Abstract
We investigated the role of the 5′-untranslated region between the primer binding site and the gag initiation codon in ovine lentivirus maedi-visna virus (MVV) genomic RNA encapsidation. We identified five computer-predicted stem-loops, three of which were highly conserved in primary sequence and structure. One stable 83-nucleotide (nt) stem-loop (SL4) was not conserved in the primary sequence, but phylogenetic analysis revealed several base pair covariations. The deletion of individual stem-loops did not markedly affect the relative encapsidation efficiency (REE). Only one mutant, carrying a disruption of a 31-nt stem-loop (SL5), had 58% REE in fetal ovine synovial (FOS) cells. A 168-nt deletion (Δ3MSD) downstream of the major splice donor (MSD) which removed three stem-loops, including SL5, resulted in 24% and 20% REE in FOS and 293T cells, respectively. A 100-nt deletion (Δ5MSD) upstream of the MSD resulted in 15-fold lower cellular genomic RNA levels than the wild-type levels in 293T cells. The Δ5MSD mutant and a double mutant (DM) (Δ5MSD and Δ3MSD) did not express detectable levels of virion proteins in 293T cells. In contrast, the region deleted in Δ5MSD was dispensable in FOS cells, and the DM had the same REE as the Δ3MSD virus. Thus, the region upstream of the MSD contains sequences critical for RNA and protein expression in a cell type-specific fashion. Our results indicate that MVV encapsidation determinants are located downstream of the MSD. These results provide comparative insight into lentiviral encapsidation and can be utilized in the design of MVV-based gene transfer vectors.
The 5′-untranslated region (5′-UTR) in retrovirus genomes contains functional cis-acting stem-loop structures that are involved in various critical steps of the retrovirus life cycle. These steps are regulation of RNA transcription, mRNA splicing, initiation of translation, dimerization of genomic RNA, encapsidation of the viral genomic RNA into virions, and initiation of reverse transcription (6, 13, 44). While transcription, splicing, and initiation of reverse transcription are associated with short and well-defined regions, translation and encapsidation determinants are less well defined in length and structure.
Retroviral genomic RNA is the predominant RNA in virions, yet it constitutes only 1% of the total RNA in the cytoplasm of the infected cell. Thus, the process of encapsidation of two copies of retroviral genomic RNA in the assembling virion is specific. Encapsidation requires interaction of the nucleocapsid (NC) region of the Gag polyprotein with conserved RNA motifs in the 5′-UTR, collectively referred to as encapsidation determinants (Ψ). In mammalian retroviruses, these RNA determinants are often located between the major splice donor (MSD) and the gag initiation codon (8). Thus, splicing to generate various retroviral mRNAs removes cis encapsidation determinants. This arrangement promotes encapsidation of genomic RNA but excludes spliced subgenomic RNAs.
Maedi-visna virus (MVV) is a retrovirus belonging to the lentivirus family. MVV infects sheep, resulting in chronic progressive pulmonary disease (maedi) and degenerative central nervous system pathology (visna) (51, 56). A recent publication by Monie et al. describes the structure of the MVV leader region and identifies the dimer initiation site (DIS) in a stem-loop located just upstream of the MSD (43). No data have been reported on MVV encapsidation determinants. Identification of the MVV encapsidation determinants is of interest for better understanding lentivirus replication and also for the design of MVV vectors suitable for gene transfer.
Genomic RNA encapsidation has been studied thoroughly in the closely related lentivirus human immunodeficiency virus type 1 (HIV-1) (35). Models of HIV-1's encapsidation determinants consist of four stem-loop structures (SL1 through SL4) in the 5′-UTR. These structures overlap the MSD and extend into the proximal gag gene (5, 15, 26). The contribution of these stem-loop structures to encapsidation has been suggested on the basis of deletion mutant analysis, biochemical analysis, phylogenetic comparisons, and computer modeling of RNA secondary structures. The HIV-1 encapsidation determinants appear to be multipartite and context dependent, and a minimal essential encapsidation determinant has not yet been defined (35). SL1 has a highly conserved six-base palindromic sequence in the terminal loop which is the DIS (7, 57). The mechanism by which dimerization initiation is believed to take place is through a “kissing-loop complex” of the two monomeric RNA strands (34, 48). The weak RNA dimer is further converted to a stable, noncovalently linked dimer as the virus particle matures. This seems to be a common feature for other retroviruses besides HIV-1, including MVV (12, 20, 21). Whether retroviral RNA dimerization and encapsidation are independent events or whether RNA dimerization is a prerequisite for HIV-1 encapsidation remains unclear (54). SL2 in HIV-1 contains the MSD in its loop and is capable of binding the NC with high affinity (2). Its contribution to encapsidation is generally thought to be minimal, as deleting it has no effect on encapsidation (27, 37-39). SL3 has a conserved GGAG motif in its loop and binds the NC with high affinity (15). It has been shown in a number of studies to be the major encapsidation determinant (reviewed in reference 18). Recently, it was reported that the SL3 structure is also required for dimerization, indicating that SL1 may constitute only part of the dimer linkage structure (53). SL4 encompasses the gag initiation codon. It binds weakly to the NC and is believed to stabilize the structure of the encapsidation site (3).
The encapsidation determinants of HIV-2 appear to be located upstream of the MSD, and sequences between the MSD and the gag initiation codon contribute negligibly to encaphpsidation (41). The selectivity of full-length RNA encapsidation, as opposed to encapsidation of spliced RNA species, is believed to be maintained by cotranslational encapsidation of the genomic RNA encoding the Gag polyprotein, with limited availability of the Gag polyprotein to capture other RNAs (23, 31, 32). Encapsidation determinants are also located upstream of the MSD in simian immunodeficiency virus (SIV) (50, 59) and in the nonprimate lentivirus feline immunodeficiency virus (FIV) (14, 33). Sequences elsewhere in some lentiviral genomes have also been shown to contribute to encapsidation, such as the TAR loop and R/U5 sequences in HIV-1 (25, 28, 40) and sequences extending into the gag open reading frame in FIV and HIV-1 (14, 33, 37, 49).
Elements of the HIV-1 UTR can fold in vitro into two alternative conformations, namely, the long-distance interaction structure (LDI) and the branched multiple-hairpin structure (BMH) (30). It has been demonstrated that an LDI-BMH riboswitch regulates genomic RNA dimerization (30), and possibly encapsidation, when the encapsidation hairpins are in the right structural context in the BMH conformation (1, 46).
Several gene transfer vectors have been constructed based on nonprimate lentiviruses, such as equine infectious anemia virus (EIAV), bovine immunodeficiency virus (BIV), and FIV (9, 45, 52). A detailed knowledge of cis encapsidation determinants is necessary to meet the requirements of a safe gene transfer system, where cis encapsidation determinants are included in the vector construct and excluded from the packaging constructs. The main emphasis in these gene transfer systems has been on including putative viral encapsidation regions in the vector construct for high-titer vector encapsidation, but less attention has been focused on determining whether the packaging construct RNA is excluded from encapsidation into viral particles. Attempts have been made to construct a gene transfer system based on MVV (10). The vectors transduced cells very inefficiently, presumably due to a block in postentry steps, such as reverse transcription or integration of the vector.
Working towards the goal of better understanding MVV biology and developing safe MVV-based vectors, we mapped MVV encapsidation determinants by computer modeling of RNA secondary structures, phylogenetic comparison, and deletion mutant analysis in the region between the primer binding site (PBS) and the gag initiation codon. We show that encapsidation determinants are located downstream of the MSD. The region upstream of the MSD contributes negligibly to encapsidation in fetal ovine synovial (FOS) cells. In contrast, this upstream region appears to contain signals for efficient viral genomic RNA expression in human 293T cells. None of the deletion mutants completely abolished encapsidation, indicating that encapsidation determinants are also located elsewhere in the MVV genome.
MATERIALS AND METHODS
Sequence analysis.
The Mfold program (64; http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html) was used to predict the most stable RNA secondary structure in the MVV 5′-UTR. Mfold is an algorithm that lists intramolecular base pairings (i.e., secondary structures) corresponding to the lowest equilibrium for free energy (ΔG). The maximum base pair distance was set at 100 nucleotides (nt). The computer program CLUSTALWPROF (http://workbench.sdsc.edu/) was used to align the sequences. The sequences of the cloned molecular isolates used in the alignments were as follows (GenBank accession numbers are given in parentheses): Icelandic ovine strain 1514 (M379772), British ovine strain EV1 (S51392), North American ovine strain NA-MVV (85/34) (AY101611), Portuguese ovine strain P10LV (AF479638), South African ovine strain SA-OMVV (M31646), and a caprine arthritis-encephalitis virus (CAEV) strain (M33677). To compare the lengths of the leader regions between the PBS and the gag initiation codon, we used published sequences of BIV (M32690), EIAV (M16575), FIV (M25381), HIV-1 (AF033819), HIV-2 (M15390), and SIV (M19499).
Construction of deletion mutants.
The infectious molecular clone LV1-1KS2 in pKS2 (58) was used as the parental wild-type virus, and its sequence was used as a reference (GenBank accession no. M37977). Numbering was relative to the first nucleotide in the viral RNA. The deletion mutants were generated by overlap extension PCR. The primers used for primary PCRs to generate two DNA fragments with overlapping ends were as follows: for deletion Δ5MSD (deletion of nt 190 to 290) (Fig. 1), forward primer V23F (5′-CCTACCCTGGAAAATGACT-3′) with reverse complementary primer V443B-DEL (5′-TCAGGCGTCATTCTTTGTCGAGCCCCACG-3′) (nucleotides in italics represent the complementary sequence for overlap) and forward complementary primer V542F-DEL (5′-ACAAAGAATGACGCCTGAAGTAAGGTAAG-3′) with reverse primer V787B (5′-TGCACTGTCTCGGGTTTCAT-3′); for deletion Δ3MSD (deletion of nt 312 to 480), forward primer V97F (5′-AGCTTGCCTGGTTATTATCG-3′) with complementary overlap primer V294B-DEL (5′-GTCTCTAGCCTCTTACCTTACTTCAGGC-3′) and forward complementary primer V481F-DEL (5′-TAAGGTAAGAGTGCTTCGCCATGTCTCTAGC-3′) with reverse primer V787B; for deletion ΔSL1 (deletion of nt 224 to 245), forward primer V97F with reverse complementary primer B-DEL-SL1 (5′-TACTCTCCTTCCGTGGTCCCATAACTC-3′) and forward complementary primer F-DEL-SL1 (5′-CTGGTGCCTAGGAGAGTAACGGACACG-3′) with reverse primer V787B; for deletion ΔSL2 (deletion of nt 282 to 301), forward primer V97F with reverse complementary primer B-DEL-SL2 (5′-TCTTACCTTTTTCTTTCACTTTTTGTC-3′) and forward complementary primer F-DEL-SL2 (5′-ACTTTCTTTAAGGTAAGAGAGACACCT-3′) with reverse primer V787B; for deletion ΔSL3 (deletion of nt 317 to 333), forward primer V97F with reverse complementary primer B-DEL-SL3 (5′-GGGCTATTCTGTCTCTCTTACCTTACT-3′) and forward complementary primer F-DEL-SL3 (5′-TCTCTCTGTGAATAGCCCTTCAGTGAA-3′) with reverse primer V787B; for deletion ΔSL4 (deletion of nt 359 to 441), forward primer V97F with reverse complementary primer B-DEL-SL4 (5′-GTTGCTCTTTTTCTCCTTCACTGAAGG-3′) and forward complementary primer F-DEL-SL4 (5′-TTCCTCTTAAGAGCAACACTGGTAAG-3′) with reverse primer V787B; and for deletion ΔSL5 (deletion of nt 445 to 479), forward primer V97F with reverse complementary primer B-DEL-SL5 (5′-TCTCTAGCTCTTGTGTCCCCTCGGCCC-3′) and forward complementary primer F-DEL-SL5 (5′-CCTGTGTTCAGCTAGAGACATGGCGAA-3′) with reverse primer V787B.
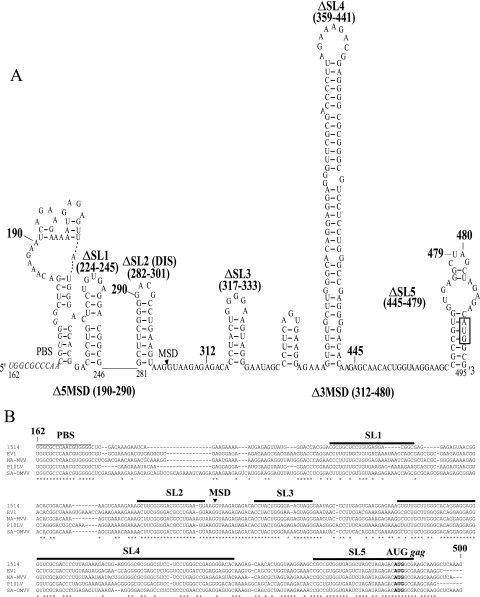
FIG. 1.
(A) Predicted stable RNA secondary structures and locations of deletion mutations in the region between the PBS and the gag initiation codon of Icelandic MVV isolate 1514 (molecular clone KS2). Folding was carried out on a fragment spanning nt 79 to 579. Nucleotides 162 to 495 are illustrated. The names and positions of the deletion mutations in the viral RNA are indicated in bold. The PBS is indicated with italic letters, the MSD is indicated by an arrowhead, and the gag initiation codon is indicated with a box. (B) Alignment analysis of the regions between the PBS and the gag initiation codon of five replication-competent MVV isolates. The 1514 sequences corresponding to the computer-predicted stem-loops deleted in this study are indicated with horizontal bars. The PBS, the MSD, and the gag start codon are shown. Conserved nucleotides are indicated by asterisks below the alignment. Numbering refers to positions in the 1514 genomic RNA.
Each reaction mix (50 μl) contained 0.1 ng of pKS2, 0.5 μM (each) primer, a 200 μM concentration of each deoxynucleoside triphosphate, 7 U of KlenTaq1 polymerase (AB Peptides), 0.05 U of PfuI polymerase (Stratagene), and 1× RDA buffer (36). Samples were denatured at 94°C for 5 min, followed by 35 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min, with a final extension step at 72°C for 7 min. The two amplified products were fractionated by electrophoresis through a 1.7% agarose gel and purified with a gel band purification kit (Amersham Biosciences Inc., Piscataway, NJ). The primary PCR products were subsequently joined in an overlap extension reaction, using 5 ng of each product, for 5 cycles at 94°C for 1 min, 50°C for 1 min, and 72°C for 1 min. Each primer (0.5 μM) was then added, and 25 cycles of amplification were performed at 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min, with a final extension step at 72°C for 7 min. The full-length overlap products were purified with a GFX PCR DNA purification kit (Amersham Biosciences Inc., Piscataway, NJ) and digested with KasI and BspEI, except for deletion Δ3MSD, which was digested with NheI and BspEI.
An intermediate plasmid to subclone the deletions was constructed by ligating a 1,605-bp EagI-SpeI fragment from pKS2, containing the complete 5′ long terminal repeat (LTR), the 5′-UTR, and the 5′ part of the gag coding region (nt 490 to 1063), into EagI-SpeI-digested pBlueScript II KS(+) (Stratagene), generating pBS-5′ KS2. The digested PCR products were then inserted into KasI-BspEI- or NheI-BspEI-digested pBS-5′KS2. Finally, the seven subcloned deletions were digested with EagI-SpeI, and the resulting fragments were used to replace a 1,605-bp EagI-SpeI fragment in pKS2, generating pΔ5MSD, pΔ3MSD, pΔSL1, pΔSL2, pΔSL3, pΔSL4, and pΔSL5 (Fig. 1A). To generate the double deletion mutant (DM [Δ5MSD and Δ3MSD]), pΔ3MSD was used as a template in primary PCRs, using forward primer V23F with reverse complementary primer V443B-DEL and forward complementary primer V542F-DEL with reverse primer V787B. The resulting PCR product from the overlap extension reaction was digested with NheI and BspEI and inserted into a corresponding site in pBS-5′ KS2. This subclone was then digested with EagI and SpeI, and the resulting fragment was used to replace a 1,605-bp EagI-SpeI fragment in pKS2 to generate pDM. All deletion mutations were confirmed by sequencing of plasmids in both the forward and reverse directions, using an ABI Prism BigDye Terminator cycle sequencing ready reaction kit (PE Biosystems, Foster City, CA) and an ABI Prism 377 DNA sequencer.
Cells and transfection.
FOS and 293T cells were grown in Dulbecco's modified Eagle's medium (Gibco; Invitrogen) supplemented with 10% fetal calf serum, 100 IU/ml penicillin-streptomycin, and 2 mM glutamine. FOS cells were transfected with 2.5 μg of each plasmid, using SuperFect transfection reagent (QIAGEN), in 6-cm-diameter dishes according to the manufacturer's instructions for primary cells. The 293T cells were transfected by the calcium phosphate precipitation method (22), with 5.0 μg of each plasmid in 6-cm-diameter dishes. Cells and supernatants were harvested at 48 h posttransfection. Transfection efficiency was measured in two ways. Firstly, the amount of transfected plasmid present in harvested cytoplasm (60 μl of purified cellular RNA, which also includes the plasmid) was measured with a real-time quantitative reverse transcriptase PCR (qRT-PCR) assay for the gag region (24). Secondly, the percentage of transfected cells in each batch of experiments was estimated by beta-galactosidase staining of cells transfected with plasmid pCH110 (Amersham Pharmacia Biotech). Cells were fixed in a 2% paraformaldehyde solution [0.1 M piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 2 mM MgCl2, 1.25 mM EDTA, pH 6.9] and then stained with X-Gal solution [1 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (Sigma) in 35 mM K3Fe(CN)6, 35 mM K4Fe(CN)6 · 3H2O, and 2 mM MgCl2].
RNA isolation.
Supernatants from transfected cells were centrifuged at 2,000 × g for 10 min and passed through 0.45-μm filters. Virions were pelleted from the cleared supernatants by ultracentrifugation through a 20% sucrose cushion in a Beckman SW27.1 rotor at 72,100 × g for 90 min. Virion RNA was purified from virion pellets by using a commercial kit (QIAamp viral RNA mini kit), and RNA was eluted in 60 μl AVE buffer from the kit. Cytoplasmic RNA was purified using an RNeasy mini kit (QIAGEN), using the manufacturer's protocol for isolation of cytoplasmic RNA only, excluding nuclear RNA. Cytoplasmic RNA was eluted in 60 μl H2O. Cytoplasmic and virion RNA samples (19 μl) were treated with 1 U (1 μl) of RNase-free DNase I (Fermentas) at 37°C for 30 min and heat inactivated at 65°C for 10 min in 0.25 mM EDTA. Elimination of plasmid DNA contamination in DNase I-treated RNA preparations was confirmed by PCR with primers for the gag region (see below).
Reverse transcription.
DNase I-treated cytoplasmic and virion RNA preparations (11 μl) were reverse transcribed with random hexamers in 20-μl reaction mixtures by using a RevertAid H Minus first-strand cDNA synthesis kit (Fermentas) according to the manufacturer's instructions (procedure number 1).
Cloning of human β-actin.
Human β-actin (hβ-actin) mRNA (x00351) was cloned into a plasmid from a 293T cell cDNA pool as previously described for ovine β-actin mRNA (11), using forward primer 5′-CCAGCACAATGAAGATCAA-3′ and reverse primer 5′-CACGAAAGCAATGCTATCA-3′, located in exons 5 and 6, respectively, to generate a 542-bp amplicon.
Quantitative real-time PCR.
Real-time quantitative PCR assays detecting MVV genomic RNA and ovine β-actin mRNA have been described previously (11, 24). These assays are based on real-time fluorescence resonance energy transfer and are performed in a LightCycler 32 instrument (Idaho Technology, Idaho Falls, ID) (62, 63).
PCR detection of vif and human β-actin mRNAs.
To detect splicing of vif mRNA, we used the forward primer 5′-AGCTTGCCTGGTTATTATCG-3′ (nt 97 to 116), located in the U5 region 5′ of the MSD, and the reverse primer 5′-TCTTTTGGTGGCGGTATGAACTTA-3′ (nt 4992 to 4969), located in the second exon of the vif transcript. These primers should amplify products of different sizes depending on which deletion mutant construct is transfected (for details, see the legend to Fig. 4C). To detect human β-actin mRNA, we used the primers described above. The PCR sample (10 μl) contained a 200 μM concentration of each deoxynucleoside triphosphate, 0.5 μM (each) primer, 0.08 U/μl KlenTaq1 polymerase (diluted in an enzyme diluent buffer comprised of 10 mM Tris, pH 8.3, and 250 μg/ml bovine serum albumin [Idaho Technology]), and reaction buffer (50 mM Tris, pH 8.3, 250 μg/ml bovine serum albumin, and 3 mM MgCl2 [vif] or 4 mM MgCl2 [hβ-actin] [Idaho Technology]). The LightCycler qualitative PCR consisted of an initial denaturation step for 2 min at 94°C, followed by 45 cycles of nonstop (0 s) denaturation at 94°C, nonstop (0 s) annealing at 40°C (vif) or 45°C (hβ-actin), and 20 s of elongation at 72°C. Products were analyzed by electrophoresis in 1.7% agarose gels.
FIG. 4.
Cellular gag RNA levels and vif mRNA splicing of the wild-type control and deletion mutants in transfected FOS cells. (A) Levels of gag RNA, as measured by real-time qRT-PCR using gag RNA-specific primers. Measurements were normalized for ovine β-actin mRNA expression. Values are expressed as copy numbers of gag RNA per 100 copies of β-actin mRNA. Means with standard errors for at least three independent experiments are shown. (B) RT-PCR products of vif mRNA visualized after electrophoresis in 1.7% agarose. Splicing of the MSD to the splice acceptor for vif mRNA generated a 356-bp product for the wild type and for mutants with deletions located downstream of the MSD, but for the upstream mutants ΔSL1, ΔSL2, Δ5MSD, and DM, the products were expected to be 334, 336, 256, and 256 bp, respectively.
Western blotting.
Supernatants were collected at 48 h posttransfection and centrifuged at 13,200 × g for 15 min at 4°C. Viral particles were pelleted from the cleared supernatants at 100,000 × g for 90 min at 4°C. The pellets were resuspended in TNE buffer, and viral particles were purified using the high-salt purification method (60). Transfected cells were collected using a rubber policeman and washed twice with phosphate-buffered saline, with brief centrifugation and removal of the supernatant. The cells and salt-purified virion pellets were suspended in 50 μl and 30 μl 4% sodium dodecyl sulfate (SDS), respectively, lysed at 100°C for 10 min, and centrifuged at 13,200 × g in a microcentrifuge for 5 min to remove insoluble proteins. The virus and cellular protein lysates were subjected to SDS-polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and probed with a 4,000-fold dilution of murine polyclonal ascites fluid raised against the whole visna virus strain 1514 as the primary antibody. To detect bound antibody, the membranes were treated with goat anti-mouse immunoglobulin G conjugated to alkaline phosphatase (DAKOCytomation, Denmark) and visualized with a colorimetric assay using 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium (BCIP/NTP) as the substrate (Roche Diagnostics GmbH).
Quantitative RT activity assay.
Supernatants from transfected cells were collected after 48 h and clarified by spinning at 2,000 × g for 30 min. The virions were pelleted by ultracentrifugation in a Beckman SW27.1 rotor at 72,100 × g for 10 min and then suspended in lysis buffer from a commercial kit (colorimetric reverse transcriptase assay; Roche Applied Science); quantification of RT activity was performed according to the kit manufacturer's protocol. Five twofold dilutions and three replicates of each virion were assayed.
Statistical analysis.
InStat (version 3.0 for Windows) was used for all statistical calculations, including determinations of means and standard deviations. P values were computed using the Dunnett multiple comparison test, which compares medians of three or more groups to a control with a 95% confidence interval.
RESULTS
Stem-loop structures between the PBS and the gag initiation codon.
We determined whether the region between the PBS and the gag initiation codon (nt 179 to 489) in the MVV 5′-UTR contained putative stable RNA secondary structures. Mfold analysis was performed on the sequence extending from nt 79 to 589 in MVV isolate 1514. This sequence extended 100 nt on each side of the PBS and the gag initiation codon. The corresponding sequences of four divergent MVV isolates and CAEV were also analyzed. Several structures of different lengths and stabilities were predicted (Fig. 1A). We identified five stem-loop structures (designated SL1 through SL5) based on conservation of the primary sequence, phylogenetic comparison of the predicted structures, base-pairing conservation in the stems of the structures, location in the leader region, and the purine content of the terminal loop. The locations of the stem-loops in the leader sequence were in the regions of nt 224 to 245 (referred to as SL1; ΔG = −11.1 kcal/mol), nt 282 to 301 (SL2 or DIS; ΔG = −9.5 kcal/mol), nt 317 to 333 (SL3; ΔG = −6.8 kcal/mol), nt 359 to 441 (SL4; ΔG = −53.3 kcal/mol), and nt 465 to 495 (SL5; ΔG = −10.4 kcal/mol). The structure predictions for SL2, SL3, SL4, and SL5 did not change when the flanking sequences 5′ or 3′ of the nt 79-to-589 fragment were included (nt 1 to 600), implying that these segments of the genome were relatively autonomous structural elements. This was true for all of the MVV isolates and CAEV. In contrast, SL1 was not predicted with the additional 5′ or 3′ sequences.
The alignment of the MVV leader sequences of various isolates is shown in Fig. 1B. In the region upstream of the MSD, the only sequences conserved were those corresponding to the PBS and SL2. SL2 of MVV 1514 had 90% conserved identity (18 of 20 nt) with those of the other isolates and had a completely conserved structure, i.e., an 8-bp stem with a GACG loop. We also identified this structure in the CAEV leader at the same location upstream of the MSD, with a 7-bp stem and the terminal tetraloop GACG (data not shown). Very recently, and concurrent with our work, this loop was identified as the DIS in MVV (43).
The proposed SL3 of strain 1514 had 14 of 17 conserved nucleotides (82%) with the other MVV isolates. The structure was always the same in each isolate, with a 6-bp stem (completely conserved base pairing) required to maintain the structure. A slight difference between isolates was present in the nucleotides of the loop. In isolate 1514, the pentaloop sequence was GGGGA, in isolate EV1 the pentaloop sequence was GGGAA, in isolate P10LV the hexaloop sequence was GGAGAG, in isolate SA-OMVV the hexaloop sequence was GGGAAA, and in isolate NA-MVV the hexaloop sequence was GGGGAG. The differences were all due to purine transitions, so the purine richness of the loop was conserved. An SL3-like structure was not predicted for the CAEV leader region, and little conserved identity was observed in the corresponding primary sequence between MVV and CAEV (data not shown).
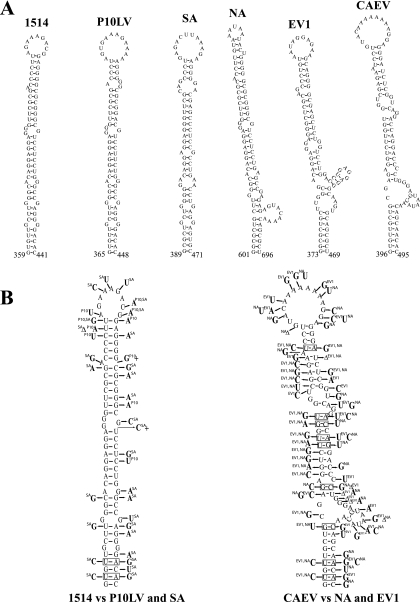
Although the primary sequence of SL4 was not highly conserved (Fig. 1B), the structure, size, and stability of this stem-loop were similar among the five MVV isolates and CAEV (Fig. 2A and Table 1). The predicted SL4 regions of isolates 1514, P10LV, and SA-OMVV were of similar sizes (83 or 84 nt) and had a high level of structural resemblance. In isolates NA, EVI, and CAEV, the predicted SL4 regions were slightly longer (96 to 100 nt) and shared a similar architecture. We performed phylogenetic comparative analysis on SL4 to support the computer-predicted RNA secondary structures. We compared the structure of SL4 predicted for isolate 1514 to the structures for P10LV and SA (Fig. 2B, left panel) and that for CAEV to the structures for NA and EV1 (Fig. 2B, right panel). The selection of isolates for comparison was due to size similarities of the predicted structures. We detected base changes on one side of the stem that were compensated for by base substitutions in the opposite strand (base pair covariation). There were two base pair covariations in the lowest part of the stem (from UA to the more stable CG) between 1514 and P10LV/SA (Fig. 2B, left panel). Sixteen base pairs were conserved, and 6 bp had neutral nucleotide changes (e.g., UG to UA). Eight base pairs had disruptive changes, and these were mainly located next to bulges or the terminal loop. Very poor conservation was detected in the primary sequence of SL4 for comparison of CAEV to EV1 and NA structures (Fig. 2B, right panel). In contrast, there was a striking conservation of regions predicted to be base paired, as evidenced by nine base pair covariations. These results imply that the structure of the predicted SL4 might serve an important biological function in MVV. The terminal loop, however, showed different sizes, with high nucleotide diversity, between the isolates. This suggests that this stem-loop might not be relevant in RNA encapsidation, since retroviral encapsidation signals are known to be hairpin structures with a highly conserved purine-rich terminal loop (8). Interestingly, the long SL4 was located between MSD and the gag initiation codon, and this region is much longer in MVV and CAEV than in the other lentiviruses (Table 2).
FIG. 2.
Phylogenetic comparison of predicted SL4 structures in different MVV isolates and CAEV. (A) Illustration of the Mfold-predicted SL4 located between the MSD and the gag initiation codon. Names of the isolates and nucleotide positions in the viral genome are indicated. (B) Phylogenetic comparison of SL4 structures. The structure of 1514 SL4 was used as a prototype and compared to those for P10LV and SA-OMVV (left panel). The structure of CAEV SL4 was also used as a prototype and compared to those for NA-MVV and EV1 (right panel). Deletions are shown with “Δ,” and insertions are shown with “+.” Nucleotide changes occurring in compared isolates are indicated, along with the isolate name in superscript. Boxes indicate base pair covariations.
TABLE 1.
Characteristics of the SL4 structure
| Isolate | ΔG (kcal/mol) | No. of nt shared with 1514 SL4/total no. of nt | Length (nt) | No. of bp | Loop size (nt) | Distance from SL3 (nt) |
|---|---|---|---|---|---|---|
| 1514 | −53.3 | 83 | 32 | 9 | 26 | |
| P10LV | −45.9 | 74/83 | 84 | 30 | 12 | 27 |
| SA | −49.8 | 55/83 | 83 | 28 | 12 | 26 |
| NA | −50.8 | 65/83 | 96 | 36 | 4 | 25 |
| EV1 | −51.2 | 53/83 | 97 | 30 | 10 | 25 |
| CAEV | −36.4 | 42/83 | 89 | 29 | 15 |
TABLE 2.
Comparison of lengths of the regions between the PBS and the gag initiation codon (AUG) in lentiviruses
| Lentivirus | Length (nt) of region
|
||
|---|---|---|---|
| PBS-AUG | PBS-MSD | MSD-AUG | |
| MVV | 312 | 126 | 186 |
| CAEV | 331 | 157 | 174 |
| BIV | 91 | 66 | 25 |
| EIAV | 123 | 119 | 4 |
| HIV-1 | 134 | 87 | 47 |
| SIV | 216 | 148 | 68 |
| HIV-2 | 227 | 153 | 74 |
| FIV | 250 | 230 | 20 |
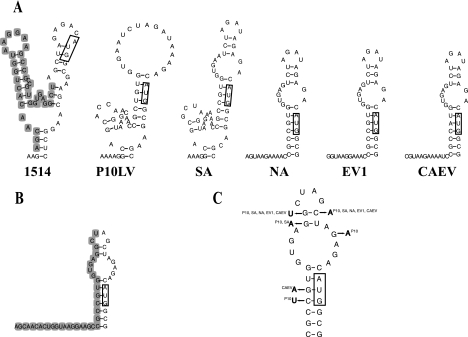
The primary sequence between SL4 and the gag initiation codon was highly conserved (Fig. 1B). Similar conservation has been described by Monie et al. (43). The computer-predicted structures in this region are illustrated in Fig. 3 for all of the isolates. The terminal loop sequence was AGAU in four of the six isolates. A 31-nt structure with a 7-bp stem encompassing the gag initiation codon appeared to be common to most of the isolates, except for isolate 1514 (Fig. 3A). Folding of only the corresponding 31-nt sequence of 1514 resulted in a similar structure to that of the other isolates (Fig. 3B). We carried out phylogenetic comparison on this structure, using the 1514 structure as a prototype (Fig. 3C). Base pairing was conserved. Three nucleotide changes were neutral with respect to base pairing, and one covariation was detected in the base pair next to the terminal loop. We assumed that this structure was significant and called it SL5.
FIG. 3.
Computer prediction of secondary structures (SL5) in the region preceding the gag initiation codon in different MVV isolates and CAEV. (A) Mfold analysis carried out with fragments corresponding to nt 79 to 589 of isolate 1514. Only sequences around the gag initiation codon are shown. Boxes indicate the gag initiation codon. The deletion of 1514 is shown with gray shading. (B) Structure prediction for isolate 1514, using only sequences corresponding to the structure common to the other isolates. (C) Phylogenetic comparison of predicted SL5 structures. The 1514 SL5 was used as a prototype. Nucleotide changes occurring in other isolates are indicated, along with the isolate name in superscript.
Genomic RNA expression of deletion mutants in FOS cells.
To test the role of the predicted stem-loop structures in MVV encapsidation, we deleted each of stem-loops SL1 to SL4 and disrupted SL5. In addition, we constructed three larger deletion mutants, including one with a deletion between nt 190 and 290 upstream of the MSD, including SL1 and SL2 (Δ5MSD); one with a deletion between nt 312 and 480 downstream of the MSD, including SL3 to SL5 (Δ3MSD); and a double mutant with both of these regions deleted (DM) (Fig. 1A). We transfected FOS cells with the wild-type control and the eight deletion mutant constructs and quantified their cellular genomic (gag) RNA expression with a real-time qRT-PCR assay (24). The transfection efficiencies were similar between the different constructs, as measured with a real-time qRT-PCR assay for plasmid content in the cytoplasm (data not shown). On average, there were 7.2 × 107 copies of plasmid per microliter of harvested cytoplasm, with a range of 4.9 × 107 to 9.8 × 107 copies. The percentage of transfected FOS cells was 15%, with a range of 10 to 20%. To correct for sample-to-sample variation of the RNA, we normalized the cellular gag RNA levels with ovine β-actin mRNA levels in each sample (11). The gag RNA levels of the deletion mutants were slightly higher than or comparable to wild-type levels, with the exception of the deletion mutant DM (Fig. 4A). This mutant showed the highest expression level, which was 7.5 times that of the wild-type virus (P < 0.05).
Deletions did not qualitatively affect mRNA splicing in FOS cells.
RNA secondary structures can influence the efficiency and specificity of mRNA splicing (4). To test whether the removal of the predicted RNA secondary structures caused aberrant mRNA splicing, we measured the vif mRNA in transfected FOS cells by RT-PCR. Correct vif mRNA splicing occurred in all of the deletion mutants, as detected by PCR products of the expected sizes upon gel electrophoresis (Fig. 4B). We detected bands of higher intensity than those of the wild-type virus for the deletion mutants Δ5MSD and DM. The reason for this difference was presumably somewhat increased splicing in these mutants, although their shorter PCR products could have been amplified with increased efficiency. For the DM mutant, this could also have reflected high cellular gag RNA levels. Furthermore, we observed that expression of wild-type and deletion mutants in FOS cells resulted in marked syncytium formation (data not shown). This fusion is known to be mediated by epitopes of the MVV envelope glycoprotein (17). Syncytium formation confirmed that splicing and translation of env took place. We conclude from these results that removal of the predicted RNA structures did not qualitatively affect splicing.
Genomic RNA contents of deletion mutant virions.
We quantified gag RNAs in wild-type and deletion mutant virions pelleted from the supernatants of transfected cells. The gag RNA levels in deletion mutant virions were higher than wild-type levels, except for the gag RNA levels of ΔSL5 and Δ3MSD (Fig. 5A). Deletion mutant ΔSL3 had the highest gag RNA level, which was 4.2 times higher than the wild-type control level (P < 0.05). In contrast, the gag RNA levels of ΔSL5 and Δ3MSD were 1.6 and 2.1 times lower, respectively, than the wild-type levels (not statistically significant). To exclude the possibility that the gag RNAs we detected were the result of cell death due to syncytium formation, we tested the pelleted virions for β-actin mRNA. Only minimal β-actin mRNA was detected in virion pellets by a very sensitive RT-PCR assay capable of detecting down to 600 mRNA copies per reaction (Fig. 5A, right panel) (11). This result confirmed that the analysis of purified pelleted virions was not confounded by contamination with cellular debris. We therefore concluded that the gag RNAs measured in supernatant pellets were virion associated. In addition, we measured vif mRNAs in pelleted wild-type and mutant virions. There was no detectable vif mRNA in the virion pellets (data not shown), thus further indicating the absence of contamination by cellular debris. This result also demonstrated that the encapsidation specificity of the deletion mutants was retained.
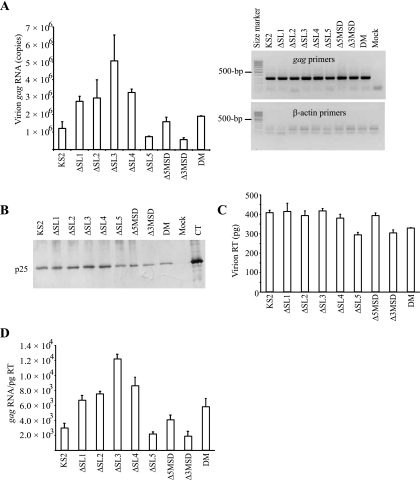
FIG. 5.
gag RNA and viral protein contents of wild-type and deletion mutant virions shed from transfected FOS cells. The names of the transfected constructs are indicated. (A) Quantification of gag RNA content in virions, calculated as gag RNA copies per volume of supernatant (left panel). The virions were purified by ultracentrifugation through a 20% sucrose cushion and measured by real-time qRT-PCR using gag RNA-specific primers. Means and standard errors for at least three independent experiments are shown on the graph. gag RNA and minimal β-actin RT-PCR products were visualized after electrophoresis in 1.7% agarose (right panel). (B) Gag protein-specific Western blot assay of virions. The virions were purified using the high-salt purification method (60). Polyclonal ascites fluid from mice immunized with whole MVV antigen was used. The position of the mature capsid (CA) protein (p25) is shown. Mock, protein from mock-transfected cells; CT, whole MVV virus preparation. (C) RT enzyme levels of virions, as measured by an enzyme activity assay and converted to pg equivalents. Means and standard errors for at least three independent experiments are shown on the graph. (D) gag RNA levels normalized for reverse transcriptase levels in virions. Means and standard errors for at least three independent experiments are shown on the graph.
Deletion mutants produced mature virions.
We detected the mature cleavage product of the MVV Gag protein (capsid protein or p25) from high-salt-purified viral particles shed from the FOS cells transfected with each deletion mutant (Fig. 5B). This indicates that the qualitative pattern of Gag protein synthesis and processing was not affected by the deletion mutants. These results were consistently observed with different stocks of plasmid preparations (data not shown).
Virion production by deletion mutants.
We also quantified virions by measuring the RT enzyme activity. RT levels were consistent with p25 levels detected by Western blotting (Fig. 5C). The RT levels of deletion mutants ΔSL5, Δ3MSD, and DM were slightly lower than the wild-type control level. We normalized the gag RNA amounts to the RT amounts in the virions (Fig. 5D). Compared to unnormalized data (Fig. 5A), the normalized data did not change significantly, although variation between samples for the same deletion mutant was decreased. These results and the Western blotting results indicate that the deletions had only limited effects on viral protein expression.
The region downstream of the MSD contributed to genomic RNA encapsidation.
We calculated the ratio of virion gag RNA (normalized for RT content) levels to cellular gag RNA levels. This ratio was then used to find the relative encapsidation efficiency (REE) by comparison with the wild-type control, whose efficiency was set at 100%. Stem-loop structures upstream of the MSD appeared to be unnecessary for MVV encapsidation in FOS cells, as measured by the REEs of ΔSL1, ΔSL2, and Δ5MSD gag RNAs (Table 3). This was expected for the nonconserved SL1 but was surprising for the highly conserved SL2 containing the MVV DIS. In fact, the ΔSL2 REE was almost twice as high as that for the wild-type control. Deletion mutations targeting SL3 or SL4, located downstream of the MSD, did not reduce the REE of the genomic RNA. In contrast, removal of the more downstream SL5 resulted in a 58% REE, suggesting that SL5 contributes to RNA encapsidation in MVV. The comparable REEs of mutants Δ3MSD and DM (24 and 26%, respectively) were lower than that for the ΔSL5 deletion mutant. These results suggest that other sequences in the region between the MSD and the gag initiation codon are also important for encapsidation.
TABLE 3.
Encapsidation efficiencies of deletion mutants in ovine and human cells
| Construct | Encapsidation efficiency (%)
|
|
|---|---|---|
| FOS cellsa | 293T cellsb | |
| KS2 | 100 | 100 |
| ΔSL1 | 84 | ND |
| ΔSL2 | 186 | ND |
| ΔSL3 | 176 | ND |
| ΔSL4 | 117 | ND |
| ΔSL5 | 58 | ND |
| Δ5MSD | 94 | 32 |
| Δ3MSD | 24 | 20a |
| DM | 26 | 0.5 |
| Mock | —c | —c |
REE was calculated as the number of virion gag RNA copies normalized for the RT amount divided by the number of intracellular gag RNA copies, expressed as a percentage of the results for wild-type KS2.
REE was calculated as the number of virion gag RNA copies divided by the number of intracellular gag RNA copies, expressed as a percentage of the results for wild-type KS2, except where indicated. ND, not determined.
—, no RNA detected.
Viral genomic RNA encapsidation in a human 293T cell line.
Retroviral genomic RNA encapsidation efficiencies can differ depending on the cell type used to produce the virus (8). To find out which sequences between the PBS and the gag initiation codon were important for encapsidation in human 293T cells, we produced deletion mutants Δ5MSD, Δ3MSD, and DM in 293T cells and measured their gag RNA REEs.
We measured the cellular gag RNA expression levels of the mutant viruses (Fig. 6A). The cellular levels of Δ5MSD gag RNA were reduced 15 times compared to the wild-type levels (P < 0.05). In contrast, expression of Δ3MSD gag RNA was slightly higher than the wild-type levels (not statistically significant), and the double mutant (DM) expressed RNA at a level comparable to that of the wild-type control. Expression of the DM gag RNA was thus intermediate between that of the Δ5MSD and Δ3MSD mutants, suggesting that the deleterious effect of the Δ5MSD mutation was compensated in part by the tendency of increased expression associated with the Δ3MSD deletion. Virion-associated gag RNA was significantly decreased for all deletion mutants compared to the wild-type level (P < 0.05) (Fig. 6B). The decreases were 48, 5, and 246 times for Δ5MSD, Δ3MSD, and DM, respectively. The effects of Δ5MSD and Δ3MSD on virion gag RNA therefore appeared to be multiplicative in the DM mutant.
FIG. 6.
Analysis of intracellular and extracellular gag RNAs and viral proteins of the wild-type and deletion mutant constructs transfected into 293T cells. Names of transfected constructs are indicated. (A) Wild-type and deletion mutant gag RNAs in the cytoplasm of transfected cells, as measured by real-time qRT-PCR and expressed as gag RNA copies per cell. All values represent means with standard errors for at least three independent experiments. (B) Levels of wild-type and deletion mutant gag RNAs in pelleted virions shed from transfected 293T cells, as measured with real-time qRT-PCR. Levels were calculated as numbers of RNA copies per ml of supernatant. Means with standard errors for at least three independent experiments are shown. (C) Pelleted virions shed from 293T cells transfected with the indicated constructs were collected and evaluated for Gag proteins by a Western blot assay, using polyclonal ascites fluid from mice immunized with whole MVV antigen. The position of the mature capsid (CA) protein (p25) is indicated. (D) RT enzyme levels of virions, as measured by an enzyme activity assay and converted to pg equivalents. Means and standard errors for at least three independent experiments are shown on the graph.
We also measured human β-actin mRNA with RT-PCR to confirm that the gag RNAs we were measuring in pellets were associated with virions but not with cellular debris. No human β-actin mRNA was detected in the pelleted virions (data not shown), indicating that the pellet was not contaminated with cellular RNA and that the gag RNA detected was virion associated.
We analyzed the virion-associated proteins by Western blotting (Fig. 6C). We detected a very faint band corresponding to the mature capsid protein for the mutant virus Δ5MSD. This was consistent with low levels of cellular Δ5MSD gag RNA. In contrast, Δ3MSD expressed wild-type levels of capsid protein in supernatants that paralleled the Δ3MSD gag RNA levels found intracellularly. We detected no capsid protein of mutant virion DM, indicating a discrepancy between the abundance of cellular gag RNA levels and virion-associated Gag proteins for the DM mutant. The RT amounts in virions were consistent with the p25 levels (Fig. 6D).
As with Western blotting, the RT assay did not detect production of the Δ5MSD and DM virions. In contrast, we measured gag RNA copies in these virions with a real-time qRT-PCR assay (Fig. 6B), which presumably reflects the higher sensitivity of this assay for detecting virion production.
The calculated REEs of Δ5MSD, Δ3MSD, and DM gag RNAs were 32%, 20%, and 0.5%, respectively (Table 3). The REEs were not corrected for RT amounts in the Δ5MSD and DM virions since we were unable to detect RT activity in some of the pellets. Therefore, the Δ5MSD and DM REE defect could have resulted either from the absence of cis encapsidation determinants or from limited production of the NC to capture gag RNA. In contrast, normal virion protein levels and a low REE of the Δ3MSD gag RNA indicate that only cis encapsidation determinants were removed in the Δ3MSD virus.
DISCUSSION
This study shows that MVV cis encapsidation determinants are located between the MSD and the gag initiation codon, as evidenced by a 168-nt deletion in this region. Conserved computer-predicted stem-loop structures in this region (SL3 and SL4) did not contribute to encapsidation in our experimental system. Disruption of one stem-loop structure (SL5) caused a modest defect in encapsidation. The region between the MSD and the gag initiation codon is not the sole determinant of MVV encapsidation.
We speculated in the beginning of our studies that the predicted SL3 and SL4 could be putative encapsidation determinants due to their conservation, purine-rich terminal loop, and location between the MSD and the gag initiation codon. The covariation of the base-paired residues of SL4, as well as the overall length of the structure, indicates functional importance. We also observed that SL4 is located in a region that is much longer in ovine and caprine lentiviruses than in other lentiviruses (Table 2). This might indicate that the predicted SL4 is unique to MVV and CAEV. Our analysis did not reveal the function of SL3 and SL4 in MVV, but we have ruled out their essential function in encapsidation, at least in our experimental system.
There has been considerable conflicting evidence in determining at which state dimerization occurs during the retroviral encapsidation pathway. We found that deletion of the MVV DIS stem-loop (SL2) did not affect the amount of gag RNA captured in virions compared to that for wild-type virus, at least when virus was produced in transfected FOS cells. Reports on MVV and other retroviruses have shown that stabilization of the dimer occurs during or after budding of the virus particle (12, 19, 21). Thus, the MVV DIS stem-loop might be active only during virus maturation or at a later stage of the virus replication cycle. Testing of an HIV-1 ΔDIS stem-loop mutant in different production cells results in either normal or decreased amounts of gag RNA captured in virions (16, 47). This suggests a cellular dependence for the production of HIV-1 virions. HIV-1 ΔDIS virions show a marked defect in infectivity (16, 47). Investigation of the infectivity and replication of the MVV deletion mutant ΔSL2 in cells for which the virus is tropic would be of interest. Although it was not tested, the ΔSL2 gag RNAs were presumably encapsidated as monomers into virions. Clever and Parslow showed that a significant amount of monomeric gag RNA can be encapsidated in HIV-1 ΔDIS virions (16). They concluded that the encapsidation of monomers was nonspecific because they also measured increased encapsidation of spliced RNA. In contrast, we detected minimal spliced viral RNA in the mutant MVV virions, thus indicating that removal of the DIS stem-loop did not result in nonspecific RNA incorporation. Other DIS loci might be located elsewhere in the MVV genome, substituting for the deleted SL2. It is notable in this regard that investigators have detected substantial amounts of stable dimeric HIV-1 genomic RNA with a mutant DIS (7, 16, 29, 55). Whether the ΔSL2 virion gag RNA is monomeric or dimeric needs to be studied in a follow-up study.
In the beginning of our studies, we aligned a 120-nt region containing the HIV-1 encapsidation determinants (NC001802) with the whole genome sequence of MVV. The HIV-1 region aligned with 43% conserved identity to the MVV sequence upstream of the MSD (data not shown). This MVV region was lacking in the Δ5MSD virus (nt 190 to 290). Interestingly, the REE of the Δ5MSD virus was defective in human 293T cells but not in ovine FOS cells. However, this mutant also showed a marked decrease in cellular gag RNA expression and virion protein levels, which could confound testing for encapsidation determinants in the region upstream of the MSD in 293T cells. In summary, we did not find unequivocal evidence that the region deleted in the Δ5MSD mutant contained cis encapsidation determinants in 293T cells.
Removal of the regions from nt 190 to 290 and nt 312 to 480 (DM) resulted in a sevenfold increase in the cellular gag RNA levels over wild-type levels in FOS cells. The molecular mechanism for this increase was not examined since it was not the goal of this paper. It could possibly have been due to altered RNA secondary structures or to the removal of regions that influence some posttranscriptional event, such as the stability of the viral genomic RNA. Elevated amounts of cellular DM gag RNA were not accompanied by a concomitant increase in virion production in either FOS or 293T cells. Since cellular Gag proteins were not measured in our study, we cannot conclude if this discrepancy was due to an assembly or translation defect. It is conceivable that we deleted or destroyed a putative internal ribosomal entry site (IRES) structure in the DM virus, resulting in impaired Gag protein production. Note that Brasey et al. recently described an IRES within the 5′ HIV-1 leader overlapping the PBS, the MSD, and the major encapsidation determinants (13). An IRES has also been described for SIV, located between the MSD and the gag initiation codon (44). Alternatively, the deletion in the DM virus might have affected viral RNA subcellular localization or resulted in an RNA transport defect. Yet another possibility is that a binding site for a chaperone protein was disrupted by the deletion.
Our results indicate that a vector construct in an MVV gene transfer system should contain the region between the MSD and the gag initiation codon for efficient encapsidation of vector RNA. In contrast, our results make the design of an MVV packaging construct more complicated. An ideal packaging construct should produce viral proteins efficiently but not encapsidate its own RNA. Therefore, packaging constructs need to be devoid of encapsidation determinants. We now know that excluding nt 312 to 480 of the MVV leader region is not enough to prevent encapsidation of the packaging construct, although gag RNA encapsidation is decreased to 20% without affecting Gag protein synthesis. Furthermore, our data suggest that packaging constructs must contain a region upstream of the MSD (nt 190 to 290) for efficient Gag protein production, at least when virus is produced in 293T cells. In contrast, we showed that this region is dispensable in FOS cells, but they might not be convenient packaging cells due to their low transfection efficiency.
The possible encapsidation of packaging construct RNA has perhaps received too little attention in the design of lentivirus-based gene transfer systems. Encapsidation determinants have not been characterized fully for the majority of lentiviruses. The main emphasis has been on including functional viral encapsidation regions in the vector construct for high-titer encapsidation. We used a novel, specific, and very sensitive assay to quantify viral gag RNA. This assay can detect as few as 60 gag RNA copies per reaction. Northern blot and RNase protection assays conventionally used for viral gag RNA quantification have less sensitivity (42, 61) and may not have detected small amounts of viral or packaging construct RNA in supernatants of producer cells.
Acknowledgments
The ascites fluid antibody was kindly provided by S. Thorsteinsdottir (Institution of Pathology, Keldur, Iceland). We thank K. Staskus and A. Haase for the generous gift of the pKS2 plasmid.
This work was funded by the Icelandic Research Council, the University of Iceland Research Fund, and the Science Fund of Landspitali-University Hospital.
Footnotes
Published ahead of print on 13 September 2006.
REFERENCES
- 1.Abbink, T. E., and B. Berkhout. 2003. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 278:11601-11611. [DOI] [PubMed] [Google Scholar]
- 2.Amarasinghe, G. K., R. N. De Guzman, R. B. Turner, K. J. Chancellor, Z. R. Wu, and M. F. Summers. 2000. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 301:491-511. [DOI] [PubMed] [Google Scholar]
- 3.Amarasinghe, G. K., J. Zhou, M. Miskimon, K. J. Chancellor, J. A. McDonald, A. G. Matthews, R. R. Miller, M. D. Rouse, and M. F. Summers. 2001. Stem-loop SL4 of the HIV-1 psi RNA packaging signal exhibits weak affinity for the nucleocapsid protein. Structural studies and implications for genome recognition. J. Mol. Biol. 314:961-970. [DOI] [PubMed] [Google Scholar]
- 4.Balvay, L., D. Libri, and M. Y. Fiszman. 1993. Pre-mRNA secondary structure and the regulation of splicing. Bioessays 15:165-169. [DOI] [PubMed] [Google Scholar]
- 5.Baudin, F., R. Marquet, C. Isel, J. L. Darlix, B. Ehresmann, and C. Ehresmann. 1993. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J. Mol. Biol. 229:382-397. [DOI] [PubMed] [Google Scholar]
- 6.Berkhout, B. 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 54:1-34. [DOI] [PubMed] [Google Scholar]
- 7.Berkhout, B., and J. L. van Wamel. 1996. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J. Virol. 70:6723-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 9.Berkowitz, R., H. Ilves, W. Y. Lin, K. Eckert, A. Coward, S. Tamaki, G. Veres, and I. Plavec. 2001. Construction and molecular analysis of gene transfer systems derived from bovine immunodeficiency virus. J. Virol. 75:3371-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berkowitz, R. D., H. Ilves, I. Plavec, and G. Veres. 2001. Gene transfer systems derived from visna virus: analysis of virus production and infectivity. Virology 279:116-129. [DOI] [PubMed] [Google Scholar]
- 11.Bjarnadottir, H., and J. J. Jonsson. 2005. A rapid real-time qRT-PCR assay for ovine beta-actin mRNA. J. Biotechnol. 117:173-182. [DOI] [PubMed] [Google Scholar]
- 12.Brahic, M., and R. Vigne. 1975. Properties of visna virus particles harvested at short time intervals: RNA content, infectivity and ultrastructure. J. Virol. 15:1222-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brasey, A., M. Lopez-Lastra, T. Ohlmann, N. Beerens, B. Berkhout, J. L. Darlix, and N. Sonenberg. 2003. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 77:3939-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Browning, M. T., F. Mustafa, R. D. Schmidt, K. A. Lew, and T. A. Rizvi. 2003. Delineation of sequences important for efficient packaging of feline immunodeficiency virus RNA. J. Gen. Virol. 84:621-627. [DOI] [PubMed] [Google Scholar]
- 15.Clever, J., C. Sassetti, and T. G. Parslow. 1995. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J. Virol. 69:2101-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clever, J. L., and T. G. Parslow. 1997. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 71:3407-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crane, S. E., J. E. Clements, and O. Narayan. 1988. Separate epitopes in the envelope of visna virus are responsible for fusion and neutralization: biological implications for anti-fusion antibodies in limiting virus replication. J. Virol. 62:2680-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.D'Souza, V., and M. F. Summers. 2005. How retroviruses select their genomes. Nat. Rev. Microbiol. 3:643-655. [DOI] [PubMed] [Google Scholar]
- 19.Feng, Y. X., T. D. Copeland, L. E. Henderson, R. J. Gorelick, W. J. Bosche, J. G. Levin, and A. Rein. 1996. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. USA 93:7577-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu, W., R. J. Gorelick, and A. Rein. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 23.Griffin, S. D., J. F. Allen, and A. M. Lever. 2001. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 75:12058-12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gudmundsson, B., H. Bjarnadottir, S. Kristjansdottir, and J. J. Jonsson. 2003. Quantitative assays for maedi-visna virus genetic sequences and mRNA's based on RT-PCR with real-time FRET measurements. Virology 307:135-142. [DOI] [PubMed] [Google Scholar]
- 25.Harrich, D., C. W. Hooker, and E. Parry. 2000. The human immunodeficiency virus type 1 TAR RNA upper stem-loop plays distinct roles in reverse transcription and RNA packaging. J. Virol. 74:5639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison, G. P., and A. M. Lever. 1992. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J. Virol. 66:4144-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison, G. P., G. Miele, E. Hunter, and A. M. Lever. 1998. Functional analysis of the core human immunodeficiency virus type 1 packaging signal in a permissive cell line. J. Virol. 72:5886-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helga-Maria, C., M. L. Hammarskjold, and D. Rekosh. 1999. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type 1 genomic RNA. J. Virol. 73:4127-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill, M. K., M. Shehu-Xhilaga, S. M. Campbell, P. Poumbourios, S. M. Crowe, and J. Mak. 2003. The dimer initiation sequence stem-loop of human immunodeficiency virus type 1 is dispensable for viral replication in peripheral blood mononuclear cells. J. Virol. 77:8329-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huthoff, H., and B. Berkhout. 2001. Two alternating structures of the HIV-1 leader RNA. RNA 7:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaye, J. F., and A. M. Lever. 1999. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J. Virol. 73:3023-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaye, J. F., and A. M. Lever. 1998. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation. J. Virol. 72:5877-5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kemler, I., R. Barraza, and E. M. Poeschla. 2002. Mapping the encapsidation determinants of feline immunodeficiency virus. J. Virol. 76:11889-11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laughrea, M., and L. Jette. 1994. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry 33:13464-13474. [DOI] [PubMed] [Google Scholar]
- 35.Lever, A. M. 2000. HIV RNA packaging and lentivirus-based vectors. Adv. Pharmacol. 48:1-28. [DOI] [PubMed] [Google Scholar]
- 36.Lisitsyn, N., and M. Wigler. 1993. Cloning the differences between two complex genomes. Science 259:946-951. [DOI] [PubMed] [Google Scholar]
- 37.Luban, J., and S. P. Goff. 1994. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J. Virol. 68:3784-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McBride, M. S., and A. T. Panganiban. 1996. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 70:2963-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McBride, M. S., and A. T. Panganiban. 1997. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J. Virol. 71:2050-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McBride, M. S., M. D. Schwartz, and A. T. Panganiban. 1997. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 71:4544-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCann, E. M., and A. M. Lever. 1997. Location of cis-acting signals important for RNA encapsidation in the leader sequence of human immunodeficiency virus type 2. J. Virol. 71:4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melton, D. A., P. A. Krieg, M. R. Rebagliati, T. Maniatis, K. Zinn, and M. R. Green. 1984. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 12:7035-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monie, T. P., J. S. Greatorex, L. Maynard-Smith, B. D. Hook, N. Bishop, L. P. Beales, and A. M. Lever. 2005. Identification and visualization of the dimerization initiation site of the prototype lentivirus, maedi visna virus: a potential GACG tetraloop displays structural homology with the alpha- and gamma-retroviruses. Biochemistry 44:294-302. [DOI] [PubMed] [Google Scholar]
- 44.Ohlmann, T., M. Lopez-Lastra, and J. L. Darlix. 2000. An internal ribosome entry segment promotes translation of the simian immunodeficiency virus genomic RNA. J. Biol. Chem. 275:11899-11906. [DOI] [PubMed] [Google Scholar]
- 45.Olsen, J. C. 1998. Gene transfer vectors derived from equine infectious anemia virus. Gene Ther. 5:1481-1487. [DOI] [PubMed] [Google Scholar]
- 46.Ooms, M., H. Huthoff, R. Russell, C. Liang, and B. Berkhout. 2004. A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 78:10814-10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paillart, J. C., L. Berthoux, M. Ottmann, J. L. Darlix, R. Marquet, B. Ehresmann, and C. Ehresmann. 1996. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J. Virol. 70:8348-8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paillart, J. C., E. Skripkin, B. Ehresmann, C. Ehresmann, and R. Marquet. 1996. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA 93:5572-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parolin, C., T. Dorfman, G. Palu, H. Gottlinger, and J. Sodroski. 1994. Analysis in human immunodeficiency virus type 1 vectors of cis-acting sequences that affect gene transfer into human lymphocytes. J. Virol. 68:3888-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel, J., S. W. Wang, E. Izmailova, and A. Aldovini. 2003. The simian immunodeficiency virus 5′ untranslated leader sequence plays a role in intracellular viral protein accumulation and in RNA packaging. J. Virol. 77:6284-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petursson, G., V. Andresdottir, O. Andresson, S. Torsteinsdottir, G. Georgsson, and P. A. Palsson. 1991. Human and ovine lentiviral infections compared. Comp. Immunol. Microbiol. Infect. Dis. 14:277-287. [DOI] [PubMed] [Google Scholar]
- 52.Poeschla, E. M., F. Wong-Staal, and D. J. Looney. 1998. Efficient transduction of nondividing human cells by feline immunodeficiency virus lentiviral vectors. Nat. Med. 4:354-357. [DOI] [PubMed] [Google Scholar]
- 53.Russell, R. S., J. Hu, V. Beriault, A. J. Mouland, M. Laughrea, L. Kleiman, M. A. Wainberg, and C. Liang. 2003. Sequences downstream of the 5′ splice donor site are required for both packaging and dimerization of human immunodeficiency virus type 1 RNA. J. Virol. 77:84-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russell, R. S., C. Liang, and M. A. Wainberg. 2004. Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology 1:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakuragi, J. I., and A. T. Panganiban. 1997. Human immunodeficiency virus type 1 RNA outside the primary encapsidation and dimer linkage region affects RNA dimer stability in vivo. J. Virol. 71:3250-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sigurdsson, B., P. Palsson, and H. Grimsson. 1957. Visna, a demyelinating transmissible disease of sheep. J. Neuropathol. Exp. Neurol. 16:389-403. [DOI] [PubMed] [Google Scholar]
- 57.Skripkin, E., J. C. Paillart, R. Marquet, B. Ehresmann, and C. Ehresmann. 1994. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA 91:4945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Staskus, K. A., E. F. Retzel, E. D. Lewis, J. L. Silsby, S. St. Cyr, J. M. Rank, S. W. Wietgrefe, A. T. Haase, R. Cook, D. Fast, et al. 1991. Isolation of replication-competent molecular clones of visna virus. Virology 181:228-240. [DOI] [PubMed] [Google Scholar]
- 59.Strappe, P. M., J. Greatorex, J. Thomas, P. Biswas, E. McCann, and A. M. Lever. 2003. The packaging signal of simian immunodeficiency virus is upstream of the major splice donor at a distance from the RNA cap site similar to that of human immunodeficiency virus types 1 and 2. J. Gen. Virol. 84:2423-2430. [DOI] [PubMed] [Google Scholar]
- 60.Torsteinsdottir, S., G. Georgsson, E. Gisladottir, B. Rafnar, P. A. Palsson, and G. Petursson. 1992. Pathogenesis of central nervous system lesions in visna: cell-mediated immunity and lymphocyte subsets in blood, brain and cerebrospinal fluid. J. Neuroimmunol. 41:149-158. [DOI] [PubMed] [Google Scholar]
- 61.Wang, T., and M. J. Brown. 1999. mRNA quantification by real time TaqMan polymerase chain reaction: validation and comparison with RNase protection. Anal. Biochem. 269:198-201. [DOI] [PubMed] [Google Scholar]
- 62.Wittwer, C. T., M. G. Herrmann, A. A. Moss, and R. P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. BioTechniques 22:130-131, 134-138. [DOI] [PubMed] [Google Scholar]
- 63.Wittwer, C. T., K. M. Ririe, R. V. Andrew, D. A. David, R. A. Gundry, and U. J. Balis. 1997. The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. BioTechniques 22:176-181. [DOI] [PubMed] [Google Scholar]
- 64.Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244:48-52. [DOI] [PubMed] [Google Scholar]