Abstract
The order Nidovirales comprises viruses from the families Coronaviridae (genera Coronavirus and Torovirus), Roniviridae (genus Okavirus), and Arteriviridae (genus Arterivirus). In this study, we characterized White bream virus (WBV), a bacilliform plus-strand RNA virus isolated from fish. Analysis of the nucleotide sequence, organization, and expression of the 26.6-kb genome provided conclusive evidence for a phylogenetic relationship between WBV and nidoviruses. The polycistronic genome of WBV contains five open reading frames (ORFs), called ORF1a, -1b, -2, -3, and -4. In WBV-infected cells, three subgenomic RNAs expressing the structural proteins S, M, and N were identified. The subgenomic RNAs were revealed to share a 42-nucleotide, 5′ leader sequence that is identical to the 5′-terminal genome sequence. The data suggest that a conserved nonanucleotide sequence, CA(G/A)CACUAC, located downstream of the leader and upstream of the structural protein genes acts as the core transcription-regulating sequence element in WBV. Like other nidoviruses with large genomes (>26 kb), WBV encodes in its ORF1b an extensive set of enzymes, including putative polymerase, helicase, ribose methyltransferase, exoribonuclease, and endoribonuclease activities. ORF1a encodes several membrane domains, a putative ADP-ribose 1"-phosphatase, and a chymotrypsin-like serine protease whose activity was established in this study. Comparative sequence analysis revealed that WBV represents a separate cluster of nidoviruses that significantly diverged from toroviruses and, even more, from coronaviruses, roniviruses, and arteriviruses. The study adds to the amazing diversity of nidoviruses and appeals for a more extensive characterization of nonmammalian nidoviruses to better understand the evolution of these largest known RNA viruses.
The order Nidovirales currently comprises three families, Coronaviridae (genera Coronavirus and Torovirus), Roniviridae (genus Okavirus), and Arteriviridae (genus Arterivirus) (52). The vast majority of nidovirus genome sequences have been reported for the genera Coronavirus and Arterivirus, whereas sequence information for toro- and okaviruses is extremely limited (12, 18, 52). Based on antigenic and genetic criteria, coronaviruses have been further subdivided into three major groups, whereas arteriviruses form as many as four (comparably distant) genetic clusters (24). Although there are huge differences in genome size, ranging from 12.7 to 31.3 kilobases, between “small” (Arteriviridae) and “large” nidoviruses (Coronaviridae and Roniviridae) and although there is no apparent relationship between the various nidovirus families in terms of virion morphology and structural proteins, the common ancestry of the three virus families has been firmly established (9, 12, 16, 22, 24). Essentially, it is based on a conserved array of functional domains in the viral replicative polyproteins as well as common transcriptional and (post)translational strategies used in viral genome expression. Thus, for example, all nidoviruses produce a 3′-coterminal (“nested”) set of subgenome-length (sg) RNAs, whose number may vary between two and nine in different nidoviruses. In their 5′-terminal genome regions, all nidoviruses possess two large open reading frames (ORFs), ORF1a and ORF1b, which together form the viral replicase gene. The downstream ORF1b is expressed by −1 ribosomal frameshifting, occurring just upstream of the ORF1a stop codon (7). The products specified by the nidovirus replicase gene are called polyprotein 1a (pp1a) (encoded by ORF1a) and pp1ab (encoded by ORFs 1a and 1b) (74, 75). ORF1b is the most conserved part of the nidovirus genome and encodes, among other domains, the principal replicative enzymes of the virus (polymerase and helicase) and two nidovirus-specific domains (a multinuclear zinc-binding domain [ZBD] and the nidoviral uridylate-specific endoribonuclease [NendoU]) (22, 30, 51, 54, 71). The conserved order of domains in the nidovirus replicase polyproteins can be described as follows (from the N to the C terminus): transmembrane domain (TM), chymotrypsin-like (3C-like) main protease (3CLpro), TM, ribosomal frameshift element, RNA-dependent RNA polymerase (RdRp) (11), ZBD, helicase, and NendoU (6, 30). Large nidoviruses (that is, members of the Coronaviridae and Roniviridae), featuring genomes of more than 26 kb, also encode 3′-to-5′ exoribonuclease (34) and ribose-2′-O-methyltransferase (54, 66) domains which reside in the C-terminal part of pp1ab. Furthermore, nidoviruses may encode additional enzymatic functions, including papain-like proteases, macro domain-related proteins with ADP-ribose/poly(ADP-ribose)-binding and/or ADP-ribose 1"-phosphatase (ADRP) activities, and cyclic nucleotide phosphodiesterase activities (25, 40, 43, 54). The individual subunits of the nidovirus replicase machinery are released from pp1a/pp1ab by autoproteolytic processing, involving viral proteases that themselves are part of the polyproteins (75).
Recently, significant progress was made in the functional and structural characterization of the nidovirus replication/transcription complex, with Severe acute respiratory syndrome coronavirus (SARS-CoV) being the most extensively studied nidovirus to date (1, 2, 4, 19, 30, 31, 34, 42, 43, 48, 57, 58, 68-70). It is now generally accepted that the enzymology involved in nidovirus replication is significantly more complex than that of other plus-strand RNA viruses. Most probably, these additional enzymes are required to replicate the exceptionally large genomes of nidoviruses and synthesize the nested set of sg RNAs from which the structural and, in some cases, several accessory proteins are expressed (37, 47). Previous studies identified conserved proteins and mechanisms mediating the replication cycle of viruses from the various nidovirus genera, but they also revealed interesting differences. Thus, for example, most, but not all, nidovirus sg RNAs contain a 5′ leader sequence derived from the 5′ end of the genomic RNA (13, 17, 56, 65). Furthermore, the number of replicase gene-encoded enzymes varies between different nidovirus genera or even between different groups of the same genus (23, 54), and also, the functional and structural properties of several key replicative proteins, including the main proteases of the various genera, are remarkably diverse (1, 3, 53, 72). Taken together, the studies have made it clear that more information is needed to understand the biological meaning of the differential conservation of specific proteins and/or enzymatic activities among nidoviruses (and other plus-strand RNA virus families). In this context, additional sequence information, particularly from nidoviruses prototyping previously unknown and distantly related genera, can be expected to provide new insights into the evolution of the Nidovirales and, potentially, even provide clues for a better understanding of the mechanisms and driving forces that have governed the separation of the Nidovirales from the bulk of plus-strand RNA viruses featuring smaller genomes and less-complex genome replication and expression strategies.
Here, we report the complete genome sequence of White bream virus (WBV), a novel plus-strand RNA virus isolated from fish (Blicca bjoerkna L.) (27). The sequence, organization, and expression of the 26.6-kb genome qualify WBV as a new member of the order Nidovirales. In common with other nidoviruses, the WBV genome contains two large ORFs (1a and 1b) in the 5′ region of the genome, with ORF1b probably being expressed by −1 ribosomal frameshifting, occurring at a putative UUUAAAC heptanucleotide “slippery” sequence located upstream of the ORF1a stop codon and a putative RNA pseudoknot structure. WBV is predicted to encode three structural proteins, spike (S) protein, membrane (M) protein, and nucleocapsid (N) protein, which are expressed from 5′ leader-containing sg RNAs as shown by Northern blotting and sequence analysis. Upstream of each of the S, M, and N ORFs, a conserved nonanucleotide sequence, CA(G/A)CACUAC, was identified which is also present near the 5′ end of the genome. By analogy with other nidoviruses, we predict this sequence to be the conserved core of the transcription-regulating sequence (TRS) elements that facilitate the template switch required to attach the complement of the 5′ leader sequence to the 3′ ends of subgenome-length minus strands, thereby producing the templates for the synthesis of 5′ leader-containing plus-strand RNAs. The phylogenetic relationship between WBV and nidoviruses is further corroborated by the presence of a conserved array of putative functional domains in pp1a, including an ADRP domain, three TMs, and the presumed viral main protease, 3CLpro, whose activity was established in this study. Furthermore, an RNA-dependent RNA polymerase domain featuring the nidovirus-specific SDD signature, as well as ZBD, helicase, exoribonuclease, NendoU, and ribose-2′-O-methyltransferase domains, were identified in the C-terminal part of pp1ab. Sequence comparisons and phylogenetic studies lead us to conclude that toroviruses (followed by coronaviruses) are the closest relatives of WBV. In terms of classification, we think the phylogenetic position of WBV would be best reflected if the virus was assigned to a yet-to-be-established new genus.
MATERIALS AND METHODS
Cells and virus.
WBV (isolate DF24/00) was propagated on epithelioma papulosum cyprini (EPC) cells at 20°C in a 2.5% CO2 atmosphere as described previously (27). EPC cells (RIE 173) were obtained from the cell line collection of the Friedrich-Loeffler-Institut, Federal Research Institute for Animal Health (Insel Riems, Germany).
Virus purification and RNA isolation.
WBV was harvested by two cycles of freezing and thawing of infected cells. Cell debris was removed by centrifugation at 4,000 rpm (SW28 rotor; Beckman) for 10 min, and the supernatant was layered onto a 15% sucrose cushion and centrifuged at 20,000 rpm for 90 min. The resulting virus pellet was resuspended in STE buffer (0.15 M NaCl, 0.01 M Tris-HCl, pH 8.3, 0.01 M EDTA, pH 8.0) and applied on a continuous gradient of 5 to 50% sucrose in STE. After centrifugation at 20,000 rpm (SW28 rotor; Beckman) for 30 min, the virus-containing band was isolated and dialyzed against STE. The virus was concentrated by sedimentation at 40,000 rpm (SW60; Beckman) for 4 h, and viral RNA was extracted using guanidinium thiocyanate, followed by centrifugation through a 5.7 M cesium chloride-EDTA cushion according to standard protocols (44).
RNA transfection.
EPC cells (5 × 105) were transfected with purified WBV genome RNA. To do this, 3 μg RNA was diluted in 50 μl diethyl pyrocarbonate-treated water containing 60 U RNasin (Promega) and mixed with 200 μl of serum-free OptiMEM (Invitrogen) containing 12.5 μl of Lipofectamine transfection reagent (Invitrogen). After incubation on ice for 5 min, the transfection mix was layered onto nearly confluent EPC cells maintained in OptiMEM. After incubation for 2 hours at 20°C, the medium was replaced by Eagle's minimal essential medium supplemented with 10% fetal bovine serum. Transfected cells were incubated for 6 days at 20°C in a 2.5% CO2 atmosphere.
Electron microscopy.
For negative staining, infected cell cultures were scraped off from the cell culture dish, pelleted by low-speed centrifugation, and resuspended in phosphate-buffered saline. Formvar-coated grids were placed for 7 min on drops of cell culture supernatant or resuspended pellet. Negative staining was performed with 2% phosphotungstic acid (pH 7.4) for 7 min. Stained grids were examined with a Philips electron microscope 400T (Eindhoven, The Netherlands).
Cloning and sequence analysis of the WBV genome.
Purified WBV genome RNA was used as a template for the construction of cDNA libraries. Viral RNA (0.5 to 1 μg) was incubated with 0.5 μg of oligo(dT)12-18 primers or 0.037 μg of random pd(N)6 hexadeoxynucleotides (TimeSaver cDNA synthesis kit; Amersham Biosciences). In the course of the study, additional cDNA libraries were generated by “primer walking,” using WBV-specific oligonucleotides (DF14, DF20, DF16, DF29, and DF5) and previously described methods (49; data not shown). To further corroborate the WBV genome sequence analysis, a series of reverse transcription (RT)-PCRs were performed to amplify overlapping fragments covering the entire genome RNA (see Fig. 2). The amplicons obtained were cloned using pGEM-T Easy vector system II (Promega) according to the manufacturer's protocols. RT-PCR 17 (see Fig. 2) was performed using the one-step RT-PCR (QIAGEN) and SuperScript III one-step RT-PCR (Invitrogen) systems. For RT-PCR 28, poly(A)-containing RNA from WBV-infected EPC cells was isolated using previously described methods (60). The poly(A) RNA was reverse transcribed using Transcriptor reverse transcriptase (Roche) and oligonucleotide JZ562 (55°C, 60 min). PCR amplification was done using the TripleMaster PCR system (Eppendorf) and oligonucleotides JZ561 and JZ564. The WBV 5′ and 3′ genome termini were determined by rapid amplification of cDNA ends (RACE) techniques as described by Hoffmann et al. (29). The sequences of the oligonucleotide primers used for cDNA synthesis and RT-PCR are available upon request. Plasmid DNAs containing cDNA and RT-PCR inserts (see Fig. 2) were sequenced using standard procedures. At least three independently derived clones were analyzed on both strands to ascertain the obtained sequence. The product from RT-PCR 28 was sequenced directly. To determine the 5′ and 3′ genome ends, at least six clones from independent RACE reactions were sequenced. Analysis of sequencing data was done using Lasergene biocomputing software (DNAStar). Multiple-sequence alignments were generated using ClustalW v1.83 (http://www.ebi.ac.uk/clustalw/) (63) and ClustalX v1.8 (62) and used as input for the ESPript (version 2.2) program (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). Transmembrane domains were predicted using the hidden Markov model-based program TMHMM v.2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) (32), and signal peptides were identified using SignalP v3.0 (http://www.cbs.dtu.dk/services/SignalP/) (5). Phylogenetic trees were generated from multiple-sequence alignments by the neighbor-joining method in ClustalX v1.8 or MEGA 3.1 (http://www.megasoftware.net), and bootstrap values were determined by 1,000 replicates. Phylogenetic trees were plotted with TreeView v1.6.6 (36) and manually edited.
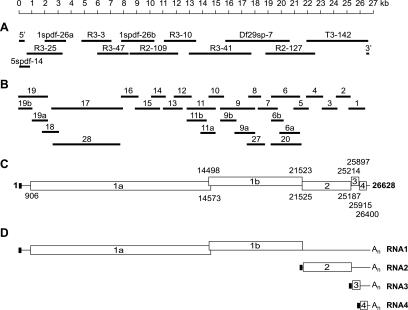
FIG. 2.
Structural organization and sequence analysis of the WBV (strain DF24/00) genome. (A) Given are the sizes and positions of cDNA clones from WBV genomic libraries that were used to determine the WBV genome sequence. Also shown are the 5′- and 3′-terminal amplicons generated by RACE. (B) Given are the sizes and positions of RT-PCR products used to ascertain the sequence derived from the cDNA clones shown in panel A. (C) Predicted functional ORFs in the WBV genome. Numbers indicate the 5′ and 3′ nucleotides, respectively, of predicted translation start and stop codons. Note that translation of ORF1b is predicted to involve a −1 ribosomal frameshift occurring just upstream of the ORF1a translation stop codon (and downstream of the most 5′-terminal AUG codon that is used here to indicate the ORF1b 5′ end) (for further details, see the text and Table 1). (D) WBV-specific RNAs as determined in this study. The little black box at the 5′ end of the genome indicates the 42-nt leader sequence, which is also present at the 5′ ends of the three subgenome-length RNAs (Fig. 3 and 4). The available evidence from other nidoviruses (37, 47) suggests that attachment of the leader sequence to the coding (body) sequences of WBV subgenome-length RNAs is due to discontinuous extension of subgenome-length minus-strand RNAs. In this process, nascent minus strands switch their template at TRSs located upstream of the S, M, and N genes and bind then to an identical sequence called leader TRS near the 5′ end of the genome, after which the leader sequence is copied to complete negative-strand synthesis (see the text and Fig. 4).
Analysis of WBV RNA synthesis in infected cells.
EPC cells (5 × 105) were infected with WBV at a multiplicity of infection of 0.01. Two days after infection, intracellular poly(A)-containing RNA from mock-infected and WBV-infected EPC cells was prepared using oligo(dT)25 Dynabeads (Dynal) as described by Thiel et al. (61). RNA was separated on a 2.2 M formaldehyde-1.3% agarose gel, blotted on a nylon membrane, and hybridized with a α-32P-multiprime-labeled DNA probe specific for WBV nts 25992 to 26582. RNAs were analyzed by autoradiography. Poly(A)-containing RNAs from human coronavirus 229E (HCoV-229E)-infected cells (60) and Rep-1 RNA (28) were used as RNA size markers in this Northern blotting experiment. pRep-1 RNA (24.4 kb) was produced by in vitro transcription as described previously (28). HCoV-229E-specific RNAs and pRep-1 RNA were visualized by including in the hybridization buffer a second α-32P-multiprime-labeled DNA probe specific for the HCoV-229E nts 26857 to 27277 (34). To determine the leader-to-body fusion sites of WBV subgenomic RNAs, reverse transcription of poly(A)-containing RNA from WBV-infected cells was primed using oligonucleotides JZ590_RT-N (5′-26628GGTAATTTTATATCATACATGGATAGTCAT26599-3′), JZ618_RT-M (5′-25419CCGACCATTGGTAATCTACCAACG25396-3′), and JZ620_RT-S (5′-21940CAGTATTGTTCGGCTGTTGTTAAC21917-3′). PCR amplification was done using the WBV 5′-end (leader)-specific oligonucleotide JZ598 (5′-1GAGATTATTACGAATTTCGTTTTAACACAC30-3′) in combination with body-specific oligonucleotides JZ593-N (5′-26582AGAAACGACATACACGCGAAATTG26559-3′), JZ619-M (5′-25390AATGTAGTAGACCAGAGTATTCAC25357-3′), and JZ621-S (5′-21790TTGTCATAGTTGAGTGTGTACGAC21767-3′). PCR products were sequenced directly using primers JZ593-N (amplicon JZ598/JZ593-N), JZ619-M (amplicon JZ598/JZ619-M), and JZ621-S (amplicon JZ598/JZ621-S). In addition, the PCR products were cloned into the unique EcoRV restriction site of pBluescript II KS(+) plasmid DNA (Stratagene). Leader-body junctions of the three subgenomic RNAs were then further analyzed by sequencing the relevant regions of individual plasmid clones, using T3 and T7 promoter-specific oligonucleotides.
Cloning, expression, and activity of the WBV 3C-like protease domain.
The predicted coding sequence of WBV 3CLpro, together with short flanking sequences, was amplified by RT-PCR using poly(A)-containing RNA isolated from WBV-infected EPC cells as a template and oligonucleotides JZ559 (5′-TCAGCATCAGAATGCATTCTGTAT-3′) and JZ560 (5′-AAAGAATTCTTATTGCATGTTGTTAACTGGTGATGTG-3′). The 961-bp PCR product was treated with T4 DNA polymerase, phosphorylated with T4 polynucleotide kinase, cleaved with EcoRI, and inserted into XmnI/EcoRI-digested pMal-c2 plasmid DNA (New England Biolabs). The resulting plasmid, pMal-WBV-3CL_559-560, encoded the WBV pp1a/pp1ab residues Ser3424 to Gln3726 fused to the Escherichia coli maltose-binding protein (MBP). As a negative control, a mutant derivative, pMal-WBV-3CL_S3589A, was generated by site-directed mutagenesis of pMal-WBV-3CL_559-560, using PCR-based methods (73). E. coli TB1 cells transformed with pMal-WBV_559-560 and pMal-WBV-3CL_S3589A were grown at 37°C in Luria-Bertani (LB) medium containing 100 μg of ampicillin per ml until they reached a culture density (A595) of 0.6. Expression of the recombinant proteins was induced by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 24°C. For analysis of recombinant protein expression, aliquots of the cell cultures were suspended in 2× Laemmli sample buffer and incubated at 94°C for 3 min, and the lysates were analyzed by electrophoresis in sodium dodecyl sulfate-polyacrylamide gels as described previously (72).
RESULTS AND DISCUSSION
WBV genome RNA is infectious.
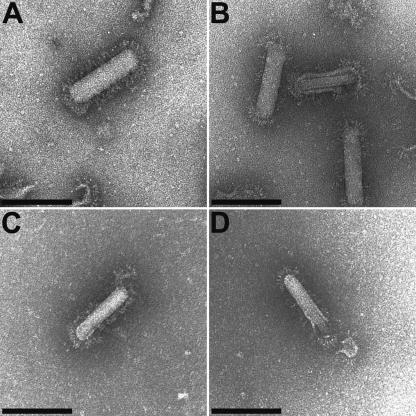
In a previous study, WBV was shown to be an enveloped RNA virus featuring a bacilliform shape and carrying coronavirus-like projections on its surface (27; Fig. 1). To further characterize the genome of this virus, we isolated viral RNA from purified WBV particles, transfected this RNA into EPC cells, and examined by electron microscopy whether virus particles were released from those cells. As Fig. 1 shows, rod-shaped virus particles identical to those produced in WBV-infected cells could be identified in tissue culture supernatants obtained from cells transfected with purified WBV genome RNA (Fig. 1C and D) but not in supernatants from mock-transfected cells (data not shown). The data provide conclusive evidence that, in the absence of viral proteins, WBV genome RNA is able to trigger a full replication cycle, implying that the genome RNA of WBV is infectious and, therefore, must be of mRNA (that is, positive) polarity.
FIG. 1.
Purified WBV genome RNA is infectious: evidence for viral particle formation and release. The cell culture supernatant from EPC cells transfected with purified WBV genome RNA was analyzed at 6 days posttransfection by electron microscopy (negative staining). (C and D). For comparison, electron micrographs taken from purified WBV virions (27) are shown in panels A and B. Representative pictures of both intact (A and C) and partially opened (B and D) virions are shown. Bar, 150 nm.
Genome sequence analysis reveals that WBV is a nidovirus.
Except for a 5′-proximal fragment (nts 3176 to 7881), which could not be stably propagated in E. coli, the complete sequence of the WBV genome was determined from a collection of cDNA clones derived from reverse-transcribed WBV genome RNA (see Materials and Methods) (Fig. 2, and data not shown). The genome ends were determined by using RACE methods, and the completeness and correctness of the obtained sequence were further corroborated by sequence analysis of a set of overlapping RT-PCR products covering the entire genome, including the 5′-proximal fragment. The WBV genome sequence was revealed to encompass 26,628 nucleotides [excluding the 3′ poly(A) tail] and has been deposited in the GenBank database (accession number DQ898157). It contains five major ORFs, ORF1a, -1b, -2, -3, and -4 (Fig. 2; Table 1), which are flanked by 5′- and 3′-terminal untranslated regions of 905 and 228 nucleotides, respectively. The nucleotide sequence and organization of the genome unambiguously identified WBV as a nidovirus. A more detailed sequence analysis showed that WBV is only distantly related to other nidoviruses and that the virus could not readily be assigned to any of the established nidovirus taxa. However, on the basis of its genome size and replicase domain structure, WBV is clearly a member of the large nidoviruses (Coronaviridae and Roniviridae) (24). Furthermore, in database searches for related sequences, a special relationship between WBV and toro- and coronaviruses became immediately evident when WBV ORF1b sequences were used as query sequences. A more detailed comparison between the replicase genes of WBV and other nidoviruses will be given below.
TABLE 1.
Predicted proteins expressed from WBV genomic and subgenomic RNAsa
| ORF | Nucleotides in the genome | Predicted protein size (amino acid residues) | mRNA predicted to be used for expression | Protein name |
|---|---|---|---|---|
| 1a | 906-14573 | 4,555 | 1 (genome) | Polyprotein 1a |
| 1b | 14498-21523 | 6,872 | 1 (genome) | Polyprotein 1abb |
| 2 | 21525-25187 | 1,220 | 2 | Spike protein |
| 3 | 25214-25897 | 227 | 3 | Membrane protein |
| 4 | 25915-26400 | 161 | 4 | Nucleocapsid protein |
By analogy with other nidoviruses, the ORF1b sequence is predicted to be expressed by (−1) ribosomal frameshifting, occurring just upstream of the ORF1a translation stop codon. Accordingly, polyprotein 1ab is encoded by two ORFs, 1a and 1b, and shares its N-terminal part with the ORF1a-encoded pp1a.
Note that ORF1b encodes only the C-terminal third of this protein, whereas its N-terminal two-thirds are encoded by ORF1a.
Putative structural proteins of WBV.
Database searches and comparative sequence analysis revealed a remote sequence similarity between the WBV ORF2 gene product and the S proteins of corona- and toroviruses (data not shown), leading us to propose that WBV ORF2 encodes the viral S protein (1,220 residues), which probably mediates receptor binding and fusion between viral and cellular membranes. Further studies (data not shown) suggest that the WBV S protein, like its homologs in corona- and toroviruses (14, 52), is a type I membrane glycoprotein, featuring (i) an N-terminal signal peptide (with a predicted cleavage site between residues Ala15 and Gln16), (ii) a putative C-terminal transmembrane anchor, and (iii) a short cytoplasmic tail.
Computer-aided sequence analysis further suggested that the WBV ORF3 gene product is a 227-residue, triple-membrane-spanning glycoprotein. Both the size of this protein and the predicted topology and length of the transmembrane regions (data not shown) link this protein to the triple-membrane-spanning M proteins of corona- and toroviruses (14, 52), even though at the primary structure level, the relationship is weak and the generation of reliable sequence alignments proved to be impossible. However, this distant relationship is not really surprising as the coronavirus and torovirus S and M proteins are only poorly conserved, with similarities being restricted mainly to general features, such as the numbers and positions of transmembrane domains, protease cleavage sites, and the general domain organization of these proteins (52).
Based on its 3′-terminal position, which in corona- and toroviruses is generally occupied by the N gene, and the quite reliable functional assignments for all of the other WBV gene products, it seemed reasonable to suggest that ORF4 specifies the WBV N protein (161 residues). The size of this protein corresponds well to that of the torovirus (but not coronavirus) N proteins, and also, some of the most conserved sequence signatures of torovirus N proteins appear to be partly conserved in the putative WBV N protein (data not shown). However, to unambiguously establish a structural relationship between the WBV and torovirus N proteins, further evidence has to be obtained. In support of this potential relationship, we note that the intracellular nucleocapsids formed by WBV and toroviruses share a rod-like structure (55) and also that the straight or slightly bent (kidney-like) rod-like morphology described for some (but not all) torovirus particles resembles the rod-like structure of WBV particles remarkably well (Fig. 1) (27). Taken together, the analysis suggests that WBV encodes three structural proteins, S, M, and N. The virus does not encode a homolog of the coronavirus E protein or a hemagglutin in esterase protein, the latter being conserved in toroviruses and several coronaviruses (52).
WBV-specific RNAs.
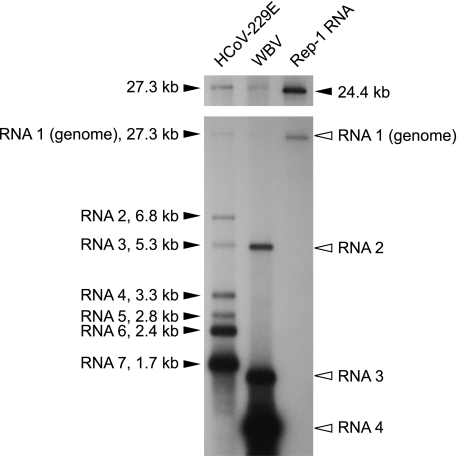
All previously characterized nidoviruses produce a 3′-coterminal nested set of sg RNAs to express their structural and, in some cases, several accessory proteins. Therefore, the identification of three putative structural protein genes in the 3′-terminal region of the genome led us to predict that WBV produces sg RNAs to express these downstream ORFs. To confirm this hypothesis, we isolated poly(A)-containing RNA from WBV-infected cells and used a 3′ terminus-specific probe to detect WBV-specific genome-length and subgenome-length RNAs. The data we obtained in Northern blotting experiments (Fig. 3 and data not shown) demonstrate that WBV produces four RNAs (genome RNA and three sg RNAs) to express its genome. To determine the approximate sizes of the WBV-specific RNAs, we used two RNA markers, namely, (i) HCoV-229E genomic and sg RNAs and (ii) a 24.4-kb HCoV-229E-derived replicon RNA called Rep-1 (28). These RNA markers were detected by including an HCoV-229E-specific probe in the hybridization buffer. The observed sizes of the three sg RNAs strongly support the idea that RNAs 2, 3, and 4 are used to express the viral S, M, and N proteins, respectively. The size of RNA 1 was confirmed to be between 27.3 kb (HCoV-229E genome) and 24.4 kb (Rep-1 RNA), providing additional evidence for a WBV genome size of ∼26.6 kilobases which we had determined by genome sequence analysis.
FIG. 3.
Detection of WBV genome- and subgenome-length RNAs in virus-infected cells. Northern blot analysis of poly(A)-containing RNA isolated from WBV-infected EPC cells (lane 2). Poly(A) RNAs isolated from HCoV-229E-infected MRC-5 cells (lane 1) and HCoV-229E-derived replicon RNA Rep-1 (lane 3) (28) were used as RNA size markers in this experiment. To detect both the HCoV-229E- and WBV-specific RNAs, a mixture of α-32P-multiprime-labeled probes specific for the 3′-terminal regions of HCoV-229E (nucleotides 26857 to 27277) and WBV (nucleotides 25992 to 26582) was used for hybridization. HCoV-229E genome- and subgenome-length RNAs and the in vitro-transcribed HCoV-229E Rep-1 RNA are indicated by black arrowheads, with sizes given in kilobases. White arrowheads indicate the four WBV-specific RNAs detected in this experiment. The longer exposure presented above shows the size of the WBV genomic RNA more clearly and allows its size to be compared with those of the 27.3- and 24.4-kb marker RNAs. The calculated sizes (Table 1 and Fig. 4) of the sg RNAs are 5,162 nts (RNA 2), 1,475 nts (RNA 3), and 774 nts (RNA 4) [including the 5′ leader but excluding the poly(A) tail].
As mentioned above, coronaviruses and arteriviruses produce a large number of sg RNAs that share a 5′ leader sequence identical to that of the 5′ end of the genome. The templates used for plus-strand sg RNA synthesis are sg minus strands carrying the complement of this leader sequence (antileader) at their 3′ ends. Attachment of this antileader requires a discontinuous step in minus-strand RNA synthesis (37, 47) and involves TRSs which are located upstream of each of the 3′ structural and accessory protein genes (body TRSs) as well as downstream of the 5′ leader sequence (leader TRS). Specific base-pairing interactions between the body TRS complements and the leader TRS have been shown for corona- and arteriviruses to guide the strand transfer of the nascent minus strand to the 5′ end of the genome, where minus-strand synthesis is completed by copying the leader sequence (38, 76). In contrast to corona- and arteriviruses, Equine torovirus (strain Berne) produces only one leader-containing sg RNA (65) and the two ronivirus sg RNAs lack a 5′ leader sequence altogether (13). In view of these differences among the various nidovirus genera, we were interested in examining whether the three sg RNAs of WBV contained 5′ leader sequences. Like researchers of a previous study (60), we designed three sets of primers to amplify by RT-PCR the (potentially existing) junctions between 5′ leader and 3′ body sequences for the three WBV-specific sg RNAs (see Materials and Methods). The upstream primer was specific for the 5′ end of the genome, and the downstream primers were specific for one of the ORFs, 2, 3, or 4. In each of the three reactions, we obtained specific amplicons whose sizes were consistent with the presence of a short 5′ leader sequence (data not shown). Direct sequence analysis of the amplicons confirmed this conclusion and revealed the precise positions of the leader-body fusion sites (Fig. 4). Double peaks identified in two of the chromatograms suggested that two alternative fusion sites were used in the case of WBV RNAs 2 and 4. To address this possibility more rigorously, we cloned the RT-PCR products into pBluescript plasmid DNA and determined the sequences of individual cDNA clones. The data from this sequence analysis (summarized in Fig. 4) lead us to suggest the following: (i) WBV RNAs 2, 3, and 4 contain a 5′ leader sequence of generally 42 nucleotides whose sequence corresponds to that of the 5′ end of the WBV genome; (ii) in RNAs 2 and 4 (but not RNA 3), alternative leader-body fusion sites located three bases (RNA 2) and two bases (RNA 4) upstream of the major fusion site were occasionally used (Fig. 4); (iii) upstream of each of the ORFs, 2, 3, and 4, and downstream of the leader, a nonanucleotide sequence, CA(G/A)CACUAC, is conserved, which, like in corona- and arteriviruses, might represent the core element of WBV TRSs. This putative core TRS could promote base pairing of as many as nine consecutive bases, thereby probably creating a very stable RNA structure. The actual fusion of leader and body sequences appears to occur outside of the base-paired region, namely, one (in a few cases, three or four) nucleotide(s) upstream of the core TRS element (plus-strand numbering). Similar observations have previously been made for several minor sg RNA species of arteriviruses (10, 15, 33). The WBV leader-body fusion site data provide additional and very strong support for the sg RNA synthesis model originally introduced for coronaviruses by Sawicki and Sawicki (45, 46) and later extended to arteriviruses by van Marle and colleagues (64). Specifically, the data argue against a “free leader”-priming model wherein a free leader of a size greater than is found on the sg mRNA is annealed, trimmed with an exonuclease back to the fusion site, and extended.
FIG. 4.
Leader-body junctions in WBV subgenomic RNAs. Shown are the junction sites between a short sequence, called leader, that is derived from the 5′ end of the genome, and the coding (so-called body) sequences of subgenomic RNAs 2 to 4. The gray box highlights a sequence, CA(G/A)CACUAC, and its negative-strand complement that we predict to act as a core TRS element in WBV. As in corona- and arteriviruses (37, 47), this leader sequence is conserved near the 5′ end of the genome (nts 44 to 52) and upstream of the translation start codon of each of the downstream ORFs specifying the viral structural proteins, S, M, and N. For each of the core TRS elements, the flanking sequences in the WBV genome RNA and the corresponding minus-strand sequence are given. Possible base-pairing interactions between the minus strand and the proposed leader TRS are indicated, and the leader-body (L-B) junction in the respective mRNA, as determined by RT-PCR and sequencing, is given below. Sequences derived from the 5′ end of the genome (leader sequence) are boldfaced and underlined. Translation start codons of the S (RNA 2), M (RNA 3), and N (RNA 4) genes are boldfaced and italicized. Please note that, with respect to the minus-strand sequence, the actual fusion appears to occur slightly downstream of the fully complementary sequence rather than within this particular sequence.
The data further suggest that, after its relocation to the leader TRS, the nascent minus-strand RNA is extended by forming a phosphodiester bond with a mismatched 5′ nucleotide, which, in most cases, is a uridylate. We also note that, in all leader-body fusion events, the first nucleotide to be incorporated by the minus-strand polymerase after the template switch is a guanylate. It remains to be seen whether this reflects specific requirements for initiation and/or reinitiation of RNA synthesis by the WBV RdRp. Notably, the 3′-terminal residue of the genome [preceding the poly(A) tail] is a cytidine, implying that minus-strand RNA synthesis (also) starts by the incorporation of a guanylate.
Taken together, these data confirm and extend the previously established models for coronavirus and arterivirus sg RNA transcription (37, 47). At the same time, they make it clear that there is no simple relationship between the number of sg RNAs produced by specific groups of nidoviruses and the presence of a 5′ leader.
Identification of a putative ribosomal frameshifting element at the ORF1a/1b junction.
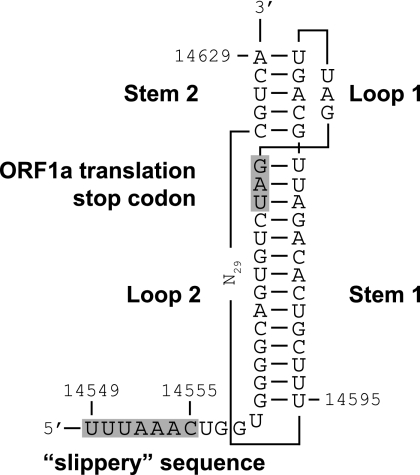
We identified a putative slippery sequence, 14549UUUAAAC14555, just upstream of the WBV ORF1a translation stop codon, and the sequence downstream of the slippery sequence could be modeled into an RNA pseudoknot structure (Fig. 5). Over the past years, coronavirus RNA pseudoknot structures have been studied extensively, both structurally and functionally, and their critical role in mediating a shift into the −1 reading frame during translation has been firmly established (8, 35). Although our study does not provide formal evidence for that, it seems reasonable to predict that, as in other nidoviruses, WBV ORF1b expression (generating the viral RdRp and other key replicative proteins) is regulated at the translational level by ribosomal frameshifting.
FIG. 5.
Model of the WBV ribosomal frameshifting element. By analogy with other nidoviruses, the element is proposed to consist of a putative RNA pseudoknot structure (comprised of two stems and two loops) and a slippery sequence (14549UUUAAAC15555) at which the actual frameshift is predicted to occur. The sequences boxed in gray indicate the predicted slippery sequence and the ORF1a translation termination codon.
Identification of a WBV ORF1a-encoded serine protease activity.
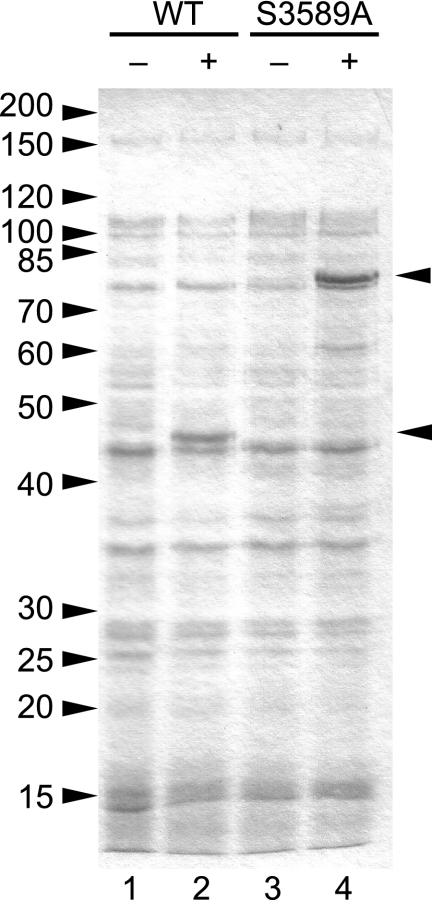
The central and C-terminal portions of nidovirus replicase polyproteins are extensively processed by ORF1a-encoded 3C-like “main” proteases (75). Within the replicase polyprotein, nidovirus 3C-like proteases are generally flanked by membrane-spanning domains. Our sequence analysis of WBV ORF1a identified a putative 3CLpro domain in the C-terminal third of pp1a. The putative WBV 3CLpro domain contained the conserved GX(S/C)G signature of chymotrypsin-like proteases and was found to be flanked by hydrophobic domains at its N- and C-terminal borders. To confirm the proteolytic activity of this domain, we expressed in Escherichia coli the WBV pp1a/pp1ab residues Ser3424 to Gln3726 fused to MBP. The predicted size of the MBP-3CLpro fusion protein was 74.9 kDa. In lysates obtained from IPTG-induced cells transformed with the appropriate expression plasmid (pMal-WBV-3CL_559-560), we observed a protein of approximately 46 kDa which was not detected in lysates obtained from noninduced cells (Fig. 6, lanes 1 and 2). The low molecular mass of the overexpressed protein was consistent with the predicted autoprocessing activity of the protein. However, we failed to obtain clear evidence for the presence of a second processing product, which might be explained by potentially comigrating E. coli proteins or the instability of the protein following autoproteolytic cleavage. Western blot analysis using an MBP-specific antiserum revealed that the 46-kDa protein was the N-terminal processing product (data not shown). MBP itself has a molecular mass of 42.4 kDa, leading us to suggest that the cleavage site was located within the N-terminal 20 to 30 residues of the expressed WBV sequence, which was consistent with our tentative delineation of the WBV 3CLpro domain. To further corroborate this interpretation of the data and to exclude that the observed cleavage resulted from an E. coli protease activity, we expressed a mutant version of MBP-3CLpro in which the presumed active-site nucleophile, Ser3589, was substituted with Ala. In this case, a protein of about 75 kDa was found to be overexpressed (Fig. 6, lanes 3 and 4), which was in agreement with the calculated mass of the MBP-3CLpro fusion protein. Taken together, the data strongly suggest that WBV encodes a 3C-like serine protease activity which, by analogy to other nidoviruses, is predicted to play a key role in the proteolytic processing of the WBV replicase polyproteins. The approximate positions of a limited number of cleavage sites can be inferred from the domain borders of the conserved ORF1b-encoded enzymes. However, we failed to identify strictly conserved sequence signatures at these interdomain junctions, suggesting that the substrate specificity of the WBV 3CLpro domain may be less well conserved than those of its coronavirus and torovirus counterparts. In order to make reliable predictions on potential 3CLpro cleavage sites, a minimum of biochemical information on the WBV 3CLpro substrate specificity needs to be obtained.
FIG. 6.
Proteolytic activity of WBV pp1a/pp1ab amino acid residues Ser3424 toGln3726. Total cell lysates from E. coli TB1 cells transformed with pMal-WBV-3CL_559-560 (WT) (lanes 1 and 2) and pMal-WBV-3CL_S3589A (S3589A) (lanes 3 and 4) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in a 12.5% polyacrylamide gel and stained with Coomassie brilliant blue R-250. The bacteria were mock induced (lanes 1 and 3) or induced with 1 mM IPTG for 3 h (lanes 2 and 4). The positions of the fusion protein and cleavage product are indicated by arrowheads. The molecular masses of marker proteins are given in kDa to the left.
Domain structure of the WBV replicase.
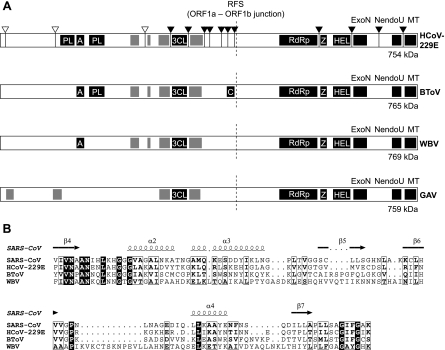
Comparative sequence analyses of the ORF1b-encoded part of the replicase polyproteins of WBV and other nidoviruses suggested that WBV is a large nidovirus (24) which is most closely related to members of the genera Torovirus and Coronavirus. This conclusion is supported by the following observations. First, unlike small nidoviruses (Arteriviridae), which encode only a limited set of domains (24), WBV encodes the full set of ORF1b domains conserved in previously characterized large nidoviruses, namely, RdRp, zinc-binding, helicase, exoribonuclease, NendoU, and putative ribose-2′-O-methyltransferase domains (Fig. 7A). Second, at least four putative membrane domains (two of them flanking 3CLpro) were identified in the ORF1a-encoded sequence. The relative positions and approximate sizes of the WBV pp1a membrane domains were found to correspond very well to those identified in toro- and coronaviruses, whereas the distributions of ronivirus pp1a membrane domains (except for the two domains flanking 3CLpro) were clearly different (Fig. 7A). Third, the special affinity between WBV and toro- and coronaviruses was supported by the identification of an ADP-ribose 1"-phosphatase domain in WBV (Fig. 7B), the conservation of which has not been reported for Arteriviridae and Roniviridae. Thus far, we failed to obtain convincing evidence for the conservation of close homologs of the papain-like proteases of corona- and toroviruses in WBV. Experimental studies are under way to get insight into the expression of the WBV N-proximal pp1a/pp1ab regions.
FIG. 7.
Domain organization of nidovirus replicase polyproteins. (A) Comparison of the WBV pp1ab domain organization with those of representative viruses from other nidovirus genera: HCoV-229E (genus Coronavirus and family Coronaviridae), bovine torovirus (BToV; genus Torovirus and family Coronaviridae), and gill-associated virus (GAV; genus Okavirus and family Roniviridae). The polyproteins are processed by viral proteases that are part of the polyprotein. The coronavirus pp1ab proteolytic processing has been characterized in considerable detail (71, 75) and is illustrated here for HCoV-229E. To produce a total of 16 nonstructural proteins, three cleavages are carried out by papain-like proteases (PL) in the N-proximal region of the polyprotein (indicated by white arrowheads), and 11 cleavages are carried out by the 3C-like protease (3CL) in the central and C-terminal parts of the polyprotein (indicated by black arrowheads). For the genera Torovirus and Okavirus, only limited information on proteases and their cleavage sites is available (53, 72). The putative proteases of BToV have not been characterized, and only a few 3C-like protease cleavage sites (not shown) have been identified for GAV (72). Proteases and other conserved enzymatic activities are indicated by black boxes. A, ADP-ribose 1"-phosphatase (ADRP) related to cellular macro domain proteins (20, 40); Z, zinc-binding domain (51); HEL, helicase domain (50); ExoN, 3′-to-5′ exoribonuclease (34); MT, putative ribose-2′-O-methyltransferase domain (21, 54); C, putative cyclic nucleotide phosphodiesterase (54); RFS, ribosomal frameshift site. Regions with predicted transmembrane domains (see Materials and Methods) are indicated by gray boxes. Note that the expression of the C-terminal part of pp1ab requires a ribosomal frameshift into ORF1b, which is predicted to occur just upstream of the ORF1a translation stop codon (Fig. 5). The sizes and positions of the polyproteins and functional domains are not precisely drawn to scale. (B) Partial sequence alignment of ADRP domains from SARS-CoV and HCoV-229E, whose activities have been characterized previously (20, 40, 41, 43), and the predicted ADRP domains from BToV (18) and WBV (this study). The alignment was generated using the ClustalX program (version 1.8). The secondary structure information was derived from the published SARS-CoV ADRP crystal structure (Protein Data Bank no. 2ACF) (43) and, together with the alignment, used as input for the ESPript program, version 2.2 (http://prodes.toulouse.inra.fr/ESPript/cgi-bin/ESPript.cgi). Sequences of the proteins were derived from the DDBJ/EMBL/GenBank database accession numbers NC_002645 (HCoV-229E pp1a/pp1ab residues Phe1299 to Lys1398), AY291315 (SARS-CoV pp1a/pp1ab residues Val1034 to Lsy1135), AY427798 (BToV [strain Breda-1] pp1a/pp1ab residues Y1668 to Ser1775), and DQ898157 (WBV pp1a/pp1ab residues Phe1667 to Lys1796). Black boxes, identical residues; white boxes, similar residues.
Phylogeny of WBV.
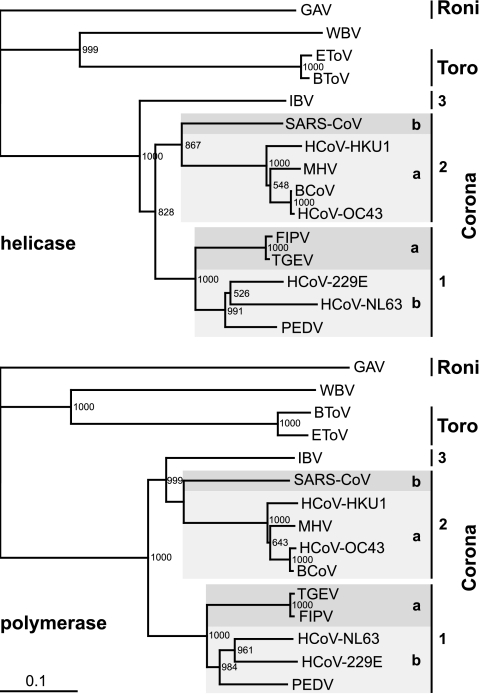
Taken together, the data provide strong evidence that WBV and other nidoviruses share a common ancestor. This hypothesis is based on multiple lines of evidence, including (i) the nidovirus-like polycistronic genome organization of WBV, (ii) the relationship between (some of) the WBV structural proteins and the respective homologs from toro- and (to a lesser extent) coronaviruses, and (iii) the use of common genome expression strategies involving regulation at the transcriptional level (i.e., production of a nested set of sgRNAs), at the translational level (use of ribosomal frameshifting to express the replicase core domains), and at the posttranslational level (evidence for proteolytic processing by a chymotrypsin-like main protease). To define the relationship of WBV with other large nidoviruses more precisely, we produced sequence alignments of conserved ORF1b domains and used them to generate phylogenetic trees (Fig. 8 and data not shown). In these analyses, WBV consistently grouped together with members of the genus Torovirus. A moderate sequence similarity between WBV and toroviruses was also evident when the viral structural proteins were compared (see above), and there are similarities between WBV and toroviruses with respect to virion morphology. It is also noteworthy that WBV and toroviruses share extremely long 5′-terminal nontranslated regions of more than 800 nucleotides which lack the small ORFs upstream of ORF1a that are conserved in coronaviruses and arteriviruses. Furthermore, the 3CLpro domains of WBV and toroviruses are more related to each other than they are to other nidovirus main proteases (R. Ulferts and J. Ziebuhr, unpublished data). In several other respects, however, the two clusters have diverged significantly. For example, (i) WBV does not encode cyclic nucleotide phosphodiesterase, which is conserved in bovine and equine toroviruses; (ii) there is a poor sequence conservation between the N-proximal pp1a/pp1ab regions of WBV and toroviruses; (iii) unlike Equine torovirus (strain Berne), in which three (out of four) sg RNAs lack a 5′ leader (65), all of the sg RNAs of WBV possess a 5′ leader sequence; (iv) in contrast to toroviruses, WBV does not encode a hemagglutinin esterase structural protein; and (v) the natural hosts of toroviruses (mammals) and WBV (fish) differ profoundly.
FIG. 8.
Phylogenetic analysis of WBV helicase and polymerase core domains. Phylogenetic trees were generated from multiple-sequence alignments of the most conserved regions of nidovirus RNA-dependent RNA polymerase (residues Thr4723 to Gln5396 in the WBV pp1ab sequence) and helicase domains (residues Ala5644 to Cys5924 in the WBV pp1ab sequence), using the neighbor-joining algorithm as implemented in the ClustalX 1.8 program (for details, see Materials and Methods). The WBV sequences were compared with those from Gill-associated virus (GAV, accession no. AF227196), Equine torovirus Berne (EToV, X52374), and Bovine torovirus Breda-1 (BToV, AY427798) as well as from viruses representing the three coronavirus groups, including the recently introduced subgroups 1a, 1b, 2a, and 2b (26). Group 1a, Transmissible gastroenteritis virus Purdue-115 (TGEV, accession no. Z34093) and Feline infectious peritonitis virus WSU 79/1146 (FIPV, DQ010921); group 1b, HCoV-229E (NC_002645), Human coronavirus NL63 Amsterdam I (HCoV-NL63, AY567487), and Porcine epidemic diarrhea virus CV777 (PEDV, AF353511); group 2a, Bovine coronavirus LUN (BCoV, AF391542), Human coronavirus OC43 serotype Paris (HCoV-OC43, AY585229), Mouse hepatitis virus A59 (MHV, NC_001846), and Human coronavirus HKU1 (HCoV-HKU1, NC_006577); group 2b, Severe acute respiratory syndrome coronavirus Frankfurt 1 (SARS-CoV, AY291315); group 3, Avian infectious bronchitis virus Beaudette (IBV, NC_001451).
Conclusion.
The study reports the complete genome sequence of WBV, the first nidovirus to be isolated from fish. A preliminary characterization of this virus and comparative sequence analysis identified toroviruses (followed by coronaviruses) as the closest known relatives of WBV. Given the limited number of published torovirus sequences (18, 53), the reconstruction of possible scenarios involved in the evolution of the WBV cluster remains speculative at the present time. Most likely, WBV (and its yet-to-be-identified close relatives) and the present-day toroviruses have evolved from a common ancestor that split off from the lineage that led to the present-day coronaviruses. Given the quite different hosts infected by WBV and toroviruses, it seems less likely that the ancestor of the WBV cluster split first from the common coronavirus-torovirus trunk to evolve then in parallel with the torovirus cluster. But clearly, additional studies and sequences of WBV- and torovirus-related viruses are required to (re)construct the most plausible scenario for the evolution of the WBV-like, coronavirus, and torovirus clusters. However, even in the absence of this information, we think the profoundly divergent evolution of the WBV cluster from the related genera Torovirus and Coronavirus and the different host ranges of these viruses would justify the introduction of a new genus, with WBV being its tentative type species. We propose the name Bafinivirus for this yet-to-be-approved nidovirus genus, referring to the bacilliform morphology of this cluster of fish nidoviruses. If approved, the new genus might then trigger a more general discussion and, possibly, revision of the current taxonomic structure of the Nidovirales.
The identification of nidoviruses in a very large number of mammalian species (39, 52, 59, 67) as well as in invertebrates (12) and fish (this study) suggests that nidoviruses, which in several respects are distinct from the huge variety of plus-strand RNA viruses (24), have managed to adapt to a remarkable diversity of biological niches. The continued sampling and characterization of nidoviruses, of which this study is a part, are anticipated to increasingly fill the major gaps that still exist between the individual clusters of nidoviruses. Additional sequence information and functional studies will help to identify the major forces and constraints that shape the evolution of nidoviruses. This information will also be required to unravel the basis for the differential conservation of specific replicase gene-encoded proteins among the various nidovirus families, genera, and species (24, 54) and help to relate these proteins to specific metabolic pathways and molecular mechanisms. More sequences and phylogenetic studies are also needed to determine the position of the WBV cluster within the nidovirus tree more precisely.
Acknowledgments
We thank Sabine Feistkorn, Petra Meyer, Mandy Jörn, and Beatrix Loth for excellent technical assistance.
The work of R.U., B.S., S.B., and J.Z. was supported in part by the Deutsche Forschungsgemeinschaft (Zi 618/3 and SFB 479-A8).
Footnotes
Published ahead of print on 20 September 2006.
REFERENCES
- 1.Anand, K., G. J. Palm, J. R. Mesters, S. G. Siddell, J. Ziebuhr, and R. Hilgenfeld. 2002. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 21:3213-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. [DOI] [PubMed] [Google Scholar]
- 3.Barrette-Ng, I. H., K. K. Ng, B. L. Mark, D. Van Aken, M. M. Cherney, C. Garen, Y. Kolodenko, A. E. Gorbalenya, E. J. Snijder, and M. N. James. 2002. Structure of arterivirus nsp4. The smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole. J. Biol. Chem. 277:39960-39966. [DOI] [PubMed] [Google Scholar]
- 4.Bartlam, M., H. Yang, and Z. Rao. 2005. Structural insights into SARS coronavirus proteins. Curr. Opin. Struct. Biol. 15:664-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 6.Bhardwaj, K., L. Guarino, and C. C. Kao. 2004. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 78:12218-12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brierley, I., M. E. Boursnell, M. M. Binns, B. Bilimoria, V. C. Blok, T. D. Brown, and S. C. Inglis. 1987. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 6:3779-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brierley, I., and F. J. Dos Ramos. 2006. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 119:29-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 10.Chen, Z., L. Kuo, R. R. Rowland, C. Even, K. S. Faaberg, and P. G. Plagemann. 1993. Sequences of 3′ end of genome and of 5′ end of open reading frame 1a of lactate dehydrogenase-elevating virus and common junction motifs between 5′ leader and bodies of seven subgenomic mRNAs. J. Gen. Virol. 74:643-659. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, A., W. Zhang, Y. Xie, W. Jiang, E. Arnold, S. G. Sarafianos, and J. Ding. 2005. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology 335:165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowley, J. A., C. M. Dimmock, K. M. Spann, and P. J. Walker. 2000. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri- and coronaviruses. J. Gen. Virol. 81:1473-1484. [DOI] [PubMed] [Google Scholar]
- 13.Cowley, J. A., C. M. Dimmock, and P. J. Walker. 2002. Gill-associated nidovirus of Penaeus monodon prawns transcribes 3′-coterminal subgenomic mRNAs that do not possess 5′-leader sequences. J. Gen. Virol. 83:927-935. [DOI] [PubMed] [Google Scholar]
- 14.de Haan, C. A., and P. J. Rottier. 2005. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 64:165-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.den Boon, J. A., M. F. Kleijnen, W. J. Spaan, and E. J. Snijder. 1996. Equine arteritis virus subgenomic mRNA synthesis: analysis of leader-body junctions and replicative-form RNAs. J. Virol. 70:4291-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.den Boon, J. A., E. J. Snijder, E. D. Chirnside, A. A. de Vries, M. C. Horzinek, and W. J. Spaan. 1991. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 65:2910-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Vries, A. A., E. D. Chirnside, P. J. Bredenbeek, L. A. Gravestein, M. C. Horzinek, and W. J. Spaan. 1990. All subgenomic mRNAs of equine arteritis virus contain a common leader sequence. Nucleic Acids Res. 18:3241-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Draker, R., R. L. Roper, M. Petric, and R. Tellier. 2006. The complete sequence of the bovine torovirus genome. Virus Res. 115:56-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egloff, M. P., F. Ferron, V. Campanacci, S. Longhi, C. Rancurel, H. Dutartre, E. J. Snijder, A. E. Gorbalenya, C. Cambillau, and B. Canard. 2004. The severe acute respiratory syndrome-coronavirus replicative protein Nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. USA 101:3792-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egloff, M.-P., H. Malet, Á. Putics, M. Heinonen, H. Dutartre, A. Frangeul, A. Gruez, V. Campanacci, C. Cambillau, J. Ziebuhr, T. Ahola, and B. Canard. 2006. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 80:8493-8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feder, M., J. Pas, L. S. Wyrwicz, and J. M. Bujnicki. 2003. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene 302:129-138. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez, J. M., P. Gomez-Puertas, D. Cavanagh, A. E. Gorbalenya, and L. Enjuanes. 2003. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 148:2207-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorbalenya, A. E. 2001. Big nidovirus genome. When count and order of domains matter. Adv. Exp. Med. Biol. 494:1-17. [PubMed] [Google Scholar]
- 24.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorbalenya, A. E., E. V. Koonin, and M. M. Lai. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 288:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorbalenya, A. E., E. J. Snijder, and W. J. Spaan. 2004. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 78:7863-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Granzow, H., F. Weiland, D. Fichtner, H. Schütze, A. Karger, E. Mundt, B. Dresenkamp, P. Martin, and T. C. Mettenleiter. 2001. Identification and ultrastructural characterization of a novel virus from fish. J. Gen. Virol. 82:2849-2859. [DOI] [PubMed] [Google Scholar]
- 28.Hertzig, T., E. Scandella, B. Schelle, J. Ziebuhr, S. G. Siddell, B. Ludewig, and V. Thiel. 2004. Rapid identification of coronavirus replicase inhibitors using a selectable replicon RNA. J. Gen. Virol. 85:1717-1725. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann, B., H. Schütze, and T. C. Mettenleiter. 2002. Determination of the complete genomic sequence and analysis of the gene products of the virus of spring viremia of carp, a fish rhabdovirus. Virus Res. 84:89-100. [DOI] [PubMed] [Google Scholar]
- 30.Ivanov, K. A., T. Hertzig, M. Rozanov, S. Bayer, V. Thiel, A. E. Gorbalenya, and J. Ziebuhr. 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA 101:12694-12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanov, K. A., V. Thiel, J. C. Dobbe, Y. van der Meer, E. J. Snijder, and J. Ziebuhr. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78:5619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 33.Meulenberg, J. J., E. J. de Meijer, and R. J. Moormann. 1993. Subgenomic RNAs of Lelystad virus contain a conserved leader-body junction sequence. J. Gen. Virol. 74:1697-1701. [DOI] [PubMed] [Google Scholar]
- 34.Minskaia, E., T. Hertzig, A. E. Gorbalenya, V. Campanacci, C. Cambillau, B. Canard, and J. Ziebuhr. 2006. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 103:5108-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Namy, O., S. J. Moran, D. I. Stuart, R. J. Gilbert, and I. Brierley. 2006. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 441:244-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 37.Pasternak, A. O., W. J. Spaan, and E. J. Snijder. 2006. Nidovirus transcription: how to make sense…? J. Gen. Virol. 87:1403-1421. [DOI] [PubMed] [Google Scholar]
- 38.Pasternak, A. O., E. van den Born, W. J. Spaan, and E. J. Snijder. 2001. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 20:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poon, L. L., D. K. Chu, K. H. Chan, O. K. Wong, T. M. Ellis, Y. H. Leung, S. K. Lau, P. C. Woo, K. Y. Suen, K. Y. Yuen, Y. Guan, and J. S. Peiris. 2005. Identification of a novel coronavirus in bats. J. Virol. 79:2001-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Putics, Á., W. Filipowicz, J. Hall, A. E. Gorbalenya, and J. Ziebuhr. 2005. ADP-ribose-1"-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 79:12721-12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Putics, Á., A. E. Gorbalenya, and J. Ziebuhr. 2006. Identification of protease and ADP-ribose 1"-monophosphatase activities associated with transmissible gastroenteritis virus non-structural protein 3. J. Gen. Virol. 87:651-656. [DOI] [PubMed] [Google Scholar]
- 42.Ratia, K., K. S. Saikatendu, B. D. Santarsiero, N. Barretto, S. C. Baker, R. C. Stevens, and A. D. Mesecar. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. USA 103:5717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saikatendu, K. S., J. S. Joseph, V. Subramanian, T. Clayton, M. Griffith, K. Moy, J. Velasquez, B. W. Neuman, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2005. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1"-phosphate dephosphorylation by a conserved domain of nsP3. Structure 13:1665-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Sawicki, S. G., and D. L. Sawicki. 1995. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 380:499-506. [DOI] [PubMed] [Google Scholar]
- 46.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-219. [DOI] [PubMed] [Google Scholar]
- 47.Sawicki, S. G., and D. L. Sawicki. 2005. Coronavirus transcription: a perspective, p. 31-55. In L. Enjuanes (ed.), Coronavirus replication and reverse genetics, vol. 287. Springer, Berlin, Germany. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sawicki, S. G., D. L. Sawicki, D. Younker, Y. Meyer, V. Thiel, H. Stokes, and S. G. Siddell. 2005. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathogens 1:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schütze, H., P. J. Enzmann, R. Kuchling, E. Mundt, H. Niemann, and T. C. Mettenleiter. 1995. Complete genomic sequence of the fish rhabdovirus infectious haematopoietic necrosis virus. J. Gen. Virol. 76:2519-2527. [DOI] [PubMed] [Google Scholar]
- 50.Seybert, A., A. Hegyi, S. G. Siddell, and J. Ziebuhr. 2000. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA 6:1056-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seybert, A., C. C. Posthuma, L. C. van Dinten, E. J. Snijder, A. E. Gorbalenya, and J. Ziebuhr. 2005. A complex zinc finger controls the enzymatic activities of nidovirus helicases. J. Virol. 79:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siddell, S. G., J. Ziebuhr, and E. J. Snijder. 2005. Coronaviruses, toroviruses, and arteriviruses, p. 823-856. In B. W. J. Mahy and V. ter Meulen (ed.), Topley and Wilson's microbiology and microbial infections, 10th ed., vol. 1. Hodder Arnold, London, United Kingdom. [Google Scholar]
- 53.Smits, S. L., E. J. Snijder, and R. J. de Groot. 2006. Characterization of a torovirus main proteinase. J. Virol. 80:4157-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snijder, E. J., and M. C. Horzinek. 1995. The molecular biology of toroviruses, p. 219-238. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 56.Spaan, W., H. Delius, M. Skinner, J. Armstrong, P. Rottier, S. Smeekens, B. A. van der Zeijst, and S. G. Siddell. 1983. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 2:1839-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su, D., Z. Lou, F. Sun, Y. Zhai, H. Yang, R. Zhang, A. Joachimiak, X. C. Zhang, M. Bartlam, and Z. Rao. 2006. Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. J. Virol. 80:7902-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sutton, G., E. Fry, L. Carter, S. Sainsbury, T. Walter, J. Nettleship, N. Berrow, R. Owens, R. Gilbert, A. Davidson, S. Siddell, L. L. Poon, J. Diprose, D. Alderton, M. Walsh, J. M. Grimes, and D. I. Stuart. 2004. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure 12:341-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang, X. C., J. X. Zhang, S. Y. Zhang, P. Wang, X. H. Fan, L. F. Li, G. Li, B. Q. Dong, W. Liu, C. L. Cheung, K. M. Xu, W. J. Song, D. Vijaykrishna, L. L. Poon, J. S. Peiris, G. J. Smith, H. Chen, and Y. Guan. 2006. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 80:7481-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 61.Thiel, V., A. Rashtchian, J. Herold, D. M. Schuster, N. Guan, and S. G. Siddell. 1997. Effective amplification of 20-kb DNA by reverse transcription PCR. Anal. Biochem. 252:62-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:12056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Vliet, A. L., S. L. Smits, P. J. Rottier, and R. J. de Groot. 2002. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 21:6571-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.von Grotthuss, M., L. S. Wyrwicz, and L. Rychlewski. 2003. mRNA cap-1 methyltransferase in the SARS genome. Cell 113:701-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woo, P. C., S. K. Lau, K. S. Li, R. W. Poon, B. H. Wong, H. W. Tsoi, B. C. Yip, Y. Huang, K. H. Chan, and K. Y. Yuen. 2006. Molecular diversity of coronaviruses in bats. Virology 351:180-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu, X., Y. Zhai, F. Sun, Z. Lou, D. Su, Y. Xu, R. Zhang, A. Joachimiak, X. C. Zhang, M. Bartlam, and Z. Rao. 2006. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J. Virol. 80:7909-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhai, Y., F. Sun, X. Li, H. Pang, X. Xu, M. Bartlam, and Z. Rao. 2005. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 12:980-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ziebuhr, J. 2004. Molecular biology of severe acute respiratory syndrome coronavirus. Curr. Opin. Microbiol. 7:412-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ziebuhr, J. 2005. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 287:57-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ziebuhr, J., S. Bayer, J. A. Cowley, and A. E. Gorbalenya. 2003. The 3C-like proteinase of an invertebrate nidovirus links coronavirus and potyvirus homologs. J. Virol. 77:1415-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ziebuhr, J., J. Herold, and S. G. Siddell. 1995. Characterization of a human coronavirus (strain 229E) 3C-like proteinase activity. J. Virol. 69:4331-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ziebuhr, J., and S. G. Siddell. 1999. Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab. J. Virol. 73:177-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]
- 76.Zúñiga, S., I. Sola, S. Alonso, and L. Enjuanes. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78:980-994. [DOI] [PMC free article] [PubMed] [Google Scholar]