Abstract
The positive-strand RNA genome of the hepatitis C virus (HCV) is flanked by 5′- and 3′-untranslated regions (UTRs). Translation of the viral RNA is directed by the internal ribosome entry site (IRES) in the 5′-UTR, and subsequent viral RNA replication requires sequences in the 3′-UTR and in the 5′-UTR. Addressing previous conflicting reports on a possible function of the 3′-UTR for RNA translation in this study, we found that reporter construct design is an important parameter in experiments testing 3′-UTR function. A translation enhancer function of the HCV 3′-UTR was detected only after transfection of monocistronic reporter RNAs or complete RNA genomes having a 3′-UTR with a precise 3′ terminus. The 3′-UTR strongly stimulates HCV IRES-dependent translation in human hepatoma cell lines but only weakly in nonliver cell lines. The variable region, the poly(U · C) tract, and the most 3′ terminal stem-loop 1 of the highly conserved 3′ X region contribute significantly to translation enhancement, whereas stem-loops 2 and 3 of the 3′ X region are involved only to a minor extent. Thus, the signals for translation enhancement and for the initiation of RNA minus-strand synthesis in the HCV 3′-UTR partially overlap, supporting the idea that these sequences along with viral and possibly also cellular factors may be involved in an RNA 3′-5′ end interaction and a switch between translation and RNA replication.
Hepatitis C virus (HCV), the main causative agent of non-A, non-B hepatitis (7), belongs to the unique genus Hepacivirus in the family Flaviviridae (1). HCV has infected about 170 million people worldwide. About 80% of the patients infected by HCV are unable to eliminate the virus, and these patients are at high risk to develop chronic liver diseases including cirrhosis and hepatocellular carcinoma (28). The recent development of the replicon system (3, 32) has greatly stimulated research on several aspects of HCV replication. However, tissue culture systems supporting a complete replication cycle of HCV are available for only a short time (30, 51, 60).
The genome of HCV is a single-stranded, positive-sense RNA of approximately 9,600 nucleotides with only one large open reading frame (ORF) that encodes a single polyprotein of 3,010 to 3,033 amino acid residues. The HCV ORF is flanked by highly conserved 5′- and 3′-untranslated regions (5′-UTR and 3′-UTR) required for viral replication. The 5′-UTR forms extensive secondary structures (6) and regulates translation initiation in an internal ribosome entry site (IRES)-dependent manner (50, 52). However, in contrast to picornaviruses (2), the HCV IRES can bind the 40S ribosomal subunit directly in the absence of any other canonical translation initiation factor (eukaryotic initiation factor, or eIF), thereby positioning the authentic initiator AUG codon of the ORF precisely at the P site of the ribosome (41). The 3′-UTR lacks a poly(A) tail and is composed of three sequence elements supposed to be involved in RNA replication: a nonconserved variable region (30 to 50 nucleotides), a poly(U · C) stretch (20 to 200 nucleotides), and a conserved 98-nucleotide sequence, termed the 3′ X region, which forms a three stem-loop (SL) structure (4, 10, 24, 25, 46, 54-57). The 3′-UTR binds various cellular proteins such as the 52-kDa La autoantigen, the 57-kDa polypyrimidine tract-binding protein, and other proteins (8, 13, 18, 33, 45, 49, 53).
Most eukaryotic mRNAs and many viral RNAs have a cap structure at the 5′ terminus and a poly(A) tail at the 3′ end which play essential roles in the regulation of translation, either individually or in concert (14, 43). Each of these elements associates with specific RNA-binding proteins and stimulates RNA translation. The translation initiation factor eIF4F binds to the 5′-terminal cap structure of cellular mRNA through its subunit eIF4E, and the poly(A) binding protein facilitates mRNA 5′-3′ end interaction by binding both to the mRNA poly(A) tail and to the eIF4G subunit of eIF4F (47). This concept of RNA 5′-3′ end interaction for translation stimulation could also apply to the HCV RNA genome, even in the absence of the terminal cap and poly(A) structures. Such an interaction of the terminal HCV genome structures, possibly facilitated by yet unknown viral and/or cellular proteins, could play a role in a switch from RNA translation to negative-strand RNA synthesis, similar to the switch reported for poliovirus (12).
Several conflicting results have been reported about the possible role of the HCV 3′-UTR in RNA translation, obtained either in vitro, in cell culture, or in transfected livers of mice. In some studies, a positive influence of 3′-UTR sequences (sometimes comprising only the 3′ X region) was shown (19, 20, 34, 35). One study reported an inhibitory effect of the 3′-UTR (37), whereas other reports claimed that there is no effect of the HCV 3′-UTR on IRES-directed translation (9, 10, 15, 26, 56).
In order to investigate the possible role of the viral 3′ terminal sequence for HCV IRES-directed translation, we have performed a series of translation assays including in vitro and cell culture systems. Our results reveal that the HCV 3′-UTR significantly enhances HCV IRES-dependent translation in human liver cell lines but has only weak enhancing effects in nonliver cell lines. Interestingly, both the transfection approach employed and the reporter construct design were found to be important for the outcome of experiments testing the role of the HCV 3′-UTR in enhancing IRES-directed translation.
MATERIALS AND METHODS
Plasmids.
The monocistronic reporter plasmid pHCV+3′-UTR (wild-type) contains the following elements in 5′ to 3′ order in a pBlueScript SK(+) vector: the cytomegalovirus (CMV) immediate early promoter, a T7 promoter, the entire 5′-UTR (nucleotides 1 to 341) plus partial core protein-encoding sequences of the HCV-1b strain, a ubiquitin sequence followed by the firefly luciferase (FLuc) gene, and the entire HCV 3′-UTR. Plasmid pHCV-Δ3′-UTR was obtained from pHCV+3′-UTR by double digestion and religation to delete the HCV 3′-UTR. A series of mutant plasmids with deletions within the 3′-UTR (see Fig. 5A) were prepared from pHCV+3′-UTR by PCR mutagenesis. The organization of the corresponding RNA transcripts is shown in Fig. 5A. Plasmid pRL-FMDV-wt-FL was derived from pD12 (39) and contains a T7 promoter, the Renilla luciferase (RLuc) sequence, the IRES of foot-and-mouth disease virus (FMDV) and the FLuc sequence. Plasmid pM12-FLuc was derived from pM12 (39). It contains the FMDV IRES and the FLuc sequence followed by 29 nucleotides of the HCV 3′-UTR variable region plus 14 unrelated linker nucleotides and a T7 promoter in reverse orientation. Plasmid pFK1-9605C1wtdg contains the complete HCV-1b genome (32) with the hepatitis delta virus (HDV) genomic ribozyme sequence immediately downstream of the HCV sequence. Plasmid pFK1-9374wtdg contains the complete HCV genome without the 3′-UTR. The corresponding plasmids with the NS5B polymerase GND mutation have an exchange of NS5B amino acid 318 (D to N) that abrogates NS5B polymerase activity (31). Plasmid pNS5B-rev contains in a pUC-based plasmid downstream of a T7 promoter sequence 15 nucleotides of linker sequence, followed by 29 nucleotides of the variable region and 298 nucleotides of NS5B sequence in antisense orientation.
FIG. 5.
Effects of deletions within the HCV 3′-UTR on translation efficiency. (A) The general structure of the reporter construct (Fig. 1A) is shown in the bottom. The HCV 3′-UTR was fused exactly to the luciferase coding sequence. Deletion mutants are drawn above. HCV+RD15 includes an artificial sequence of 15 random nucleotides (open bar). (B) FLuc activities expressed in the cytoplasm of Huh-7 cells were measured 4 h after liposome-mediated transfection with deletion mutant RNAs specified on the left. The FLuc activity of HCV+3′-UTR wild type was set to 100%. (C) RNA stability control. Cytoplasmic RNA from Huh-7 cells transfected with the RNAs used in panel B was harvested 4 h after liposome-mediated transfection, purified, and analyzed by RNase protection assay using 0.35 pmol of the [α-32P]UTP-labeled antisense FLuc RNA probe (A). As a control, the same cytoplasmic RNA samples were analyzed using GAPDH mRNA antisense RNA probe (lower panel). In lanes 1 and 16, 10% of the undigested antisense RNA probes was loaded. (D) RNA stability control of selected RNAs harvested 4 h after transfection of Huh-7 cells by using electroporation. UC, poly(U · C) tract; wt, wild type.
RNA synthesis.
HCV-FLuc reporter plasmid DNAs were linearized with BamHI downstream of the reporter construct cassette. These linearized DNAs were also used as templates to amplify PCR fragments of interest. The HCV+RD15 RNA template was obtained from pHCV+3′-UTR by PCR amplification using an oligonucleotide which includes the 3′ terminus of the firefly luciferase-coding sequence plus 15 unrelated nucleotides. The amplified DNA fragments were gel purified, extracted with phenol-chloroform, ethanol precipitated, dissolved in water, and used for transcription using T7 RNA polymerase (New England Biolabs). pRL-FMDV-wt-FL was linearized with XbaI downstream of the RLuc sequence and transcribed with T7 RNA polymerase to obtain RLuc control RNA for cotransfections. Reactions were treated with RNase-free DNase I to digest DNA templates, and RNA was purified with RNeasy kits (QIAGEN). The integrity of RNA was checked by denaturing gel electrophoresis. RNA concentrations were determined by gel images and a photometer. Plasmid pM12-FLuc was linearized with EcoRV and transcribed with T7 RNA polymerase. The resulting antisense FLuc RNA contains 14 unrelated linker nucleotides and 29 nucleotides of the HCV variable region plus 314 nucleotides of antisense FLuc sequence starting from the FLuc coding sequence 3′ end. Plasmids pFK1-9605C1wtdg and pFK1-9374wtdg were linearized with SpeI downstream of the HDV ribozyme sequence, and authentic 3′ ends were generated by autocatalytic cleavage by the ribozyme. Plasmid pNS5B-rev was linearized with KpnI in the NS5B region. The resulting RNA contains 15 nucleotides of linker sequence, 29 nucleotides of the variable region, and 298 nucleotides of NS5B sequence in reverse orientation. Plasmid pXCM-hu-GAPDH (kindly provided by H. Kleinert) was linearized with HindIII and transcribed with T3 RNA polymerase to yield glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) antisense RNA. The fragment protected in the RNase protection assay is 105 nucleotides long. Labeled RNAs were synthesized using T7 RNA polymerase in the presence of 2.5 μM [α-32P]UTP (400 Ci/mmol; Amersham) plus 10 μM nonradioactive labeling nucleotide and checked on 8 M urea-10% polyacrylamide gels.
In vitro translation.
Reactions were carried out in S7 nuclease-treated rabbit reticulocyte lysates (RRL; Promega) (40, 44). Briefly, different amounts of RNA as indicated were used in 10-μl reaction mixtures containing 4.4 μl of RRL and 0.2 μl of [35S]methionine. In addition to the 50 mM endogenous potassium acetate that is added to the RRL by the supplier, KCl was added to a final potassium concentration of 130 mM. Reactions were incubated at 30°C for 60 min. Aliquots of the translation products were separated on 12% sodium dodecyl sulfate-polyacrylamide gels (SDS-PAGE) and analyzed by autoradiography.
Cells and transfections.
Cells were maintained in standard Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% penicillin (10 U/ml)-streptomycin (10 μg/ml) and 1% l-glutamine. For liposome-mediated transfection, cells were split into 12-well plates and grown in DMEM with 10% FBS and antibiotics to 80 to 90% confluence. One day before transfection, the medium was changed to DMEM plus FBS but without antibiotics. For DNA transfections, 0.5 μg of plasmid or PCR fragment, respectively, and 1 μl of Lipofectamine (Invitrogen) were diluted separately in each 25 μl of DMEM (without FBS and antibiotics) and incubated at room temperature. Within 5 min, both dilutions were mixed together, incubated for another 25 min at room temperature, and then added to the cells. For RNA transfections, approximately 0.8 μg of each in vitro transcribed RNA was used for transfection of the cells using a TransMessenger transfection kit (QIAGEN). After the time as indicated in the figures, cells were washed in phosphate-buffered saline (PBS), and then PBS was aspirated, 150 μl of passive lysis buffer (Promega) was added to each well, and the cells were lysed by gentle agitation. Lysates were centrifuged for 1 min at 13,000 × g, and 15 μl of supernatant was used for measuring firefly luciferase activity as described before (38) or Renilla luciferase as described by the supplier (Promega). For electroporation, two samples (10 μg each) of in vitro transcript were mixed with 400 μl of a suspension of 107 Huh-7 cells per ml in a cuvette with a gap width of 0.4 cm (Bio-Rad, Munich, Germany). After one pulse at 975 μF and 270 V with a Gene Pulser system II (Bio-Rad), cells were immediately transferred into 20 ml of complete Dulbecco's modified Eagle medium. Aliquots of the cell suspension were seeded in 6-cm-diameter culture dishes and harvested at various time points.
RNase protection assay.
For the short reporter RNAs, cells were washed in PBS 4 h after transfection and lysed in passive lysis buffer, and nuclei removed by centrifugation. For the HCV full-length genome RNAs, cells were washed with PBS and lysed in 7 M guanidinium-isothiocyanate and 10% 2-mercaptoethanol. Total RNA was isolated using RNeasy kits (QIAGEN) and dissolved in 0.5% SDS. All samples were treated with proteinase K in the presence of 0.5% SDS and 1 mM CaCl2 at 50°C for 2 h, phenol-chloroform extracted, and ethanol-precipitated. Residual DNA was removed by treatment with DNase I. RNase protection assays were performed essentially as described previously (21) using [α-32P]UTP-labeled antisense RNA probes transcribed from plasmids pM12-FLuc, pNS5B-rev, and pXCM-hu-GAPDH. After hybridization, samples were treated first with RNase A and then with proteinase K, phenol-chloroform extracted, and ethanol precipitated. Samples were separated on 8 M urea-10% polyacrylamide gels and analyzed by autoradiography.
Western blotting.
Cells were lysed with PBS containing 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride and 0.1 μg/ml aprotonin. Samples were run on SDS 12%-polyacrylamide gels, blotted to nitrocellulose, and stained with anti-NS5B or anti-actin antibodies by standard procedures.
RESULTS
Conflicting results have been reported on the effect of the HCV 3′-UTR on IRES-directed translation (9, 10, 15, 19, 20, 26, 34, 35, 37, 56). The reasons for these discrepancies are not directly evident. Some researchers reported that different concentrations of translation factors in the employed translation assay systems, different intracellular mRNA concentrations, and/or misfolding of in vitro generated RNAs could contribute to the discrepancies (26). However, we noted (i) that some of these studies concentrated only on a possible role of the HCV 3′ X region but not the entire 3′-UTR and (ii) that the HCV RNA molecules used in most studies did not have precise 3′ termini. We hypothesized that these experimental variations may have caused the discrepancies and therefore compared different reporter constructs and translation systems.
Effect of the HCV 3′-UTR on IRES-mediated translation in RRL.
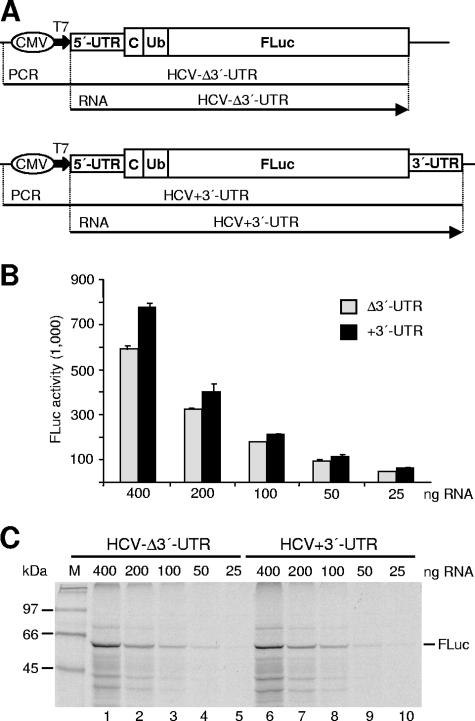
First, we designed in vitro translation experiments using two different monocistronic reporter mRNAs containing the entire HCV 5′-UTR sequence plus partial core protein coding sequences and the FLuc reporter gene, either ending exactly at the 3′ end of the FLuc coding sequence or additionally including the complete HCV 3′-UTR (Fig. 1A). Templates for in vitro transcription of the reporter RNAs were generated by PCR to obtain runoff transcripts with precise 3′ ends. These RNAs were in vitro translated in RRL at a final potassium concentration of 130 mM, which is close to physiological salt conditions. The translational products of these RNAs were examined by measuring FLuc activity (Fig. 1B) and by analyzing FLuc protein amounts by SDS-PAGE and autoradiography (Fig. 1C). No significant difference in translation efficiency was detected between these two RNAs, even when different amounts of RNA were used to rule out the possible influence of saturation effects in the reticulocyte lysate. These results indicate that the HCV 3′-UTR has no obvious effect on HCV IRES-directed translation in RRL.
FIG. 1.
Analysis of the effect of the HCV 3′-UTR on translation efficiency in vitro. (A) The HCV reporter constructs. RNAs (arrows) were obtained by in vitro transcription using T7 RNA polymerase either from linearized plasmids or from PCR fragments (lines). CMV, CMV immediate-early promoter; T7, T7 RNA polymerase promoter; C, partial core protein encoding sequences; Ub, ubiquitin sequences. (B) FLuc activities obtained with various amounts of RNA translated in RRL at physiological salt conditions (130 mM K+). Columns and bars represent means and standard deviations of three independent experiments. (C) [35S]methionine-labeled proteins translated from the given amounts of RNA were separated by 12% SDS-PAGE. The position of the firefly luciferase protein (62 kDa) is marked on the right. M, 14C-labeled marker proteins.
Influence of reporter construct design and transfection conditions on translation enhancement by the 3′-UTR.
Since HCV replicates primarily in human liver cells, several groups examined a possible effect of the HCV 3′-UTR on RNA translation by using transfected human hepatoma cell lines. However, results are controversial (10, 15, 20, 26, 34). Most of these studies used transfection and transient expression of complete circular or linearized reporter plasmids. In only one study were reporter RNAs transfected into Huh-7 cells, but no enhancement of translation by the HCV 3′-UTR was detected (10). In an attempt to eliminate experimental parameters that hamper a possible enhancing action of the 3′-UTR on HCV IRES-dependent translation, Huh-7 cells were transfected with circular plasmid DNAs of two types of the HCV-IRES/luciferase constructs, i.e., either containing or lacking the entire 3′-UTR (pHCV+3′-UTR and pHCV-Δ3′-UTR, respectively) (Fig. 1A). However, when FLuc activities were measured at different time points after transfection, no significant differences could be observed (Fig. 2A).
FIG. 2.
Significance of the type of transfected reporter nucleic acid and of precise 3′ ends for the enhancement of RNA translation by the HCV 3′-UTR. (A to C) Translation efficiencies of HCV reporter constructs (as shown in Fig. 1A) after transfection of circular plasmids (A), PCR fragments with exact 3′ ends (B), or RNA with exact 3′ ends (C). Cells were harvested at different times as indicated and FLuc activities were measured. Columns and bars represent means and standard deviations, respectively, of at least three independent transfections. (D) Organization of the antisense FLuc RNA probe for the RNase protection assay shown in panel E. (E) RNA stability control. Cytoplasmic RNA from cells transfected with reporter RNAs as in panel C, i.e., HCV-Δ3′-UTR (lane 2) and HCV+3′-UTR (lane 3), was reextracted from the cells 4 h after transfection, purified, and analyzed by RNase protection assay using the [α-32P]UTP-labeled antisense FLuc RNA probe. In lane 1, 5% of the undigested antisense FLuc RNA probe was loaded. As a control, the same cytoplasmic RNAs reextracted from the cells transfected with HCV-Δ3′-UTR (lane 5) and HCV+3′-UTR (lane 6) were analyzed with an RNA probe hybridizing to GAPDH mRNA (5% loaded in lane 4). (F and G) Cotransfection controls. HCV+3′-UTR and HCV-Δ3′-UTR reporter RNAs (Fig. 1A) were cotransfected together with capped and polyadenylated Renilla luciferase RNA in different amounts into Huh-7 cells. Cells were harvested after 4 h and firefly luciferase (F) and Renilla luciferase (G) activities were determined. Δ3′-UTR, HCV-Δ3′-UTR; +3′-UTR, HCV+3′-UTR; Ub, ubiquitin; C, partial core protein.
To analyze if the exact 3′ end of the HCV 3′-UTR is required for enhancing RNA translation, we generated two types of PCR fragments that included the CMV promoter and either terminate at the luciferase stop codon or additionally included the HCV 3′-UTR (Fig. 1A). Upon transfection of Huh-7 cells with these PCR fragments, significant differences in FLuc expression were observed (Fig. 2B). At early time points after transfection, FLuc expression from the PCR fragment containing the HCV 3′-UTR was up to threefold higher than that from the PCR fragment without the 3′-UTR, while this relative difference decreased at later time points. Thus, assaying reporter gene activity in a time course after transfection may be important for the outcome of experiments testing a possibly enhancing effect of the 3′-UTR on HCV translation.
In contrast to the DNA transfection experiments, a very strong stimulating effect of the HCV 3′-UTR on translation was found when reporter RNAs with the precise 3′ end (Fig. 1A) were transfected into Huh-7 cells (Fig. 2C). Translation efficiency of the RNA containing the 3′-UTR (HCV+3′-UTR) was 10- to 25-fold higher than that of the RNA without the 3′-UTR (HCV-Δ3′-UTR). This enhancement is much higher than the one found in previous reports (20, 35) or detected in our PCR fragment transfection experiments (Fig. 2B). Thus, the HCV 3′-UTR can strongly enhance translation directed by the HCV IRES.
Expression differences are not due to differences in RNA stability or transfection efficiency.
SL1 in the 3′ X region forms a stable secondary RNA structure (see Fig. 5A) that may protect the 3′ end of the reporter RNA against degradation. To rule out the possibility that differences in translation efficiency are due to differences in RNA stability, we tested the integrity of the reporter RNAs after extraction from the transfected cells in RNase protection assays (Fig. 2E) using the antisense Fluc probe described in Fig. 2D. The results show that with both reporter constructs, the same amounts of protected fragments are detected, indicating similar half-lives of the RNAs independent of the presence or absence of the 3′-UTR. Thus, the differences in translation efficiency are not due to differences in RNA stability.
To further rule out the possibility that the differences in FLuc expression are due to variations in transfection efficiency, we cotransfected the HCV reporter RNAs together with capped and polyadenylated RNA containing the Renilla luciferase gene. Again, HCV IRES-directed FLuc expression was much more efficient in the presence of the HCV 3′-UTR (Fig. 2F), while the cotransfected Renilla reporter RNA was expressed with similar efficiency in both cases (Fig. 2G). Moreover, translation stimulation by the HCV 3′-UTR was similar after transfection of different amounts of reporter RNA, ruling out any possible saturation effects.
Additional nucleotides at the 3′ end of the 3′-UTR interfere with stimulation.
From the above results, we concluded that three parameters have an impact on the outcome of experiments testing the translation enhancement by the HCV 3′-UTR: (i) the time course of measuring reporter gene activity reveals that the stimulatory action of the 3′-UTR is more evident at shorter times after transfection (Fig. 2B and C); (ii) the nature of transfected nucleic acid is important since translation stimulation by the 3′-UTR was remarkably more evident when RNA rather than DNA was used (Fig. 2C); and (iii) the nature of the 3′ end of the reporter construct plays a role since a PCR fragment providing an exact end of the 3′-UTR resulted in translation stimulation (Fig. 2B) while transfection of circular plasmid DNA did not (Fig. 2A).
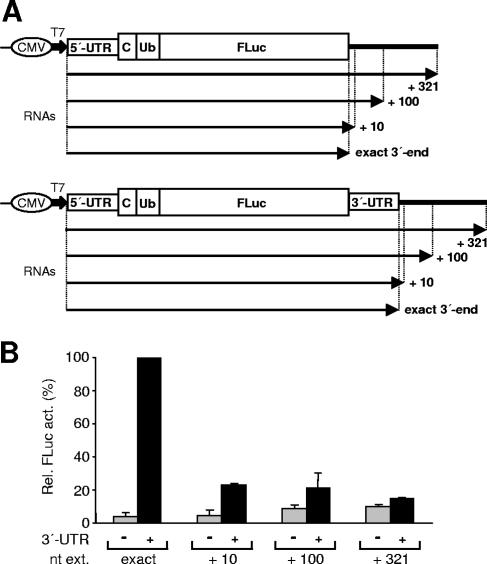
To further test whether efficient translation stimulation requires a precise 3′ end of the HCV 3′-UTR, we generated additional reporter construct templates having unrelated vector sequences at the 3′-UTR end (Fig. 3A). When the corresponding RNAs with artificially extended 3′ ends were compared with the RNA having an authentic 3′-UTR, translation enhancement was strongest with the latter (Fig. 3B). This confirms that a precise 3′ end of the HCV 3′-UTR (corresponding to that of the authentic viral RNA genome) is important for translation enhancement.
FIG. 3.
Additional nucleotides at the RNA 3′-terminal end impair translation enhancement by the 3′-UTR. (A) The different RNAs (arrows) without or with the HCV 3′-UTR at their 3′ ends either have the additional nucleotide extensions derived from vector sequence as indicated or terminate exactly either at the FLuc stop codon or at the authentic 3′ end of the HCV 3′-UTR. All templates were generated by PCR. (B) Relative activities of FLuc (Rel. Fluc act.) in Huh-7 cell lysates harvested at 4 h after transfection with the reporter RNAs specified in panel A. nt ext, nucleotide extensions; Ub, ubiquitin; C, partial core protein sequences.
The HCV 3′-UTR enhances IRES-dependent translation preferentially in human liver-derived cell lines.
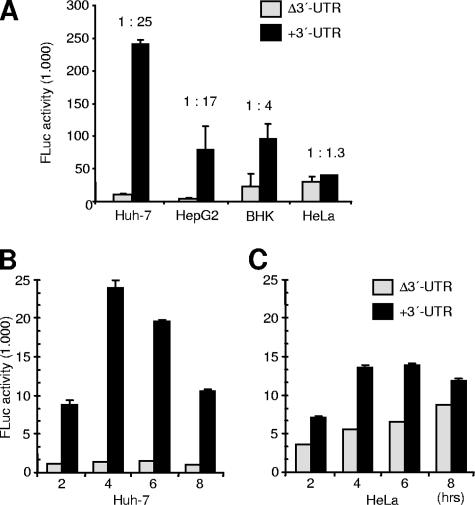
The results described so far suggest that the HCV 3′-UTR can remarkably enhance IRES-mediated translation in a human liver cell-based translation system but not in a nonliver system such as RRL, consistent with the fact that HCV preferentially infects liver cells. Nevertheless, the HCV IRES is active also in nonliver-derived cells (22, 42, 52). Therefore, we analyzed whether the stimulatory effect of the 3′-UTR is restricted to human hepatoma cells such as Huh-7 or HepG2 or occurs also in nonhepatoma cells such as HeLa or BHK-21 cells. Reporter RNAs HCV-Δ3′-UTR and HCV+3′-UTR (Fig. 1A) were transfected into these cell lines, and luciferase activities were determined at different times after transfection. The result shows that the 3′-UTR enhances translation only 4-fold in BHK cells and 1.3-fold in HeLa cells but 25-fold in Huh-7 cells and 17-fold in HepG2 cells (Fig. 4A), indicating that the 3′-UTR preferentially enhances HCV IRES-dependent translation in a cellular environment derived from human liver cells. In a time course experiment, translation enhancement in the presence of the HCV 3′-UTR was maximally 2-fold in HeLa cells but up to 25-fold in Huh-7 cells (Fig. 4B and C).
FIG. 4.
Effect of the 3′-UTR on HCV translation in different cells. (A) FLuc activities were determined 4 h after transfection of the HCV reporter RNAs with or without 3′-UTR (Fig. 1A) in different cell lines. (B and C) FLuc activities were determined at different times (as indicated) after transfection of Huh-7 cells (B) and HeLa cells (C). The columns and bars represent the means and standard deviations, respectively, of two independent experiments.
Effects of deletions in the 3′-UTR on HCV IRES-mediated translation.
To determine the contribution of the three elements of the 3′-UTR—the variable region (VR), the poly(U · C) tract, and the 3′ X region with its three SLs—to translation stimulation, we generated a series of deletion mutants in the 3′-UTR sequence (Fig. 5A). FLuc activities expressed from these reporter RNAs after transfection in Huh-7 cells were measured over a time range of 2 to 12 h after transfection (data not shown). Since the effect of each mutation in question appeared to be similar at all time points analyzed, we display here only the values determined at 4 h posttransfection (Fig. 5B).
First, we tested if efficient reporter translation requires additional sequences downstream of the luciferase gene stop codon. Therefore, the translation efficiency of a mutant in which the luciferase stop codon was followed by a random sequence of 15 nucleotides (Fig. 5A, HCV+RD15) was compared to the translation efficiency of the Δ3′-UTR mutant RNA that directly terminates at the luciferase stop codon. After transfection into Huh-7 cells, the HCV-Δ3′-UTR RNA was translated with a similar efficiency as the HCV+RD15 RNA (Fig. 5B), indicating that the luciferase gene can be correctly translated from an RNA ending exactly at the luciferase stop codon.
Deletion of any of the three sequence elements of the HCV 3′-UTR, the VR, the poly(U · C) tract, or the 3′ X region (RNAs HCV-ΔVR, -ΔUC, and -ΔX, respectively), drastically reduced translation efficiencies down to 12 to 17% of the wild type. Also, a combination deletion of the variable region and the pyrimidine tract (RNA HCV-ΔVUC) resulted in seriously decreased translation efficiency.
When the three predicted SL structures of the 3′ X region were successively deleted (Fig. 5A), a gradual decline of RNA translation was found (Fig. 5B). Deletion of SL1 allows only 28% residual translation activity, whereas deletion of SL2 results in 50% and deletion of SL3 results in 78% activity. With combination deletions of the three stem-loops of the 3′ X region we obtained intermediate results. When SL1 was deleted in combination together with SL2, the translation efficiency was 20%. Similarly, deletion of SL1 together with SL3 resulted in 25% activity. These results suggest that the combination of SL1 together with the other SLs may contribute to translation stimulation cooperatively.
The 3′ ends of the various deletion mutant RNAs described above differ in their predicted secondary structures. Only some of these reporter RNAs have very stable RNA secondary structures at their 3′ ends, which may protect the RNA in question from degradation by exonucleases. We therefore determined the amounts of intracellular reporter RNA by an RNase protection assay (Fig. 5C). All transfected RNAs were detected in comparable amounts, indicating that differences in RNA stability do not account for the differences in translation efficiency. Moreover, since it could be argued that after liposome-mediated transfection a significant proportion of the RNA may remain associated with the lipid reagent after cellular uptake and may thereby also be protected against degradation by RNases, we verified the results after transfection by electroporation (Fig. 5D). Only slight variations of the amounts of intracellular reporter RNAs were found that could not account for the up to 20-fold difference detected in the translation assays (Fig. 5B). Thus, differences in translation efficiency are not due to different half-lives of transfected reporter RNAs.
The 3′-UTR stimulates translation of unmodified full-length HCV genomes.
To demonstrate that the results obtained with the short reporter RNAs which contain only the HCV IRES and 3′-UTR sequences flanking a heterologous reporter gene are biologically relevant, we used full-length HCV RNA genomes. Genomic RNAs of the isolate Con1 with or without the 3′-UTR (Fig. 6A) were generated in vitro by transcription and transfected into Huh-7 cells by electroporation. Precise 3′ ends were generated by autocatalytic cleavage mediated by the HDV ribozyme present immediately downstream of the HCV sequence. When the expression of the NS5B protein from the full-length HCV genomes was analyzed at different time points after transfection by Western blotting (Fig. 6B, upper panel), a significantly higher expression of NS5B was detected with RNAs carrying the 3′-UTR (lanes 1, 3, and 5) compared to RNAs lacking the 3′-UTR (lanes 2, 4, and 6). Detection of actin in the same protein samples was used to confirm equal protein loads in each lane (Fig. 6B, lower panel). RNase protection assays (Fig. 6C, upper panel) using an antisense RNA probe complementary to a short stretch of the variable region of the 3′-UTR and to about 300 nucleotides of the NS5B coding region show that different RNA amounts do not account for the differences in NS5B expression observed in the Western blots. An RNase protection assay with GAPDH antisense RNA probe was used as internal control (Fig. 6C, lower panel).
FIG. 6.
The HCV 3′-UTR enhances translation of full-length genomes. (A) Structure of the transfected RNAs. Full-length HCV genome RNAs including the 3′-UTR (upper panel) or not (lower panel) were in vitro transcribed using T7 RNA polymerase. Precise 3′ ends were generated after transcription by autocatalytic RNA cleavage mediated by the HDV genomic ribozyme fused downstream to the HCV sequence. (B) Western blots with extracts of Huh-7 cells harvested at different times after electroporation transfection with the genomic HCV RNAs shown in panel A. Proteins were detected with antibodies directed against HCV NS5B protein (upper panel) or actin (lower panel). (C) RNA stability control. Total RNA was harvested from Huh-7 cells at the indicated times after electroporation as in panel B, purified, and analyzed by RNase protection assay using 0.35 pmol of the [α-32P]UTP-labeled antisense NS5B RNA probe specified in panel A. It contains 15 nucleotides of linker sequence, 29 nucleotides of the variable region, and 298 nucleotides of NS5B sequence (upper panel). As a control, the same RNA samples were analyzed using GAPDH antisense RNA (lower panel). In lanes 1, 10% of the undigested antisense RNA probes was loaded. (D) Western blotting as in panel B with antibodies directed against HCV NS5B protein (upper panel) or actin (lower panel). However, the HCV genomes used were the same as shown in panel A but with an NS5B polymerase GND mutation. hrs p.t., hours posttransfection.
Although previous studies indicated that it is unlikely that replication adds to the higher NS5B levels obtained with replicon-competent HCV RNA that contains the 3′-UTR (27, 56, 57), we wanted to rule out that possibility. Therefore, we repeated the assay by using full-length HCV RNA genomes of the same structure (Fig. 6A) but with a mutation to render NS5B polymerase inactive. In these mutants, the GDD motif (NS5B amino acids 317 to 319) was changed to GND, abrogating NS5B polymerase activity (31). Also with these RNAs, higher amounts of NS5B were detected after transfection of HCV RNA genomes with the 3′-UTR (Fig. 6D, lanes 4 to 6) compared with those lacking the 3′-UTR (lanes 1 to 3).
These results confirm that the 3′-UTR enhances RNA translation from the HCV IRES.
DISCUSSION
In this study, we demonstrate that the 3′-UTR of HCV has a strong stimulatory effect on HCV IRES-mediated translation. By employing transfection of monocistronic reporter RNAs with precise 3′ ends into a human liver-derived cell line, we show that all three sequence elements of the HCV 3′-UTR—the VR, the poly(U · C) tract, and the highly conserved 3′ X region—contribute to efficient translation stimulation. This stimulation by the HCV 3′-UTR was also observed in full-length HCV RNA genomes.
Several groups have investigated a possible modulating effect of the HCV 3′-UTR on translation directed by the HCV IRES. However, a controversial situation arose from the results obtained up to now. Some studies reported a stimulatory effect of the 3′-UTR on HCV translation (19, 20, 34, 35); others showed that the HCV 3′-UTR has no stimulatory influence on HCV IRES-directed translation (9, 10, 15, 26, 56). One study even reported an inhibitory effect of the 3′-UTR (37).
As shown here, some of these discrepancies may be explained by reporter construct design, time points of analysis, and expression systems. We found that efficient translation stimulation depends strictly on a precise 3′ terminus of the HCV 3′-UTR, whereas additional 3′-terminal nucleotides reduce translation enhancement (Fig. 2 and 3). We argue that previous studies did not detect translation stimulation by the 3′-UTR because either circular plasmid DNA or plasmids linearized not exactly at the HCV 3′ end were used (9, 15, 26, 34). Moreover, nuclear processing such as capping, splicing, and polyadenylation of cellular polymerase II transcripts may have profound effects on their biological activity, consistent with our observation that translation stimulation by the 3′-UTR is much stronger when RNA rather than DNA is used for transfection (Fig. 2B and C). The requirement for a precise 3′ end of reporter constructs is also strikingly obvious from the observation that an in vitro transcribed replicon RNA with additional nucleotides at the 3′ terminus regained the original HCV 3′ end by the loss of these additional nucleotides after transfection into cells and replication passages (56).
Another common feature of the previous studies reporting a translation enhancement by the 3′-UTR (19, 20, 34, 35) is the use of monocistronic reporter constructs similar to those used here. In contrast, the studies which did not reveal translation stimulation (10, 56) were based on dicistronic reporter constructs employing an additional (picornavirus) IRES element to direct translation of HCV nonstructural proteins. The reason that translation stimulation is not found with such dicistronic reporter constructs is not clear. Nevertheless, future studies investigating the effects of mutations in the 3′-UTR in RNA translation should be performed with monocistronic reporter systems.
By using authentic HCV full-length RNA genomes, we confirmed that the 3′-UTR is required for translation stimulation and found that viral sequences or gene products do not inhibit RNA translation, at least at an early stage of genome RNA utilization. This finding does not, however, exclude the possibility that viral sequences such as the E2 coding region or E2 itself may influence translation, as shown in an in vitro system using RRL (36), although these authors found virtually no stimulation of translation by the 3′-UTR in the absence of E2 sequences.
HCV replicates preferentially in human hepatocytes (28), suggesting that the liver cell may provide factors supporting HCV RNA translation and replication. Such a tropism could be conferred by proteins binding to the 3′-UTR and leading to translation enhancement, e.g., by an RNA 3′-5′ end interaction. Even though a certain degree of translation stimulation by the 3′-UTR was reported also for nonliver-derived systems like RRL (19, 20, 35) and HeLa cells (34), we observed that translation enhancement by the 3′-UTR is much stronger in hepatoma cell lines such as Huh-7 and HepG2 cells compared to nonhepatoma cells. Thus, unknown factors differing in amounts between liver cells and nonliver cells may be required for efficient HCV RNA translation. The mechanism by which RNA translation is enhanced thus far is not known. However, while this article was under preparation, Bradrick and coworkers reported that translation stimulation by the 3′-UTR may be brought about by facilitating translation termination and, eventually, subsequent ribosome recycling for additional rounds of translation (5).
The cis-acting signals in the 3′-UTR that we found here to be involved in translation regulation partially coincide with the signals required for RNA replication (i.e., the initiation of viral minus-strand synthesis) (10, 25, 55-57). All three regions, the VR, the poly(U · C) tract, and the 3′ X region appear to contribute to both processes, replication and translation stimulation. However, for replication the variable region is less important than the pyrimidine stretch and the 3′ X region (10, 55, 56). Of the three highly conserved SL structures of the 3′ X region, SL2 only moderately contributes to translation enhancement, and SL3 contributes even less. In contrast, for replication (i.e., RNA minus-strand synthesis) both SL2 and SL3 are essential. Also the most 3′-terminal SL1 is indispensable for RNA synthesis (10, 56, 57) and enhancement of RNA translation. This observation suggests that SL1 may be involved in the regulation of a switch from translation of the viral RNA genome to RNA minus-strand synthesis such as has been shown for poliovirus (12). This process may require long-range RNA-RNA interactions and even genome circularization, involving RNA sequences and/or secondary structures not only in the 3′-UTR but also in the NS5B coding region and in the 5′-UTR as well as viral proteins such as NS5B (11, 29, 48, 58).
In the closely related bovine viral diarrhea virus, the sequences and secondary structures of the RNA signals in the 3′-UTR are quite different from those in the HCV 3′-UTR. The two conserved SLs of bovine viral diarrhea virus that are located most 3′-terminal are required for RNA minus-strand synthesis (59). In contrast, the upstream variable region binds NFAR (nuclear factor associated with double-stranded RNA) proteins and may be involved in a switch between translation and replication (16, 17). Extending this idea to the situation with HCV, several questions arise: (i) are factors such as the NFAR group proteins involved in HCV translation enhancement by mediating long-range interactions between the 3′- and the 5′-UTR; (ii) are there synergistic interactions of NFAR proteins with other proteins binding to the 3′-UTR or with NSAP1 binding near the 5′-UTR (23); (iii) are there factors in human liver-derived cells that confer at least a certain degree of tissue specificity to HCV translation by interacting preferentially with the 3′-UTR; and (iv) are such proteins involved in a switch between translation and replication during the life cycle of the virus.
Acknowledgments
We thank Volker Lohmann for stimulating discussions and Sven-Erik Behrens and Hartmut Kleinert for plasmids.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 535 and GK 370 to M.N.) and a grant of the Bristol-Myers Squibb foundation (to R.B.).
Footnotes
Published ahead of print on 13 September 2006.
REFERENCES
- 1.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. [DOI] [PubMed] [Google Scholar]
- 2.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Blight, K. J., and C. M. Rice. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 71:7345-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradrick, S. S., R. W. Walters, and M. Gromeier. 2006. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 34:1293-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, E. A., H. Zhang, L. H. Ping, and S. M. Lemon. 1992. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 20:5041-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 8.Chung, R. T., and L. M. Kaplan. 1999. Heterogeneous nuclear ribonucleoprotein I (hnRNP-I/PTB) selectively binds the conserved 3′ terminus of hepatitis C viral RNA. Biochem. Biophys. Res. Commun. 254:351-362. [DOI] [PubMed] [Google Scholar]
- 9.Fang, J. W., and R. W. Moyer. 2000. The effects of the conserved extreme 3′ end sequence of hepatitis C virus (HCV) RNA on the in vitro stabilization and translation of the HCV RNA genome. J. Hepatol. 33:632-639. [DOI] [PubMed] [Google Scholar]
- 10.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friebe, P., J. Boudet, J. P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gontarek, R. R., L. L. Gutshall, K. M. Herold, J. Tsai, G. M. Sathe, J. Mao, C. Prescott, and A. M. Del Vecchio. 1999. hnRNP C and polypyrimidine tract-binding protein specifically interact with the pyrimidine-rich region within the 3 ′ NTR of the HCV RNA genome. Nucleic Acids Res. 27:1457-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hentze, M. W. 1997. eIF4G: a multipurpose ribosome adapter? Science 275:500-501. [DOI] [PubMed] [Google Scholar]
- 15.Imbert, I., M. Dimitrova, F. Kien, M. P. Kieny, and C. Schuster. 2003. Hepatitis C virus IRES efficiency is unaffected by the genomic RNA 3′ NTR even in the presence of viral structural or non-structural proteins. J. Gen. Virol. 84:1549-1557. [DOI] [PubMed] [Google Scholar]
- 16.Isken, O., C. W. Grassmann, R. T. Sarisky, M. Kann, S. Zhang, F. Grosse, P. N. Kao, and S. E. Behrens. 2003. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 22:5655-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isken, O., C. W. Grassmann, H. Yu, and S. E. Behrens. 2004. Complex signals in the genomic 3′ nontranslated region of bovine viral diarrhea virus coordinate translation and replication of the viral RNA. RNA 10:1637-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, T., and M. M. Lai. 1997. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J. Virol. 71:8698-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito, T., and M. M. C. Lai. 1999. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology 254:288-296. [DOI] [PubMed] [Google Scholar]
- 20.Ito, T., S. M. Tahara, and M. M. C. Lai. 1998. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J. Virol. 72:8789-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Junker-Niepmann, M., R. Bartenschlager, and H. Schaller. 1990. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 9:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamoshita, N., K. Tsukiyama Kohara, M. Kohara, and A. Nomoto. 1997. Genetic analysis of internal ribosomal entry site on hepatitis C virus RNA: implication for involvement of the highly ordered structure and cell type-specific transacting factors. Virology 233:9-18. [DOI] [PubMed] [Google Scholar]
- 23.Kim, J. H., K. Y. Paek, S. H. Ha, S. Cho, K. Choi, C. S. Kim, S. H. Ryu, and S. K. Jang. 2004. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol. Cell. Biol. 24:7878-7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong, L. K., and P. Sarnow. 2002. Cytoplasmic expression of mRNAs containing the internal ribosome entry site and 3′ noncoding region of hepatitis C virus: effects of the 3′ leader on mRNA translation and mRNA stability. J. Virol. 76:12457-12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 29.Lee, H., H. Shin, E. Wimmer, and A. V. Paul. 2004. cis-acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J. Virol. 78:10865-10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 31.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 33.Luo, G. 1999. Cellular proteins bind to the poly(U) tract of the 3′ untranslated region of hepatitis C virus RNA genome. Virology 256:105-118. [DOI] [PubMed] [Google Scholar]
- 34.McCaffrey, A. P., K. Ohashi, L. Meuse, S. Shen, A. M. Lancaster, P. J. Lukavsky, P. Sarnow, and M. A. Kay. 2002. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol. Ther. 5:676-684. [DOI] [PubMed] [Google Scholar]
- 35.Michel, Y. M., A. M. Borman, S. Paulous, and K. M. Kean. 2001. Eukaryotic initiation factor 4G-poly(A) binding protein interaction is required for poly(A) tail-mediated stimulation of picornavirus internal ribosome entry segment-driven translation but not for X-mediated stimulation of hepatitis C virus translation. Mol. Cell. Biol. 21:4097-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morikawa, K., T. Ito, H. Nozawa, M. Inokuchi, M. Uchikoshi, T. Saito, K. Mitamura, and M. Imawari. 2006. Translational enhancement of HCV RNA genotype 1b by 3′-untranslated and envelope 2 protein-coding sequences. Virology 345:404-415. [DOI] [PubMed] [Google Scholar]
- 37.Murakami, K., M. Abe, T. Kageyama, N. Kamoshita, and A. Nomoto. 2001. Down-regulation of translation driven by hepatitis C virus internal ribosomal entry site by the 3′ untranslated region of RNA. Arch. Virol. 146:729-741. [DOI] [PubMed] [Google Scholar]
- 38.Niepmann, M., A. Petersen, K. Meyer, and E. Beck. 1997. Functional involvement of polypyrimidine tract-binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 71:8330-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ochs, K., R. C. Rust, and M. Niepmann. 1999. Translation initiation factor eIF4B interacts with a picornavirus internal ribosome entry site in both 48S and 80S initiation complexes independently of initiator AUG location. J. Virol. 73:7505-7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochs, K., A. Zeller, L. Saleh, G. Bassili, Y. Song, A. Sonntag, and M. Niepmann. 2003. Impaired binding of standard initiation factors mediates poliovirus translation attenuation. J. Virol. 77:115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynolds, J. E., A. Kaminski, H. J. Kettinen, K. Grace, B. E. Clarke, A. R. Carroll, D. J. Rowlands, and R. J. Jackson. 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 14:6010-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sachs, A. B., P. Sarnow, and M. W. Hentze. 1997. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell 89:831-838. [DOI] [PubMed] [Google Scholar]
- 44.Song, Y., E. Tzima, K. Ochs, G. Bassili, H. Trusheim, M. Linder, K. T. Preissner, and M. Niepmann. 2005. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA 11:1809-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spangberg, K., L. Goobar-Larsson, M. Wahren-Herlenius, and S. Schwartz. 1999. The La protein from human liver cells interacts specifically with the U-rich region in the hepatitis C virus 3′ untranslated region. J. Hum. Virol. 2:296-307. [PubMed] [Google Scholar]
- 46.Tanaka, T., N. Kato, M. J. Cho, K. Sugiyama, and K. Shimotohno. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 70:3307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarun, S. Z. Jr., and A. B. Sachs. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15:7168-7177. [PMC free article] [PubMed] [Google Scholar]
- 48.Thurner, C., C. Witwer, I. L. Hofacker, and P. F. Stadler. 2004. Conserved RNA secondary structures in Flaviviridae genomes. J. Gen. Virol. 85:1113-1124. [DOI] [PubMed] [Google Scholar]
- 49.Tsuchihara, K., T. Tanaka, M. Hijikata, S. Kuge, H. Toyoda, A. Nomoto, N. Yamamoto, and K. Shimotohno. 1997. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J. Virol. 71:6720-6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Kräusslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, C., P. Sarnow, and A. Siddiqui. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wood, J., R. M. Frederickson, S. Fields, and A. H. Patel. 2001. Hepatitis C virus 3′X region interacts with human ribosomal proteins. J. Virol. 75:1348-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamada, N., K. Tanihara, A. Takada, T. Yorihuzi, M. Tsutsumi, H. Shimomura, T. Tsuji, and T. Date. 1996. Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology 223:255-261. [DOI] [PubMed] [Google Scholar]
- 55.Yanagi, M., M. St Claire, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yi, M., and S. M. Lemon. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yi, M., and S. M. Lemon. 2003. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9:331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.You, S., D. D. Stump, A. D. Branch, and C. M. Rice. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 78:1352-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu, H., C. W. Grassmann, and S. E. Behrens. 1999. Sequence and structural elements at the 3′ terminus of bovine viral diarrhea virus genomic RNA: functional role during RNA replication. J. Virol. 73:3638-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]