicing triggered by JEV NS2B-3 (Fig. 4A and C). Even at an earlier time point (12 h posttransfection), JNS2B-3 was still not able to trigger XBP1 splicing (Fig. 4A, right). It appears that the XBP1 pathway is induced by flaviviral proteins mainly associated with the ER, such as glycoproteins and ER membrane-anchored proteins. In addition, DEN-2 NS2B-3 triggers XBP1 splicing. The mechanism of this induction is unclear, but it appears not to depend on the known enzymatic activities of NS2B-3.
Reduced XBP1 enhances cytopathic effects induced by flavivirus infection.
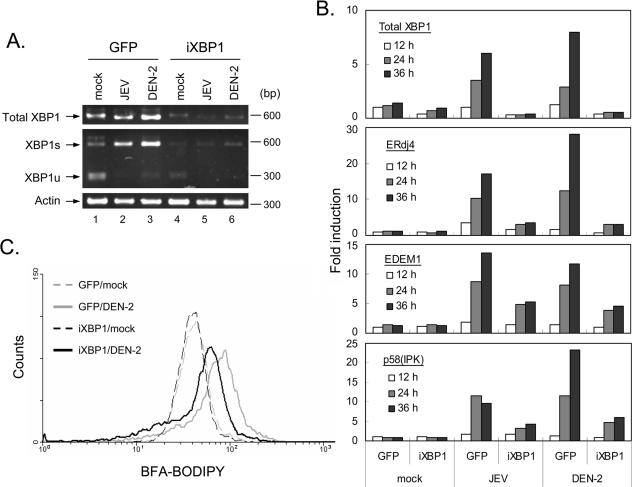
As the XBP1 pathway is activated by flaviviruses, we asked how this pathway might function in the flavivirus life cycle. To test the effect of XBP1 on flaviviral infection, we reduced XBP1 RNA expression using an RNA interference technique (5, 22, 32). A GFP-expressing VSV-G protein-pseudotyped lentiviral vector transcribing an siRNA targeting nt 738 to 756 of human XBP1s (also targeting nt 723 to 741 of mouse XBP1s) from the H1 promoter was constructed (Fig. 5A). The effectiveness of this siRNA in reducing XBP1 expression was tested by cotransfection with a plasmid encoding a C-terminal DsRed-tagged XBP1s (pXBP1s-DsRed) (Fig. 5A). The pTY-EF-eGFP vector control had no effect on XBP1s-DsRed expression (Fig. 5B, top), whereas the red fluorescence of XBP1s-DsRed was diminished in pTY-iXBP1-eGFP-transfected N18 cells (Fig. 5B, bottom). We then generated recombinant lentivirus expressing XBP1 siRNA (iXBP1) and used it to transduce N18 cells. The endogenous XBP1 RNA (data not shown) and protein (Fig. 5C) levels induced by tunicamycin were reduced in iXBP1-transduced N18 cells. The levels of XBP1 mRNA and extents of XBP1 splicing induced by JEV and DEN-2 infections were greatly reduced in iXBP1-transduced N18 cells (Fig. 6A). The induction of UPR downstream genes were also attenuated in these XBP1 knockdown cells (Fig. 6B). To explore the effects of virus and XBP1 on ER expansion, we labeled the cells with fluorescent-dye-conjugated brefeldin A (16) and then analyzed the cells by flow cytometry (52). DEN-2 infection of N18 cells transduced with GFP control induced ER expansion, whereas the level of DEN-2-induced ER expansion was reduced in iXBP1-transduced cells (Fig. 6C), suggesting that XBP1 is involved in flavivirus-induced ER expansion. Since a low level of ER expansion still occurred in these XBP1 knockdown cells, we suspect that the residual amount of XBP1s (Fig. 6A) or other unidentified factors might be involved.
FIG. 5.
Reducing XBP1 by RNA interference. (A) Schematic diagram of the pTY-iXBP1-eGFP construct containing siRNA sequences targeting XBP1 and the reporter pXBP1s-DsRed. Note that expression of GFP reflects delivery of siRNA. (B) N18 cells were cotransfected with 0.75 μg of pTY-iXBP1-eGFP or the vector control plus pXBP1s-DsRed (0.75 μg) overnight. The cells were observed and photographed under a Leica DMIRB inverted microscope. eGFP, enhanced GFP. (C) N18 cells transduced with lentivirus expressing GFP or iXBP1-GFP were treated with tunicamycin (Tu) (2.5 μg/ml) for 6 h or untreated. Endogenous XBP1s protein induction was analyzed by immunoblotting as described in the legend to Fig. 1.
FIG. 6.
siRNA targeting of XBP1 mRNA attenuated virus-induced ER expansion and XBP1 downstream gene induction. (A) Stable N18 cell populations transduced with lentivirus expressing GFP or iXBP1-GFP were mock infected or infected with JEV or DEN-2 (MOI, 5) for 24 or 30 h, respectively, before XBP1 mRNA expression and splicing were analyzed by RT-PCR and PstI digestion, as described in the legend to Fig. 1. (B) Cells were harvested at 12, 24, and 36 h post-virus infection (MOI, 5) for real-time RT-PCR analysis using primer sets specific for the indicated genes, as described in the legend to Fig. 1. (C) N18 cells transduced with lentivirus encoding GFP or iXBP1-GFP were mock infected or infected with DEN-2 (MOI, 5) for 30 h. Flow cytometry analysis of the ER contents was then performed with brefeldin A and BODIPY 558/568 conjugate staining, followed by analysis using FACSort (Becton Dickinson) and CellQuest software.
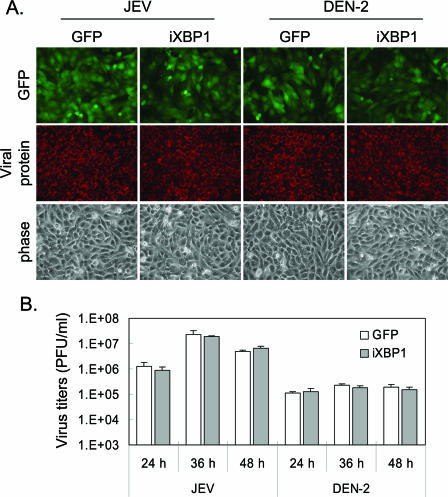
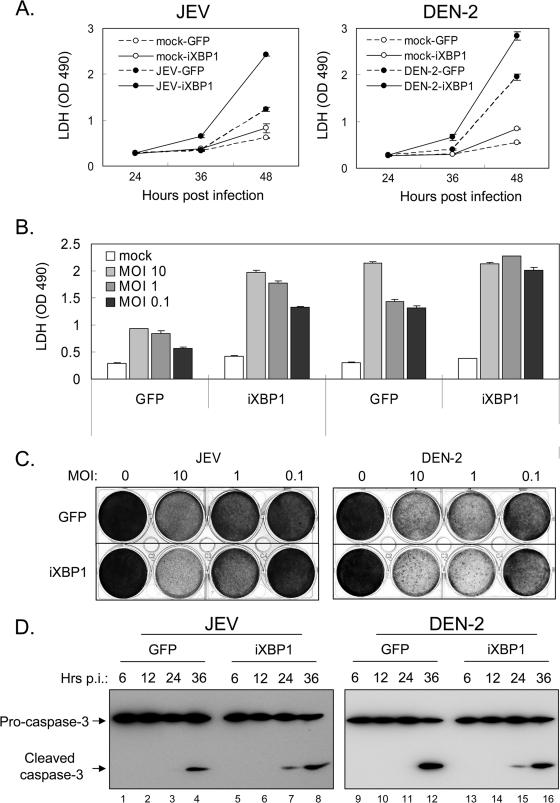
These cells were then tested for their susceptibility to flavivirus infection. As shown in Fig. 7A, viral-protein expression detected by anti-JEV NS1 or anti-DEN-2 NS1 antibody was readily observed in all infected cells (Fig. 7A). The levels of virus production from N18 cells transduced with GFP or iXBP1, as determined by plaque-forming assays, were also quite similar at several time points post-JEV or -DEN-2 infection (Fig. 7B). It is not clear whether the low level of XBP1s (Fig. 6) was enough to support viral replication or whether XBP1 was not essential for flavivirus production. Since the role of XBP1 in the UPR is to remedy ER stress, we then determined the levels of CPE induced by flaviviral infections, during which the levels of XBP1 were reduced. We measured CPE both by crystal violet staining of surviving cells and by assaying the release of the cytoplasmic enzyme LDH, as leaking of this LDH enzyme is a measurement of cytotoxicity. As shown in Fig. 8, upon JEV or DEN-2 infection, cells impaired in XBP1 exhibited a CPE much more severe than the CPE shown by control GFP cells at several time points postinfection (Fig. 8A) in a multiplicity of infection (MOI)-dependent manner, especially for JEV infection (Fig. 8B and C). Furthermore, cleavage of pro-caspase 3, an apoptosis marker, also occurred at the earlier time points post-JEV and -DEN-2 infection in iXBP1-transduced cells (Fig. 8D). Our results thus suggest that XBP1 activation may mediate protection against ER stress-mediated apoptosis triggered by flavivirus infection.
FIG. 7.
Downregulating XBP1 in N18 cells does not affect the susceptibility and virus production of flaviviruses. (A) N18 cells stably transduced with GFP or iXBP1-GFP lentivirus were infected with JEV or DEN-2 at an MOI of 3 for 24 h before immunofluorescence assay with anti-NS1 plus Alexa Fluor 568-conjugated secondary antibody. (B) The culture medium was collected for virus titration at the indicated time points after JEV or DEN-2 (MOI, 1) infection. The data shown are means plus standard deviations from three independent experiments.
FIG. 8.
Functional impairment of XBP1 in N18 cells exacerbates CPE induced by JEV and DEN-2. (A) N18 cells stably transduced with GFP or iXBP1-GFP lentivirus were mock infected or infected with JEV or DEN-2 at an MOI of 1. The culture supernatants were collected for LDH release assay at the indicated time points. The data shown are means ± standard deviations from three independent experiments. OD 490, optical density at 490 nm. (B and C) The cells described for panel A were mock infected or infected with JEV or DEN-2 at various doses (MOIs as indicated) for 48 h. The culture supernatants were collected for LDH release assay (B), and the surviving cells were fixed and stained with crystal violet (C). The LDH readings are the means plus standard deviations from two independent experiments. (D) Cells were infected with JEV or DEN-2 (MOI, 5) and harvested at the indicated time points for caspase 3 activation analysis by immunoblotting. p.i., postinfection.
DISCUSSION
Flavivirus replication and maturation are highly associated with the host ER membranes, and flavivirus infections are known to induce ER stress (27, 58, 60). In this study, we found that flavivirus infections trigger the IRE1-XBP1 pathway of the UPR, as XBP1 splicing was detected in both JEV- and DEN-2-infected neuronal N18 cells and in mouse brains inoculated with JEV (Fig. 1). Unlike the situation with HCV, where XBP1 trans-activation is repressed (59), we found that the XBP1 downstream genes ERdj4, EDEM1, and p58(IPK) were induced in JEV- and DEN-2-infected N18 cells (Fig. 1B and C), indicating that the IRE1-XBP1 pathway is activated in cells infected with JEV/DEN-2. The discrepancy in XBP1 induction between these viruses might reflect the fact that HCV usually causes chronic infection whereas JEV/DEN-2 readily kills the infected cells. The possible roles of XBP1 in flavivirus infection were further studied, and it was found that XBP1 may help the cells to cope with ER stress induced by flaviviral infection, as reduction of XBP1 leads to greater cytotoxicity in response to JEV or DEN-2 infection (Fig. 8). Our study thus provides a good example of how a virus takes advantage of a cellular signaling event to benefit its own survival.
For a flavivirus, whose life cycle is largely dependent on cellular membrane networks, the ER is the major battlefield between the host and the virus. With a positive-sense RNA genome, the flaviviral life cycle begins with translation of the incoming genomic RNA to yield a polyprotein that is processed into viral proteins (40). A membrane-associated viral replication complex then copies the genomes for virion production and for more mRNA for viral-protein synthesis. Using easily monitored reporter systems, we conducted a comprehensive search for flaviviral proteins triggering the XBP1 splicing event. ER-associated flaviviral proteins, such as the glycoproteins and the ER-anchored small hydrophobic proteins, were identified as triggering XBP1 splicing (Fig. 2 and 3). As no similarity between IRE1 and any flaviviral proteins was noticed, it is less likely that flaviviral proteins might act directly on XBP1 splicing. These viral proteins may activate the XBP1 pathway through the IRE1-mediated ER stress machinery; however, this notion will remain tentative pending further study of the IRE1-deficient cells. The molecular mechanisms for XBP1 activation by flaviviral glycoproteins probably involve an accumulation of misfolded proteins in the ER, whereas that for small hydrophobic flaviviral proteins is likely dependent on ER membrane association. With protein NS4B, retention of the 2K signal peptide targeting the ER membrane (37) increased the ability of the protein to cause XBP1 splicing compared with the mature form of the protein (2K-NS4B versus NS4B) (Fig. 3B). The possibility that these proteins modify membrane permeability (10), in turn leading to a calcium imbalance affecting XBP1 splicing, requires further study. The finding that DEN-2 NS2B-3, but not JEV NS2B-3, is a potent ER stress inducer (Fig. 4) is rather interesting. As the protease-dead S135A and the helicase/NTPase-dead K199A mutants of DEN-2 NS2B-3 were still able to trigger XBP1 splicing (Fig. 4), we suspect that DEN-2 NS2B-3 may interact with and modulate the functions of certain host factors involved in the UPR. Further characterization of the cellular proteins associating with DEN-2 NS2B-3 might illuminate the molecular mechanism.
XBP1 was first discovered as a transcription factor in B cells and was later found to be essential for B-cell differentiation into plasma cells (23, 24). A professional antibody-secretory cell needs to accommodate a large quantity of immunoglobulin, and ER expansion involving coordinated biosynthesis of ER components (both proteins and lipids) has been well demonstrated in the plasma cell. Recent studies have linked XBP1 from plasma cell differentiation to the UPR, as well as membrane biogenesis (50). Cells overexpressing XBP1s exhibit elevated levels of membrane phospholipid, an activated cytidine diphosphocholine pathway, and increased ER volume (57). Our finding that impairment of XBP1 had no effect on flavivirus production (Fig. 7) suggests that XBP1 splicing is beneficial, but may not be essential, for flavivirus production. Instead, the more important role of XBP1 may be to alleviate ER stress induced by flavivirus infection, as increased CPE was noticed in cells with reduced levels of XBP1 (Fig. 8). As intense or prolonged ER stress often leads to cell death (4, 21), apoptotic cell death associated with ER stress also contributes to the cytotoxicity of JEV infection (58). A properly functioning XBP1 pathway would not only enhance protein-folding ability by increasing expression of chaperones, but would also reduce ER loads by facilitating protein degradation. It has been reported that the ER stress induced by a glycosylation inhibitor is exacerbated by functional impairment of XBP1 (32). Here, we provide evidence that cell death induced by flaviviral infection was more extensive in cells with disrupted XBP1, suggesting that flavivirus-induced ER stress may be alleviated by the IRE1-XBP1 branch of the UPR.
Viruses, as absolutely intracellular parasites, rely on cellular machinery to complete their life cycles and thus face various cellular stress responses (21). Initiation of ER stress may be a host defense against flavivirus infection. To cope with ER stress, however, flaviviruses have evolved to activate the IRE1-XBP1 arm of the UPR. In turn, this may expand both the time and space available for flavivirus replication. Although the exact contributions of host and virus to XBP1 splicing and UPR induction remain unclear, we show here that flaviviruses initiate XBP1 signaling and benefit from this cellular event. We have also established reporter systems that work to visualize XBP1 splicing in individual cells and to quantify the splicing level in a cell population. These systems will be powerful research tools in future studies.
Acknowledgments
We are grateful to David Ron (New York University) for the XBP1 antibody, information on the target of XBP1 siRNA, and helpful discussions. We also thank L. J. Chang (University of Florida) for the lentiviral vector system and Diane E. Griffin (Johns Hopkins University) for N18 cells.
This work was supported by grants awarded to Y.-L.L. from the National Science Council (NSC-94-2320-B-001-008 and NSC-95-2320-B-001-031-MY3), National Health Research Institute (NHRI-EX94-9433SI), and Academia Sinica, Taiwan, Republic of China.
Footnotes
Published ahead of print on 20 September 2006.
REFERENCES
- 1.Amano, T., E. Richelson, and M. Nirenberg. 1972. Neurotransmitter synthesis by neuroblastoma clones (neuroblast differentiation-cell culture-choline acetyltransferase-acetylcholinesterase-tyrosine hydroxylase-axons-dendrites). Proc. Natl. Acad. Sci. USA 69:258-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benali-Furet, N. L., M. Chami, L. Houel, F. De Giorgi, F. Vernejoul, D. Lagorce, L. Buscail, R. Bartenschlager, F. Ichas, R. Rizzuto, and P. Paterlini-Brechot. 2005. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene 24:4921-4933. [DOI] [PubMed] [Google Scholar]
- 3.Bertolotti, A., Y. Zhang, L. M. Hendershot, H. P. Harding, and D. Ron. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326-332. [DOI] [PubMed] [Google Scholar]
- 4.Boyce, M., and J. Yuan. 2006. Cellular response to endoplasmic reticulum stress: a matter of life or death. Cell Death Differ. 13:363-373. [DOI] [PubMed] [Google Scholar]
- 5.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 6.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1125. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott-Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 7.Calfon, M., H. Zeng, F. Urano, J. H. Till, S. R. Hubbard, H. P. Harding, S. G. Clark, and D. Ron. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92-96. [DOI] [PubMed] [Google Scholar]
- 8.Chan, S. W., and P. A. Egan. 2005. Hepatitis C virus envelope proteins regulate CHOP via induction of the unfolded protein response. FASEB J. 19:1510-1512. [DOI] [PubMed] [Google Scholar]
- 9.Chang, L. J., V. Urlacher, T. Iwakuma, Y. Cui, and J. Zucali. 1999. Efficacy and safety analyses of a recombinant human immunodeficiency virus type 1 derived vector system. Gene Ther. 6:715-728. [DOI] [PubMed] [Google Scholar]
- 10.Chang, Y. S., C. L. Liao, C. H. Tsao, M. C. Chen, C. I. Liu, L. K. Chen, and Y. L. Lin. 1999. Membrane permeabilization by small hydrophobic nonstructural proteins of Japanese encephalitis virus. J. Virol. 73:6257-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman, R., C. Sidrauski, and P. Walter. 1998. Intracellular signaling from the endoplasmic reticulum to the nucleus. Annu. Rev. Cell Dev. Biol. 14:459-485. [DOI] [PubMed] [Google Scholar]
- 12.Chen, L. K., Y. L. Lin, C. L. Liao, C. G. Lin, Y. L. Huang, C. T. Yeh, S. C. Lai, J. T. Jan, and C. Chin. 1996. Generation and characterization of organ-tropism mutants of Japanese encephalitis virus in vivo and in vitro. Virology 223:79-88. [DOI] [PubMed] [Google Scholar]
- 13.Cox, J. S., C. E. Shamu, and P. Walter. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73:1197-1206. [DOI] [PubMed] [Google Scholar]
- 14.Cox, J. S., and P. Walter. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87:391-404. [DOI] [PubMed] [Google Scholar]
- 15.Cui, Y., T. Iwakuma, and L. J. Chang. 1999. Contributions of viral splice sites and cis-regulatory elements to lentivirus vector function. J. Virol. 73:6171-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng, Y., J. R. Bennink, H. C. Kang, R. P. Haugland, and J. W. Yewdell. 1995. Fluorescent conjugates of brefeldin A selectively stain the endoplasmic reticulum and Golgi complex of living cells. J. Histochem. Cytochem. 43:907-915. [DOI] [PubMed] [Google Scholar]
- 17.Gubler, D. J. 1998. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 11:480-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harding, H. P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18:575-599. [DOI] [PubMed] [Google Scholar]
- 19.Hase, T., P. L. Summers, P. Ray, and E. Asafo-Adjei. 1992. Cytopathology of PC12 cells infected with Japanese encephalitis virus. Virchows Arch. B 63:25-36. [DOI] [PubMed] [Google Scholar]
- 20.Haze, K., H. Yoshida, H. Yanagi, T. Yura, and K. Mori. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol Cell 10:3787-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, B. 2006. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 13:393-403. [DOI] [PubMed] [Google Scholar]
- 22.Huang, Z. M., T. Tan, H. Yoshida, K. Mori, Y. Ma, and T. S. Yen. 2005. Activation of hepatitis B virus S promoter by a cell type-restricted IRE1-dependent pathway induced by endoplasmic reticulum stress. Mol. Cell. Biol. 25:7522-7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwakoshi, N. N., A. H. Lee, and L. H. Glimcher. 2003. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol. Rev. 194:29-38. [DOI] [PubMed] [Google Scholar]
- 24.Iwakoshi, N. N., A. H. Lee, P. Vallabhajosyula, K. L. Otipoby, K. Rajewsky, and L. H. Glimcher. 2003. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat. Immunol. 4:321-329. [DOI] [PubMed] [Google Scholar]
- 25.Iwakuma, T., Y. Cui, and L. J. Chang. 1999. Self-inactivating lentiviral vectors with U3 and U5 modifications. Virology 261:120-132. [DOI] [PubMed] [Google Scholar]
- 26.Iwawaki, T., R. Akai, K. Kohno, and M. Miura. 2004. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat. Med. 10:98-102. [DOI] [PubMed] [Google Scholar]
- 27.Jordan, R., L. Wang, T. M. Graczyk, T. M. Block, and P. R. Romano. 2002. Replication of a cytopathic strain of bovine viral diarrhea virus activates PERK and induces endoplasmic reticulum stress-mediated apoptosis of MDBK cells. J. Virol. 76:9588-9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211-1233. [DOI] [PubMed] [Google Scholar]
- 29.Kozutsumi, Y., M. Segal, K. Normington, M. J. Gething, and J. Sambrook. 1988. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332:462-464. [DOI] [PubMed] [Google Scholar]
- 30.Kurisu, J., A. Honma, H. Miyajima, S. Kondo, M. Okumura, and K. Imaizumi. 2003. MDG1/ERdj4, an ER-resident DnaJ family member, suppresses cell death induced by ER stress. Genes Cells 8:189-202. [DOI] [PubMed] [Google Scholar]
- 31.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 32.Lee, A. H., N. N. Iwakoshi, K. C. Anderson, and L. H. Glimcher. 2003. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc. Natl. Acad. Sci. USA 100:9946-9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, A. H., N. N. Iwakoshi, and L. H. Glimcher. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, K., W. Tirasophon, X. Shen, M. Michalak, R. Prywes, T. Okada, H. Yoshida, K. Mori, and R. J. Kaufman. 2002. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 16:452-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung, D., K. Schroder, H. White, N. X. Fang, M. J. Stoermer, G. Abbenante, J. L. Martin, P. R. Young, and D. P. Fairlie. 2001. Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J. Biol. Chem. 276:45762-45771. [DOI] [PubMed] [Google Scholar]
- 36.Liberman, E., Y. L. Fong, M. J. Selby, Q. L. Choo, L. Cousens, M. Houghton, and T. S. Yen. 1999. Activation of the grp78 and grp94 promoters by hepatitis C virus E2 envelope protein. J. Virol. 73:3718-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, C., S. M. Amberg, T. J. Chambers, and C. M. Rice. 1993. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J. Virol. 67:2327-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, R. J., B. L. Chang, H. P. Yu, C. L. Liao, and Y. L. Lin. 2006. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 80:5908-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, Y. L., C. L. Liao, L. K. Chen, C. T. Yeh, C. I. Liu, S. H. Ma, Y. Y. Huang, Y. L. Huang, C. L. Kao, and C. C. King. 1998. Study of dengue virus infection in SCID mice engrafted with human K562 cells. J. Virol. 72:9729-9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: The viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott-Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 41.Liu, C. Y., M. Schroder, and R. J. Kaufman. 2000. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J. Biol. Chem. 275:24881-24885. [DOI] [PubMed] [Google Scholar]
- 42.Makarova, O., E. Kamberov, and B. Margolis. 2000. Generation of deletion and point mutations with one primer in a single cloning step. BioTechniques 29:970-972. [DOI] [PubMed] [Google Scholar]
- 43.Matusan, A. E., M. J. Pryor, A. D. Davidson, and P. J. Wright. 2001. Mutagenesis of the dengue virus type 2 NS3 protein within and outside helicase motifs: effects on enzyme activity and virus replication. J. Virol. 75:9633-9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mori, K., W. Ma, M. J. Gething, and J. Sambrook. 1993. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74:743-756. [DOI] [PubMed] [Google Scholar]
- 45.Ng, D. T., E. D. Spear, and P. Walter. 2000. The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 150:77-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pahl, H. L. 1999. Signal transduction from the endoplasmic reticulum to the cell nucleus. Physiol. Rev. 79:683-701. [DOI] [PubMed] [Google Scholar]
- 47.Reimold, A. M., A. Etkin, I. Clauss, A. Perkins, D. S. Friend, J. Zhang, H. F. Horton, A. Scott, S. H. Orkin, M. C. Byrne, M. J. Grusby, and L. H. Glimcher. 2000. An essential role in liver development for transcription factor XBP-1. Genes Dev. 14:152-157. [PMC free article] [PubMed] [Google Scholar]
- 48.Rice, C. M. 1996. Flaviviridae: the viruses and their replication, p. 931-959. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology. Lippincott-Raven Publisher, Philadelphia, Pa.
- 49.Robertson, S. E., B. P. Hull, O. Tomori, O. Bele, J. W. LeDuc, and K. Esteves. 1996. Yellow fever: a decade of reemergence. JAMA 276:1157-1162. [PubMed] [Google Scholar]
- 50.Ron, D., and R. Y. Hampton. 2004. Membrane biogenesis and the unfolded protein response. J. Cell Biol. 167:23-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rutkowski, D. T., and R. J. Kaufman. 2004. A trip to the ER: coping with stress. Trends Cell Biol. 14:20-28. [DOI] [PubMed] [Google Scholar]
- 52.Shaffer, A. L., M. Shapiro-Shelef, N. N. Iwakoshi, A. H. Lee, S. B. Qian, H. Zhao, X. Yu, L. Yang, B. K. Tan, A. Rosenwald, E. M. Hurt, E. Petroulakis, N. Sonenberg, J. W. Yewdell, K. Calame, L. H. Glimcher, and L. M. Staudt. 2004. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21:81-93. [DOI] [PubMed] [Google Scholar]
- 53.Shen, X., R. E. Ellis, K. Lee, C. Y. Liu, K. Yang, A. Solomon, H. Yoshida, R. Morimoto, D. M. Kurnit, K. Mori, and R. J. Kaufman. 2001. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107:893-903. [DOI] [PubMed] [Google Scholar]
- 54.Shen, X., K. Zhang, and R. J. Kaufman. 2004. The unfolded protein response—a stress signaling pathway of the endoplasmic reticulum. J Chem. Neuroanat. 28:79-92. [DOI] [PubMed] [Google Scholar]
- 55.Solomon, T. 2004. Flavivirus encephalitis. N. Engl. J. Med. 351:370-378. [DOI] [PubMed] [Google Scholar]
- 56.Solomon, T., R. Kneen, N. M. Dung, V. C. Khanh, T. T. Thuy, D. Q. Ha, N. P. Day, A. Nisalak, D. W. Vaughn, and N. J. White. 1998. Poliomyelitis-like illness due to Japanese encephalitis virus. Lancet 351:1094-1097. [DOI] [PubMed] [Google Scholar]
- 57.Sriburi, R., S. Jackowski, K. Mori, and J. W. Brewer. 2004. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J. Cell Biol. 167:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Su, H. L., C. L. Liao, and Y. L. Lin. 2002. Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J. Virol. 76:4162-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tardif, K. D., K. Mori, R. J. Kaufman, and A. Siddiqui. 2004. Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J. Biol. Chem. 279:17158-17164. [DOI] [PubMed] [Google Scholar]
- 60.Tardif, K. D., K. Mori, and A. Siddiqui. 2002. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J. Virol. 76:7453-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tirasophon, W., A. A. Welihinda, and R. J. Kaufman. 1998. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 12:1812-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomori, O. 1999. Impact of yellow fever on the developing world. Adv. Virus Res. 53:5-34. [DOI] [PubMed] [Google Scholar]
- 63.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249-258. [DOI] [PubMed] [Google Scholar]
- 64.van Huizen, R., J. L. Martindale, M. Gorospe, and N. J. Holbrook. 2003. P58IPK, a novel endoplasmic reticulum stress-inducible protein and potential negative regulator of eIF2α signaling. J. Biol. Chem. 278:15558-15564. [DOI] [PubMed] [Google Scholar]
- 65.Wang, X. Z., H. P. Harding, Y. Zhang, E. M. Jolicoeur, M. Kuroda, and D. Ron. 1998. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 17:5708-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, J., and R. J. Kaufman. 2006. From acute ER stress to physiological roles of the Unfolded Protein Response. Cell Death Differ. 13:374-384. [DOI] [PubMed] [Google Scholar]
- 67.Wu, S. F., C. J. Lee, C. L. Liao, R. A. Dwek, N. Zitzmann, and Y. L. Lin. 2002. Antiviral effects of an iminosugar derivative on flavivirus infections. J. Virol. 76:3596-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoshida, H., T. Matsui, N. Hosokawa, R. J. Kaufman, K. Nagata, and K. Mori. 2003. A time-dependent phase shift in the mammalian unfolded protein response. Dev. Cell 4:265-271. [DOI] [PubMed] [Google Scholar]
- 69.Yoshida, H., T. Matsui, A. Yamamoto, T. Okada, and K. Mori. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881-891. [DOI] [PubMed] [Google Scholar]
- 70.Yoshida, H., M. Oku, M. Suzuki, and K. Mori. 2006. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 172:565-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zolotukhin, S., M. Potter, W. W. Hauswirth, J. Guy, and N. Muzyczka. 1996. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J. Virol. 70:4646-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]