Abstract
The proteins encoded by the UL34 and UL31 genes of herpes simplex virus are conserved among herpesviruses. They form a complex that is essential for the egress of the herpesvirus nucleocapsids from the nucleus. In previous work on the homologous protein complex in murine cytomegalovirus (MCMV), we defined their mutual binding domains. Here, we started to map binding domains within the UL34/UL31 proteins of alpha-, beta-, and gammaherpesviruses and to locate other functional properties. A protein complementation assay (PCA) using the TEM-1 β-lactamase fragments fused to UL31 and UL34 protein homologues was used to study protein-protein interactions in cells. Wild-type MCMV M50 and M53 provided a strong reaction in the PCA, whereas mutants unable to form a complex did not. The homologous pairs of herpes simplex virus type 1, pseudorabies virus, human cytomegalovirus (HCMV), Epstein-Barr virus (EBV), and murine herpes virus 68 proteins also reacted, with the exception of the EBV proteins. Cross-complementation was found to be positive only within the same herpesvirus subfamily. Moreover, the HCMV homologues rescued replication-defective MCMV genomes lacking one or the other gene. We identified the binding site of M53 for M50 in the first conserved region (CR1) (M. Loetzerich, Z. Ruzsics, and U. H. Koszinowski, J. Virol. 80:73-84). Here we show that the CR1 of all tested UL31 proteins contains the UL34 binding site, and chimeric proteins carrying the subfamily-specific CR1 rescued the ability to cross-complement in the PCA.
Human herpesviruses cause highly prevalent infections associated with usually mild symptoms resulting in lifelong latency. However, they can provoke fatal disease in immune-compromised and immune-immature patients (12, 23, 41). In spite of their medical importance, at this time herpesvirus infections can be controlled only by antiviral therapy targeting viral DNA replication (7, 15). Within the last few years, promising new agents that affect protein-protein interactions involved in herpesvirus replication have been identified by high-throughput screening (14, 36). Such screening requires the identification of proteins that interact in an important or essential fashion.
Recently the interaction map, the so-called interactome, of the two herpesviruses Kaposi's sarcoma-associated herpesvirus and varicella-zoster virus was annotated (38). The protein networks predicted by yeast two-hybrid screening suggest a common core set of herpesvirus protein interactions. Predicted interactions need to be validated in the natural context. Here we have tested a protein complementation assay (PCA) (10, 37, 40), which can be used to characterize viral protein-protein interactions in cells. In principle, the same assay is applicable to the screening (26) and validation of compounds interfering with the interactions in question. Among other procedures to measure and monitor protein-protein interactions, the PCA, evolved from the classical yeast two-hybrid approach (8), detects protein-protein interactions by a direct readout in real time independently of the subcellular compartment of the interaction. In contrast to what is the case for the yeast two-hybrid system, different organisms can host the same PCA. A PCA in mammalian cells based on TEM-1 β-lactamase has been established recently (10, 40). The TEM-1 β-lactamase of Escherichia coli (EC 3.5.2.6.) is monomeric, of only 27 kDa in size, not toxic, suitable for the design of fragments, and possible to express in eukaryotic cells as well as in bacteria (10, 26, 40). Since there are no orthologous of the β-lactamase in eukaryotic cells, the complementation can be easily detected with a low background.
A number of proteins are conserved among all herpesviruses. We studied the interaction of two of these core proteins to define the preservation of binding sites. As the target we chose the herpesvirus UL34 and UL31 protein family members. Proteins of these two conserved families were studied in the alphaherpesviruses herpes simplex virus type 1 (HSV-1) (32), HSV-2 (42), pseudorabies virus (PrV) (9), and equine herpesvirus 1(25), in the betaherpesviruses murine cytomegalovirus (MCMV) (4, 16) and human cytomegalovirus (HCMV) (6), and in the gammaherpesvirus Epstein-Barr virus (EBV) (13). By a nonobligatory interaction, the proteins form a complex referred to as the nuclear egress complex (NEC), which is required for the export of viral capsids from the nucleus. We reported recently that the interaction between M50 and M53, the UL34 and UL31 family members of MCMV, is essential for productive virus infection. M50 is a membrane-anchored protein which is synthesized in the endoplasmic reticulum late in infection and translocated to the inner nuclear membrane by diffusion. Its partner, M53, is expressed with similar kinetics and targeted to the nucleus through the nuclear pores. In the nucleus, M50 binds to M53, forming the NEC at the inner nuclear membrane (4, 16, 24). The NEC is involved in destabilization of the nuclear lamina and/or remodeling of the nuclear membranes, preparing them for primary budding of viral capsids. The NEC proteins also guide the nuclear egress process of viral capsids (21).
Here we established a NEC-specific PCA for the MCMV proteins that, after validation, was extended to other members of the UL34 and UL31 protein families of all three herpesvirus subfamilies. We show that the interaction domains in UL31 proteins are conserved but that the actual interaction sites have diverged during evolution.
MATERIALS AND METHODS
Cells and viruses.
NIH 3T3 murine fibroblasts (ATCC CRL 1658), M2-10B4 bone marrow stroma cells (ATCC CRL 1972), and mouse embryonic fibroblasts (MEFs) were propagated as described previously (20). 293T cells (ATCC CRL 11268) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 0.3 mg/ml glutamine. Wild-type (WT) (pSM3fr [5]) and mutant MCMVs were reconstituted by transfection of the respective bacterial artificial chromosome (BAC) DNAs into MEFs by use of Superfect transfection reagent (QIAGEN) according to the manufacturer's instructions. All virus stocks were propagated on M2-10B4 cells (17) and titrated on MEFs by use of a previously described plaque assay (29). To analyze the growth of the mutant MCMVs, NIH 3T3 fibroblasts were infected at a multiplicity of infection of 0.1. Cells were seeded on 12-well plates (Nunc) 1 day before infection to obtain cells numbers around 3 × 105 per well on the day of infection. Cells in each well were overlaid for 1 h with 1 ml of media containing the respective viruses. Supernatants of the infected cultures were collected after infection and then daily for 5 days. The released viruses were quantified by plaque assay on MEFs (29).
Plasmids.
To construct the fusion vector, two complementary synthetic oligonucleotide pairs (S1/S2 and S3/S4) coding for a 17-amino-acid (aa) Gly/Ser spacer [(S1G4)2S2G4S] were annealed and joined by XbaI digestion followed by ligation. The resulting DNA fragment was then inserted into the Litmus 28 vector (NEB) by BglII and AflII. The resulting construct was cut with BspEI and AflII, and the annealed HA1 and HA2 oligonucleotides coding for the hemagglutinin (HA) tag were inserted, generating the cloning vector pL-HA-Spacer.
The coding sequences for the N- and C-terminal parts of TEM-1 β-lactamase (Bla; EC 3.5.2.6) were PCR amplified from the pUC19 vector (NEB) by use of the BlaNfor/BlaNrev and BlaCfor/BlaCrev primer pairs, respectively. The coding sequences for the N-terminal (aa 24 to aa 194) and C-terminal (aa 196 to aa 286) fragments of Bla were cloned into pL-HA-Spacer by BspEI/NheI and AflII/BspEI and designated as pLHA-N and pLHA-C, respectively.
The M50 mutant M50DM was generated by inverse PCR on pOriR6K-zeo-M50 (4) using the primers delmo-NheI-for and delmo2-Nhe-rev, resulting in pOriR6K-zeo-ie-M50DM. For construction of the BlaN-M50 fusions, the WT and mutant M50 fragments were PCR amplified from the corresponding expression plasmids pOriR6K-zeo-ie-M50, pOriR6K-zeo-ie-M50-aa52, pOriR6K-zeo-ie-M50-aa114 (4), and pOriR6K-zeo-ie-M50DM by use of the primer pair M50for and M50rev. The obtained PCR fragments were brought into pLHA-N by SapI and AgeI. To generate the BlaC-M53 fusion, the M53 open reading frame (ORF) was amplified from pOriR6K-zeo-ie-M53 (16) by use of the primers M53for and M53rev and inserted into pLHA-C by BamHI and ApaI. Fusion constructs were then isolated from the Litmus vectors after AflII and BspEI digestion, filled in, and cloned into expression plasmid pO6T (GenBank accession number DQ867321) by EcoRV and AflII, generating pO6T-N-M50 (or corresponding mutants) and pO6T-C-M53.
The members of the UL34 family were fused to the N-terminal Bla fragment and the members of the UL31 family to the C-terminal Bla fragment. UL34 of PrV (Kaplan strain; viral DNA, kind gift of P. Sondermeier), UL50 of HCMV (AD169 BAC [3]), Orf67 of murine herpes virus 68 (MHV68) (MHV68 BAC [1]) and BFLF1 of EBV (strain B95-8; subcloned ORFs, kind gift of Jürgen Haas) were PCR amplified from BAC or plasmid DNA with the primer pairs UL34PrVfor/UL34PrVrev, UL50for/UL50rev, ORF67for/ORF67rev, and BFRF1for/BFRF1rev, respectively. The resulting fragments were inserted into the pLHA-N vector by BamHI and AgeI. UL34 of HSV-1 (strain 17; subcloned ORFs, kind gift of Jürgen Haas) was PCR amplified with the primers HSVUL34for and HSVUL34rev, cleaved with XhoI and AgeI, and inserted into the accordingly cleaved pLHA-N. The fusion fragments were then transferred into pO6T by AgeI and NheI, generating pO6T-N-UL34(HSV), pO6T-N-UL34(PrV), pO6T-N-UL50, pO6T-N-Orf67, and pO6T-N-BFRF1.
UL31 of PrV, UL53 of HCMV, Orf69 of MHV68, and BFLF2 of EBV were PCR amplified with the primer pairs UL31PrVfor/UL31PrVrev, UL53for/UL53rev, ORF69for/ORF69rev, and BFLF2for/BFLF2rev, respectively. The resulting fragments were inserted into pLHA-C by BamHI and ApaI. UL31 of HSV-1 was PCR amplified with the primer pair HSVUL31for/HSVUL31rev and cloned into pLHA-C with BamHI and NotI. The fusion fragments were then isolated by AflII/BspEI cleavage, filled in, and inserted into pO6T with EcoRV/AflII, generating pO6T-C-UL31(HSV), pO6T-C-UL31(PrV), pO6T-C-UL53, pO6T-C-Orf69, and pO6T-C-BFLF2.
Chimeric UL31 fusions were created by interchange of CR1 and CR2 to CR4 (CR2-CR4) (16) of M53, UL31(PrV), and Orf69. CR1 of M53 (aa 1 to aa 175) was combined with CR2-CR4 of UL31 (aa 92 to aa 271) and Orf69 (aa 112 to aa 292). The generated chimeras were named C-MP and C-MG, respectively. For C-MP, an M53 fragment was PCR amplified from pO6T-C-M53 with the primer pair MP-Mfor/MP-Mrev and cleaved with BsrDI. The PrV UL31 fragment was PCR amplified from pO6T-C-UL31(PrV) with the primer pair MP-Pfor/MP-Prev and cleaved with BsrDI and NotI. Both fragments were ligated to the vector fragment of pO6T-C-M53 after BsrDI and NotI cleavage, resulting in pO6T-C-MP. pO6T-C-MG, coding for C-MG, was generated by inserting a BstAPI/NotI-cleaved PCR product, obtained with MG-Gfor and MG-Grev from pO6T-C-Orf69, into the BstAPI/NotI-treated pO6T-C-M53. CR1 of UL31 (aa 1 to aa 91) and CR1 of Orf69 (aa 1 to aa 111) were fused to CR2-CR4 of M53 (aa 176 to aa 333) and named C-PM and C-GM, respectively. For that procedure, the fragments obtained by use of PM-Pfor/PM-Prev on pO6T-C-UL31 and GM-Gfor/GM-Grev from pO6T-C-Orf69 were digested with BspMI and BsrDI and inserted into pO6T-C-M53 between the BspMI and BsrDI sites.
For all cloning procedures, restriction enzymes and the T4 ligase of NEB were used, and the manufacturer's protocols were followed. Oligonucleotide sequences can be found at http://www.lmb.uni-muenchen.de/mainframes/research/koszinowski.htm. All constructs were checked for correctness of sequence.
Generation of recombinant viral BACs.
The ORFs coding for UL34(HSV1), UL34(PrV), and UL50 were amplified by PCR and cloned into the pOriR6k-ie-zeo rescue plasmid (4) by KpnI and EcoRV. The pOriR6k-ie-zeo constructs and pO6T-N-M50 were inserted into pSM3fr-16FRT17 (WT) (5) and pSMfr3-ΔM50 BAC (4) by use of FLP-mediated recombination in E. coli (4), generating ΔM50/N-M50, ΔM50/UL50, WT/UL34(HSV), WT/UL34(PrV), ΔM50/UL34(HSV), and ΔM50/UL34(PrV). In the same way, UL31(HSV1), UL31(PrV), and Orf69 were amplified by PCR and cloned into pOriR6K-ie-zeo. The pOriR6k-ie-zeo constructs and pO6T-C-M53 were inserted into pSM3fr-16FRT17 (WT) and pSMfr3-ΔM53 BAC (16), resulting in ΔM53/C-M53, ΔM53/UL53, WT/UL31(HSV), WT/UL31(PrV), ΔM53/UL31(HSV), and ΔM53/UL31(PrV). MP and MG were introduced in the pOriR6k-ie-zeo background by the SalI and NotI sites of pOriR6K-zeo-ie-M53 (16) and then inserted into pSM3fr-16FRT17 (WT) and pSMfr3-ΔM53 BAC, giving rise to WT/MP, WT/MG, ΔM53/MP, and ΔM53/MG. Oligonucleotide sequences for the generation of recombinant viral BACs can be found at http://www.lmb.uni-muenchen.de/mainframes/research/koszinowski.htm.
Colorimetric β-lactamase PCA.
All assays were done in triplicate and repeated at least three times. 293T cells were split 1:2.5 on six-well plates 24 h before transfection. For cotransfection experiments, cells were transiently transfected with 2.5 μg of pO6T-N-UL34 homologue and 2.5 μg of pO6T-C-UL31 homologue or 2.5 μg of pO6T-N-UL34 homologue and 2.5 μg of pO6T-C chimera by use of Superfect transfection reagent (QIAGEN) according to the manufacturer's instructions. For the competition experiment (see Fig. 2C and D), a total amount of 6 μg DNA comprising 1.5 μg of pO6T-N-M50, 1.5 μg of pO6T-C-M53, the indicated amount of pOriR6K-zeo-M50 (coding for WT M50), and the corresponding amount of pO6-IET-gfp (34) as carrier DNA was transfected. Each combination was transfected in quadruplicate. Twenty-four hours after transfection, cells were trypsinated, pooled, and washed with phosphate-buffered saline. One-fourth of the cell suspension was used for Western blot analysis, whereas the major part was lysed in 225 μl luciferase reporter lysis buffer (Promega). After a 30-min incubation on ice, cell debris was removed by centrifugation for 30 min at full speed and at 4°C. To measure the β-lactamase activity, three 50-μl aliquots of the cell lysates were transferred to a 96-well plate (Nunc) and mixed with 120 μl phosphate buffer (0.1 M, pH 7), 15 μl H2O, and 15 μl of 500 mg/ml nitrocefin (Oxoid). In a Versamax plate reader (Molecular Devices), the change in absorption at 495 nm, a measure for the hydrolization of nitrocefin, was observed over 20 min at 37°C. The maximal change in absorption (Vmax) was determined by the data points in the linear range and expressed in milliabsorption units/minute. Data were normalized by the protein content of the cell lysates determined by the Bradford assay. Hydrolysis rates of nitrocefin in cell lysates after coexpressions as well as mean virus titers were subjected to analysis of variance tests.
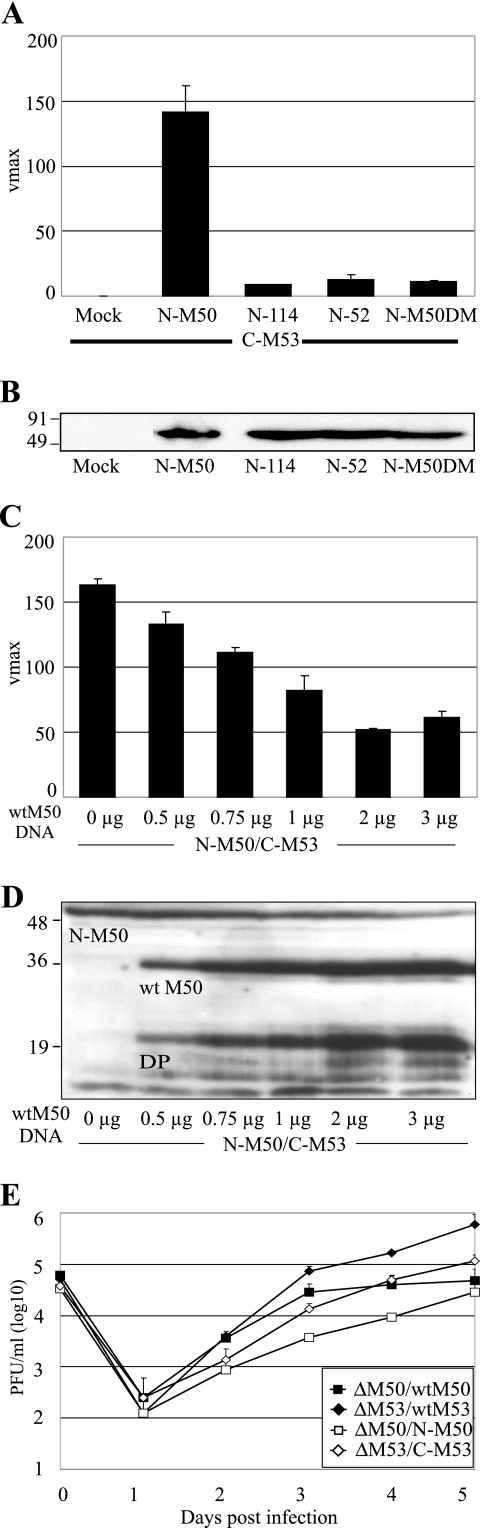
FIG. 2.
Validation of the NEC PCA. (A) Background activity of the PCA. C-M53 was coexpressed with N-M50 and with N-M50 mutants (N-52, N-114, and N-M50DM) which are deficient in binding to M53. Cell lysates were prepared 24 h posttransfection, and the lactamase substrate nitrocefin was added. Mean nitrocefin hydrolysis rates were determined and are shown as Vmax in milliabsorption units/min. (B) Expression of N-M50 and N-M50 mutants detected by HA-specific Western blotting after isolated expression. Numbers indicate the positions and sizes of the protein marker in kDa. All constructs were detected at the predicted size of 56 kDa. (C) Competition assay. N-M50 and C-M53 (1.5 μg of each DNA) were coexpressed together with increasing amounts of WT M50. Carrier DNA was added to maintain a constant amount of 6 μg total DNA. Cell lysates were prepared 24 h posttransfection, and Bla activity was determined. (D) The same cell lysates were used to show the coexpression of N-M50 and WT M50 by Western blotting with an M50-specific antiserum. N-M50 (56 kDa), WT M50 (35 kDa), and smaller degradation products (DP) of M50 are indicated. Numbers on the left indicate the positions and sizes of the protein marker in kDa. (E) Functionality of the fusion proteins in the virus context. Multistep growth curves on NIH 3T3 cells are shown for MCMV mutants which either express WT M50 or N-M50 but lack the native M50 (ΔM50/WTM50 and ΔM50/N-M50) or express WT M53 or C-M53 but lack the native M53 (ΔM53/WTM53 and ΔM53/C-M53). Cells were infected with the respective viruses at a multiplicity of infection of 0.1 (day 0), and input virus was removed after 1 hour. Newly released virus in the supernatant of infected cells was quantified daily by a plaque assay. At day 5 after infection, mean titers of ΔM50/WTM50 and ΔM50/N-M50 and of ΔM53/WTM53 and ΔM53/C-M53 showed no significant differences. Error bars indicate the standard deviation.
Western blot analysis.
For the expression control of fusion proteins, subconfluent 293T cells on 6-cm dishes were transfected with 6 μg of DNA by Ca2PO4 precipitation (35). After 24 h the medium was exchanged, and after 48 h cells were harvested, washed, and lysed in 350 μl total lysis buffer (62.5 mM Tris [pH 6.8], 2% sodium dodecyl sulfate [vol/vol], 10% glycerol [vol/vol], 6 M urea, 5% β-mercaptoethanol [vol/vol], 0.01% bromophenol blue [wt/vol], 0.01% phenol red [wt/vol]). For the expression control in the competition experiment (see Fig.2C), cells were transfected in four wells of a six-well plate as described for the analysis in the β-lactamase PCA (see above) and harvested and washed 24 h posttransfection, and then one-fourth of the cell suspension was lysed in 200 μl total lysis buffer. Fifteen microliters of the samples was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the proteins were transferred from the gel onto Hybond-P membranes (Amersham Biosciences) in the presence of blotting buffer (25 mM Tris, 192 mM glycine, 20% [vol/vol] methanol [pH 8.3]). Membranes were blocked in TBS-T (Tris-buffered saline, 0.05% Tween 20) containing 5% nonfat dry milk overnight at 4°C. To detect the constructs, the membrane was then incubated at room temperature for 1 hour with TBS-T containing anti-HA-peroxidase high-affinity antibody (1:5,000; Roche); alternatively, a specific polyclonal rabbit antiserum, anti-M50, was used (24). For M50 detection, the blots were incubated with a secondary anti-rabbit horseradish peroxidase-conjugated antibody (Dianova, Hamburg, Germany) after being washed with TBS-T. Membranes were washed with TBS-T, and proteins were visualized with an ECL-Plus Western blot detection system (Amersham Biosciences).
RESULTS
Establishment and validation of the MCMV NEC PCA.
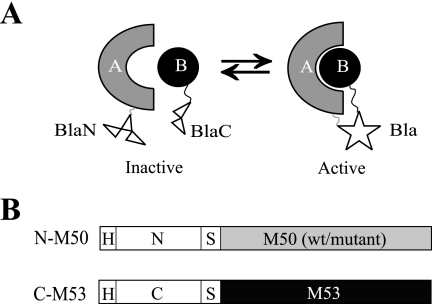
In a PCA, two nonfunctional fragments of a reporter protein are fused to the two proteins of interest. If the fragments reach proximity due to the interaction of the proteins to which they are fused, folding of the native conformation leads to reconstitution of reporter activity (Fig. 1A). The E. coli TEM-1 β-lactamase (Bla) was split in two parts, representing the N terminus from aa 24 to aa 194 (N) and the C terminus from aa 196 to aa 286 (C) (10). The HA-tagged Bla fragments were then fused N terminally to the MCMV NEC proteins M50 and M53, respectively (Fig. 1B).
FIG. 1.
Establishment and validation of the NEC PCA. (A) Simplified scheme of a PCA. The N- and C-terminal parts of a reporter enzyme (white), here the TEM-1 β-lactamase (Bla), are fused to two interacting proteins (shapes labeled A and B). If proteins A and B interact, the proximity of the enzyme fragments BlaN and BlaC allows the folding of the active enzyme (star). (B) Schematic representation of the used HA (H)-tagged constructs. The N-terminal part of Bla, representing residues 24 to 194 (N), and the C-terminal part, representing the Bla residues 196 to 286 (C), were fused to the MCMV proteins M50 (WT or mutant) and M53, linked by a glycine/serine spacer (S).
First, N-M50 and C-M53 were coexpressed to test for the reconstitution of Bla activity. In lysates of cotransfected cells, the hydrolysis rate of a Bla substrate, nitrocefin, was more than 130-fold higher than in lysates of mock-infectedcells (Fig. 2A). The coexpression of N-M50 with M53-C, bearing the C-terminal Bla fragment at the C terminus, also led to an equally strong signal in the nitrocefin assay (data not shown).
The coexpression of C-M53 with the N fusions of M50 mutants, which cannot bind to M53 (4), resulted in a nitrocefin hydrolysis rate of less than 10% of the wild type N-M50/C-M53 complementation and reflected their inability to bind (Fig. 2A). To confirm that the low signal detected in PCAs with nonbinding mutants was not due to poor protein expression, the cell lysates after isolated expression were probed with an anti-HA-antibody in a Western blot. All mutants were detected at the predicted molecular mass of 56 kDa and expressed to the same level as N-M50 (Fig. 2B). The basal hydrolysis rate detected after the cotransfection of the binding-deficient N-M50DM and C-M53 served as the background control for subsequent NEC PCAs.
Next, the specificity of the N-M50/C-M53 PCA was tested in the presence of increasing amounts of WT M50, which binds C-M53 but cannot complement due to the lack of the Bla fusion fragment. Accordingly, the PCA signal decreased in a WT M50 concentration-dependent manner (Fig. 2C). The initial hydrolysis rate (Vmax) of 170 absorption units/min in the absence of WT M50 was gradually reduced by up to more than threefold, reflecting the relative proportions of tagged and nontagged M50. N-M50 and WT M50 were visualized with an anti-M50-probed Western blot and detected at the predicted sizes of 56 kDa and 35 kDa, respectively (Fig. 2D). Degradation products of M50 with lower molecular masses were detected in all lanes except for the first lane, in which only N-M50 and C-M53 were present. This and the slight decrease of detected N-M50 in the presence of the increasing amounts of WT M50 visualized in the Western blot are presumably due to the known instability of M50 in the absence of the binding partner (4). WT M53 stabilizes WT M50, and this is also seen for N-M50 if C-M53 is present. If the C-M53 protein partner is withdrawn by WT M50 excess, N-M50 becomes unstable.
Finally, N-M50 and C-M53 were studied for functionality in the context of MCMV replication to test whether the essential protein functions are kept upon fusion to the Bla fragments. To this end, the fusion constructs were introduced into the MCMV genome by use of site-specific recombination (4). Deletion of either M50 or M53 from the MCMV genome prevents reconstitution of infectious particles, and the ectopic reinsertion of WT M50 or WT M53 into the respective deletion genomes rescues the null phenotype (4, 16). The N-M50 and C-M53 genes were inserted into ΔM50 and ΔM53 genomes, respectively. After transfection of MEFs, both the ΔM50/N-M50 and the ΔM53/C-M53 BACs gave rise to infectious progeny. Under multistep growth conditions, the growth levels of ΔM50/N-M50 and ΔM53/C-M53 were comparable (P > 0.05) to those of ΔM50/WTM50 (4) and ΔM53/WTM53 (16) (Fig. 2E).
These experiments validated the PCA for the NEC proteins M50 and M53. The background of the N-M50/C-M53 PCA was low, due to negligible spontaneous folding of the Bla fragments. The specificity of the PCA signal was confirmed by the competition experiment, and the functionality of the Bla fusion proteins was proven by their ability to replace the WT NEC proteins in the virus. Thus, we considered the NEC PCA as a tool to monitor and characterize the interaction of M50 and M53 as well as that of homologous proteins.
Visualization of the conserved UL34/UL31 interaction by NEC PCA.
Homologues of M50, i.e., the UL34 protein family, and of M53, i.e., the UL31 protein family, are highly conserved among all herpesviruses. All studied members have been shown to interact and play an important role in the nuclear egress of herpesvirus capsids (4, 9, 13, 32). We next wanted to test whether the PCA established for the M50/M53 interaction can be applied to other members of the UL34/UL31 protein families.
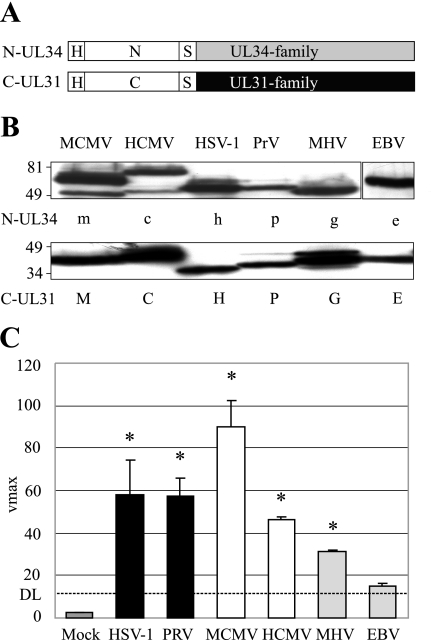
The M50 and M53 homologues of HSV-1, PrV, HCMV, MHV68, and EBV were fused to the Bla fragments in the same manner as M50 and M53 (Fig. 3A). In a Western blot, N-UL34 proteins were detected at the predicted molecular masses between 50 and 80 kDa (Fig. 3B, upper panel). C-UL31 proteins were detected at the predicted molecular masses between 33 and 50 kDa (Fig. 3B, lower panel). N-M50 and N-UL50 show degradation products of lower molecular mass (4). The NEC PCA was performed by the coexpression of the N and C fusions of each virus (Fig. 3C). Specific signals were obtained for the NEC proteins of HSV-1, PrV, MCMV, HCMV, and MHV68. For the gammaherpesvirus MHV68, a low but significant signal was detectable, but in the case of EBV, the complementation-induced hydrolysis rate was not significantly higher than the detection limit (DL) (P = 0.22). The high hydrolysis rate obtained with the MCMV proteins, in contrast to the lower signal of the homologues, probably reflects the optimization of the assay towards that particular interaction.
FIG. 3.
Interaction of UL34 and UL31 family members of different viruses. (A) Schematic representation of the HA (H)-tagged constructs used. The N- and C-terminal parts of the Bla were fused to members of the UL34 and UL31 families, linked by a glycine/serine spacer (S). (B) Expression of N-UL34 and C-UL31 fusion proteins detected by HA-specific Western blotting. N-UL34 fusion proteins were N-M50 (m), N-UL50 of HCMV (c), N-UL34 of HSV-1 (h), N-UL34 of PrV (p), N-Orf67 of MHV68 (g), and N-BFRF1 of EBV (e). C-UL31 fusion proteins were C-M53 (M), C-UL53 of HCMV (C), C-UL31 of HSV-1 (H), C-UL31 of PrV (P), C-Orf69 of MHV68 (G), and C-BLFL2 of EBV (E). N-UL34 fusion proteins were detected at the predicted molecular masses between 50 and 80 kDa (upper panel). C-UL31 fusion proteins were detected at the predicted molecular masses between 33 and 50 kDa. Numbers indicate the positions and sizes of the protein marker in kDa. (C) Interaction of N-UL34 and C-UL31. N-UL34 and C-UL31 of the same virus were coexpressed in 293T cells. Shown from left to right are N-UL34/C-UL31 (HSV-1), N-UL34/C-UL31 (PrV), N-M50/C-M53 (MCMV), N-UL50/C-UL53 (HCMV), N-Orf67/C-Orf69 (MHV68), and N-BFRF1/C-BFLF2 (EBV). Cell lysates were prepared 24 h posttransfection, and the lactamase substrate nitrocefin was added. Mean nitrocefin hydrolysis rates were determined and are shown as Vmax in milliabsorption units/min. The DL is defined by the hydrolysis of nitrocefin after the coexpression of C-M53 and N-M50DM. Error bars indicate the standard deviation, and asterisks indicate the statistical significance.
Cross-complementation between members of the UL34 and UL31 protein families.
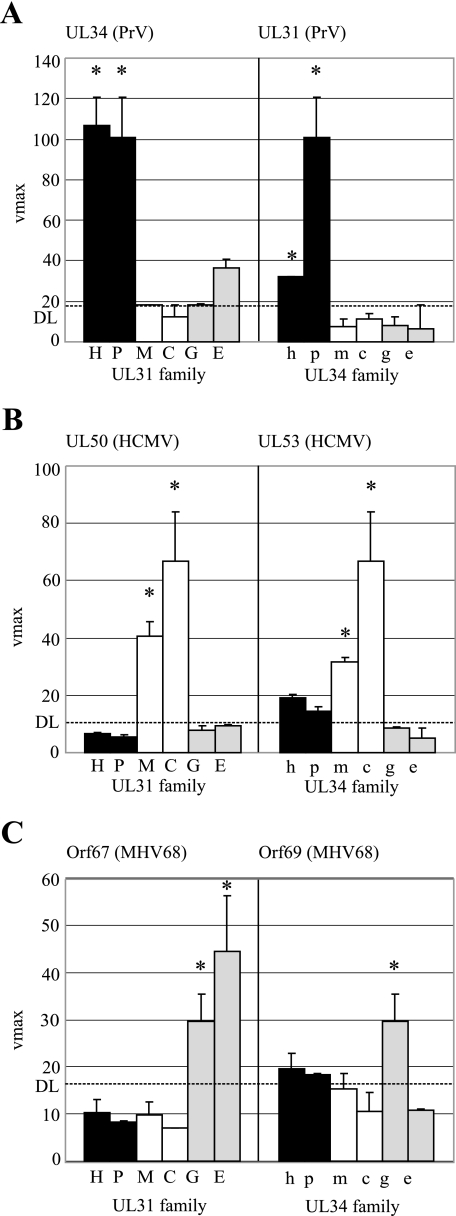
Since the PCA could be applied to other members of the UL34/UL31 protein families, we wanted to analyze to what extent the different conserved NEC proteins can bind to each other. For that purpose, the Bla fusions of UL34 and UL31 family members from three viruses, PrV, HCMV, and MHV68, representing the three herpesvirus subfamilies, were coexpressed with all six potential partners, which were tested by PCA (Fig. 3C). To measure the degree of cross-complementation, the N-UL34 homologue of PrV, HCMV, or MHV68 was coexpressed with each C-UL31 homologue and the corresponding C-UL31 homologue was coexpressed with all six N-UL34 homologues (Fig. 4).
FIG. 4.
Cross-complementation of UL34/UL31 proteins. Members of both protein families were coexpressed in heterologous combinations. (A) Representing the alphaherpesviruses, N-UL34 of PrV (p) was cross-complemented with all six C-UL31 fusion constructs (left). Accordingly, C-UL31 of PrV (P) was cross-complemented with all six N-UL34 fusion constructs (right). (B and C) The Bla fusion constructs of HCMV (B) served as representatives of the betaherpesviruses, and the Bla fusion constructs of MHV68 (C) served as representatives of the gammaherpesvirus subfamily. N-UL50, C-UL53, N-Orf67, and C-Orf69 were cross-complemented with the respective partners of the six assayed viruses. The fusion constructs were coexpressed in 293T cells. Cell lysates were prepared 24 h posttransfection, and the lactamase substrate nitrocefin was added. Mean nitrocefin hydrolysis rates were determined and are shown as Vmax in milliabsorption units/min. In the left part of the three graphs (panels A to C), the partners in the coexpressions were N-UL34 (p), N-UL50 (c), and N-Orf67 (g) in combination with C-UL31 of HSV-1 (H), C-UL31 of PrV (P), C-M53 (M), C-UL53 (C), C-Orf69 (G), and C-BFLF2 (E). In the right part of the three graphs, the partners in the coexpressions were C-UL31 of PrV (P), C-UL53 (C), and C-Orf69 (G) in combination with N-UL34 of HSV-1 (h), N-UL34 of PrV (p), N-M50 (m), N-UL50 (c), N-Orf67 (g), and N-BFRF1 (e). The indicated DL is defined by the hydrolysis rate of nitrocefin after the coexpression of C-M53 and N-M50DM. Error bars indicate the standard deviation and asterisks the significance calculated for the triplicates.
N-UL34 of PrV resulted in a high Bla activity in combination with C-UL31 of either HSV-1 or PrV. However, no significant signal was obtained in combination with C fusions to UL31 homologues of other subfamilies represented by MCMV, HCMV, MHV68, or EBV proteins (Fig. 4A, left). This shows that N-UL34 of PrV is complemented by the UL31 family members of the alphaherpesvirus subfamily only. Accordingly, the reaction of C-UL31 of PrV with the six N fusions to UL34 family members was positive only in combination with N-UL34 of either PrV or HSV-1 (Fig. 4A, right). A similar pattern was observed when either N-UL50 or N-Orf67 was coexpressed with the six C fusions (Fig. 4B and C, left) and C-UL53 or C-Orf69 with the six N fusion constructs (Fig. 4B and C, right). When either N-UL50 or N-Orf67 was kept as a constant in the experiment, the natural interaction partners, C-UL53 and C-Orf69, could be complemented only by the homologues of the virus of the same subfamily. C-Orf69 was not cross-complemented by any of the N fusions (Fig. 4C, right).
In summary, the PCA revealed a cross-complementation within the herpesvirus subfamilies. The N fusions interacted with the tested C fusions of the same subfamily to nearly the same extent as they did with the natural NEC partner. Beyond the subfamilies, no cross-complementation was detectable.
UL50 replaces M50 in the virus context.
Since the isolated NEC proteins of viruses from the same subfamily interacted in the PCA, which reflects only protein binding, we wanted to test whether the observed physical interaction would suffice for a functional replacement of NEC proteins by a homologue in the virus context. For this purpose, representative UL34 and UL31 family members were introduced into the WT MCMV genome and the deletion genomes ΔM50 and ΔM53, respectively. UL50 and UL53 were introduced into the MCMV BACs, and virus could be reconstituted in the absence of the respective MCMV gene (Table 1). Virus titers of ΔM50/UL50 were compared to those of WT MCMV under multistep growth conditions and showed no significant attenuation: the WT MCMV titer of 1.8 × 105 PFU/ml at day 5 was only slightly higher than the titer of the mutant, 1.5 × 105 PFU/ml.
TABLE 1.
Progeny characteristics of complemented WT and deletion genomes in this study
| Inserted ORF coding for: | Deletion genome
|
WT genome
|
||
|---|---|---|---|---|
| BAC | Progenya | BAC | Progenya | |
| N-M50 | ΔM50/N-M50 | +++ | NDb | NA |
| C-M53 | ΔM53/C-M53 | +++ | NDb | NA |
| UL50 | ΔM50/UL50 | +++ | NDb | NA |
| UL34(HSV-1) | ΔM50/UL34(HSV-1) | − | WT/UL34(HSV-1) | +++ |
| UL34(PrV) | ΔM50/UL34(PrV) | − | WT/UL34(PrV) | +++ |
| UL53 | ΔM53/UL53 | +++ | NDb | NA |
| UL31(HSV-1) | ΔM53/UL31(HSV-1) | − | WT/UL31(HSV-1) | +++ |
| UL31(PrV) | ΔM53/UL31(PrV) | − | WT/UL31(PrV) | +++ |
| Orf69 | ΔM53/Orf69 | − | WT/Orf69 | +++ |
| MP | ΔM53/MP | − | WT/MP | − |
| MG | ΔM53/MG | − | WT/MG | + |
+++, reconstituted as WT; +, delayed reconstitution; −, no progeny; NA, not applicable.
ND, not done.
In contrast, UL34 and UL31 homologues of HSV-1, PrV, and MHV68 could not functionally replace the NEC proteins in the deletion genomes. Virus reconstitution was successful only if M50 or M53 was still present in the mutant MCMV genome, indicating that the replication deficiency of ΔM50 expressing UL34 family members or of ΔM53 expressing UL31 family members is due to the lack of complementation rather than the toxicity of the ectopically expressed proteins (Table 1).
Thus, UL50 and UL53 of HCMV can functionally replace M50 and M53 of MCMV, whereas homologues of other herpesvirus subfamilies cannot.
The binding domain of UL31 to UL34 family members is located in the first conserved region of the protein.
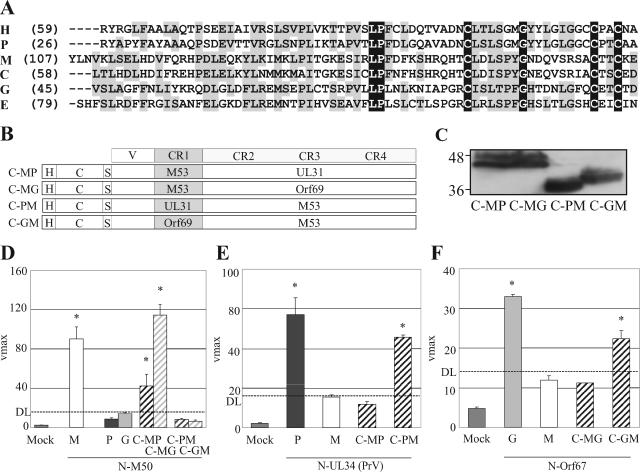
We recently located the binding site of M53 for M50 in the first conserved region (CR1) of the protein (16). For other UL31 family members, binding to the UL34 homologues has been reported (9, 13, 32), but the binding domain has not been defined. Based on the findings with M53 and the similarity of the protein sequences (Fig. 5A), we had predicted that the CR1 might harbor the binding site for UL34 family proteins in all three herpesvirus subfamilies. To this end, chimeric C fusion genes combining the variable region and the CR1 of one UL31 family member and the three subsequent conserved regions (CR2-CR4) of another were constructed. The variable region is essential only for nuclear targeting and is completely irrelevant for binding (16). CR1 of M53 was fused to CR2-CR4 of UL31 of PrV or Orf69 of MHV68 (C-MP and C-MG). Accordingly, the CR1 of UL31 or Orf69 was fused to CR2-CR4 of M53 (C-PM and C-GM) (Fig. 5B). The chimeric proteins were detected at the predicted molecular masses between 41 kDa and 53 kDa (Fig. 5C). The chimeric C fusion proteins were then coexpressed with the N fusions to UL34 family members of PrV, MCMV, and MHV68 and tested for Bla activity. As expected, a high hydrolysis rate was obtained when the NEC proteins of MCMV were coexpressed. Accordingly, no cross-complementation of N-M50 was observed in the combination of C-UL31 or C-Orf69. However, if N-M50 was coexpressed with the chimeric C fusions, bearing the CR1 of M53, binding was restored (Fig. 5D; C-MP and C-MG). In contrast, the chimeric C fusion constructs bearing the heterologous CR1s failed to complement N-M50 (Fig. 5D; C-PM and C-GM). These data confirm that the CR1 of M53 is necessary and sufficient for the binding to M50. These results also apply to the respective chimeras in combination with NEC proteins from alphaherpesviruses (Fig. 5E) and gammaherpesviruses (Fig. 5F). This shows that in all three subfamilies the UL34 binding site in the UL31 members is located within CR1.
FIG. 5.
CR1 of UL31 family members binds to UL34 family members. (A) Alignment of the first conserved region (CR1) of the studied UL31 family members. Black shading indicates the identity, and gray the similarity, of residues. (B) Schematic representation of the HA (H)-tagged constructs used. The C-terminal part (C) of the Bla was fused to chimeras consisting of the variable region (V) and the CR1 of one UL31 family member and the CR2-CR4 of another. The CR1 of M53 was combined with the CR2-CR4 of UL31 of PrV (C-MP) or the CR2-CR4 of Orf69 (C-MG). The CR1 of UL31 of PrV and the CR1 of Orf69 were combined with the CR2-CR4 of M53 (C-PM and C-GM, respectively). (C) Expression of the chimeric fusion proteins detected by HA-specific Western blotting after isolated expression. Numbers indicate the position and size of the protein marker in kDa. All constructs were detected at the predicted size between 41 kDa and 53 kDa. (D) Bla activity in cell lysates after coexpression in 293T cells. N-M50 (m) was coexpressed with C-M53 (M), C-UL31 of PrV (P), C-Orf69 (G), and the chimeric fusion constructs C-MP, C-MG, C-PM, and C-GM. (E) Bla activity in cell lysates after coexpression in 293T cells. N-UL34 (p) was coexpressed with C-UL31 (PrV) (P), C-M53 (M), and the chimeric fusion constructs C-MP and C-PM. (F) Bla activity in cell lysates after coexpression in 293T cells. N-Orf67 (g) was coexpressed with C-Orf69 (G), C-M53 (M), and the chimeric fusions C-MG and C-GM. Cell lysates were prepared 24 h posttransfection, and the lactamase substrate nitrocefin was added. Mean nitrocefin hydrolysis rates were determined and are shown as Vmax in milliabsorption units/min. The DL is defined by the hydrolysis of nitrocefin after cotransfection of C-M53 and N-M50DM. Error bars indicate the standard deviation, and asterisks indicate the statistical significance.
Binding of the chimeric UL31 proteins to M50 is not sufficient for functional replacement of M53.
The authentic CR1 of UL31 proteins is sufficient for binding to members of the UL34 family. The UL31 of HSV-1 and PrV could not functionally replace M53 in the virus context, indicating that as a minimum, the lack of binding to M50 explains the replication deficiency of these recombinant MCMVs. We asked if the restored binding capacity of the chimeric proteins to N-M50 would suffice for the complementation of the M53 null phenotype. Therefore, UL31 chimeras were constructed by a fusion of the variable region and the CR1 of M53 to the CR2-CR4 of UL31 and Orf69 (MP and MG). These chimeras were introduced into the ΔM53 BAC, and the recombinants were transfected into MEFs. Neither ΔM53/MP nor ΔM53/MG led to infectious progeny, indicating that the binding to M50 is not sufficient for virus morphogenesis (Table 1). Moreover, no infectious progeny was observed when the chimeric protein carrying the C-terminal CRs of UL31 of PrV was introduced into the WT MCMV BAC in which the WT M53 gene was still present in the genome (WT/MP). If the C-terminal CRs of Orf69 were introduced into the WT MCMV BAC (WT/MG), virus reconstitution was delayed and viral plaques were observed only after 5 weeks rather than after less than 1 week as in the case of WT MCMV. This indicated that the chimeric proteins bind to M50 in the virus context as well and compete with the formation of the WT NEC between M50 and M53.
DISCUSSION
A recent study of the herpesviral interactome (38) and sequence analyses predicted that the morphogenesis of herpesviruses meets the requirements of an conserved interaction network. We have established a cell-based PCA to study the structural/functional conservation between UL34 and UL31 family members, two key proteins of herpesvirus morphogenesis (21).
PCAs have been established with a range of reporter proteins; one such instance is the PCA involving the complementation of the murine dihydrofolate reductase (mDHFR), where fluorescein-conjugated methotrexate binds the reconstituted mDHFR (37). The mDHFR PCA allows a direct readout, and interactions can be monitored in real time; however, due to the lack of signal amplification, detection requires a high amount of interacting protein. The β-galactosidase PCA benefits from enzymatic signal amplification, but the active enzyme is a homotetramer with large individual fragments (80 kDa), and the size may cause steric problems (2, 33). Fluorescent reporter proteins, such as green fluorescent protein and yellow fluorescent protein, reassemble irreversibly (18, 27), which can provide qualitative evidence of transient interactions but may interfere negatively with the function of dynamic complexes. We chose the TEM-1 β-lactamase of E. coli (Bla) as the reporter enzyme (10) to avoid these disadvantages. The signal of the Bla PCA is amplified enzymatically, and thus only a few interacting proteins are required for detection (30, 31). Furthermore, the reversibility of the interaction allows a quantitative assessment of the interaction under study. Finally, Bla accepts numerous substrates. For example, the chromogenic substrate nitrocefin (28) and the fluorescent substrate CCF2/AM (43) have already been tested successfully for use in cell lysates and intact cells (10, 40). Nitrocefin is cost-effective and sensitive enough. The detection procedure does not require expensive instrumentation.
PCAs were applied to elucidate signal pathways involved in apoptosis or stress response (27, 37) and as a tool for mapping biological networks, either to detect proteins participating in a network or to serve as a biological sensor to detect the influence of agents on a network (22). In most previous studies, the PCA was used for an interaction screen or for the purpose of validating protein-protein interactions. Here the PCA was applied to evaluate the conservation of the interactions within UL34 and UL31 protein families and to map binding domains within the interacting proteins. This adds another PCA application to the broad field.
The MCMV proteins M50 and M53 served as the template to establish the Bla PCA for the UL34/UL31 interaction because the mutual binding sites are identified in both proteins (4, 16). Furthermore, the reverse genetics procedures for MCMV allowed the analysis of mutants in the virus context.
It was possible to extend the results gained with MCMV to study similar interactions in the UL34 and UL31 families of herpesvirus proteins in general. Most tested members of the UL34 and UL31 protein families reacted in the PCA and confirmed the described (4, 9, 32) or predicted (HCMV and MHV68) interactions. For the homologues in EBV, the signal obtained in the PCA was not significant, although their interaction was shown by other means (11, 13). This might be due to technical reasons, such as an unfavorable impact of the Bla tags on the EBV proteins or the limited sensitivity of the nitrocefin-based assay. The interaction of MHV68 proteins was significant but weaker than any other interaction that tested positive. Perhaps the NEC in gammaherpesviruses needs other factors to facilitate or stabilize the complex.
The cross-complementation study revealed that the interaction domains are still conserved but that the actual interaction sites have diverged during the evolution of herpesviruses. Moreover, the data indicate an as-yet-unknown and apparently subfamily-specific functional domain of the UL31 protein family. The chimeric UL31 proteins which fulfill the function of binding to M50 act in a dominant negative fashion and inhibit the function of the native M53. Apparently, these chimeras can form a complex with M50 via their N termini. However, they lack a function because they bear the heterologous C-terminal part of UL31 of PrV or Orf69 of MHV68, which is apparently nonfunctional in the MCMV context. Interestingly, this inhibitory effect of the MHV68 Orf69 CR2-CR4 was less stringent, which may reflect a weaker binding activity of the chimera to M50 or confirm that gammaherpesviruses are evolutionary less diverged from betaherpesviruses than alphaherpesviruses are(19). Notably, in the PCA the gammaherpesvirus chimera binds better to N-M50 than the α-chimera does (Fig. 5D).
UL53 and UL50, the HCMV homologues of M50 and M53, complemented the null phenotype of the respective deletions in MCMV. To our knowledge, this is the first example of a successful exchange of essential genes in a betaherpesvirus. For the nonessential nucleoside kinase UL97 of HCMV, we saw only a partial functional replacement in MCMV (39). MCMV, as a close relative of HCMV, provides a useful model for studying various aspects of CMV biology both in vitro and in vivo. Our data presented here indicate that certain genes of HCMV can be transferred to the MCMV background. These and comparable recombinants could be used for inhibitor and drug resistance screening in vivo. Perhaps the functional replacement of conserved proteins has a potential role in the study of proteins from viruses lacking a suitable animal model. We speculate that this complementarity can be found more frequently within the herpesvirus core genes.
The presented data show that a cell-based PCA is able to identify crucial interactions between herpesvirus proteins. The assay is simple, and the readout is at least semiquantitative; these qualities lend the NEC PCA to high-throughput inhibitor screening.
.
Acknowledgments
We thank Paul Sondermeier and Jürgen Haas for providing the subcloned ORFs of UL34 and UL31 homologues of HSV-1 and EBV and the viral DNA of PrV. We also thank S. Boos for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft through SFB 455, “Viral Functions and Immune Modulation.” M.S. was supported by GK 303, “Infection and Immunity.”
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blakely, B. T., F. M. Rossi, B. Tillotson, M. Palmer, A. Estelles, and H. M. Blau. 2000. Epidermal growth factor receptor dimerization monitored in live cells. Nat. Biotechnol. 18:218-222. [DOI] [PubMed] [Google Scholar]
- 3.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bubeck, A., M. Wagner, Z. Ruzsics, M. Lotzerich, M. Iglesias, I. R. Singh, and U. H. Koszinowski. 2004. Comprehensive mutational analysis of a herpesvirus gene in the viral genome context reveals a region essential for virus replication. J. Virol. 78:8026-8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bubic, I., M. Wagner, A. Krmpotic, T. Saulig, S. Kim, W. M. Yokoyama, S. Jonjic, and U. H. Koszinowski. 2004. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J. Virol. 78:7536-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dal Monte, P., S. Pignatelli, N. Zini, N. M. Maraldi, E. Perret, M. C. Prevost, and M. P. Landini. 2002. Analysis of intracellular and intraviral localization of the human cytomegalovirus UL53 protein. J. Gen. Virol. 83:1005-1012. [DOI] [PubMed] [Google Scholar]
- 7.De Clercq, E., and A. Holy. 2005. Acyclic nucleoside phosphonates: a key class of antiviral drugs. Nat. Rev. Drug Discov. 4:928-940. [DOI] [PubMed] [Google Scholar]
- 8.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galarneau, A., M. Primeau, L. E. Trudeau, and S. W. Michnick. 2002. Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nat. Biotechnol. 20:619-622. [DOI] [PubMed] [Google Scholar]
- 11.Gonnella, R., A. Farina, R. Santarelli, S. Raffa, R. Feederle, R. Bei, M. Granato, A. Modesti, L. Frati, H. J. Delecluse, M. R. Torrisi, A. Angeloni, and A. Faggioni. 2005. Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: interactions with BFRF1 and with the nuclear lamina. J. Virol. 79:3713-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Hagerstown, Md.
- 13.Lake, C. M., and L. M. Hutt-Fletcher. 2004. The Epstein-Barr virus BFRF1 and BFLF2 proteins interact and coexpression alters their cellular localization. Virology 320:99-106. [DOI] [PubMed] [Google Scholar]
- 14.Loregian, A., and D. M. Coen. 2006. Selective anti-cytomegalovirus compounds discovered by screening for inhibitors of subunit interactions of the viral polymerase. Chem. Biol. 13:191-200. [DOI] [PubMed] [Google Scholar]
- 15.Loregian, A., and G. Palu. 2005. Disruption of the interactions between the subunits of herpesvirus DNA polymerases as a novel antiviral strategy. Clin. Microbiol. Infect. 11:437-446. [DOI] [PubMed] [Google Scholar]
- 16.Lotzerich, M., Z. Ruzsics, and U. H. Koszinowski. 2006. Functional domains of murine cytomegalovirus nuclear egress protein M53/p38. J. Virol. 80:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lutarewych, M. A., M. R. Quirk, B. A. Kringstad, W. Li, C. M. Verfaillie, and M. C. Jordan. 1997. Propagation and titration of murine cytomegalovirus in a continuous bone marrow-derived stromal cell line (M2-10B4). J. Virol. Methods 68:193-198. [DOI] [PubMed] [Google Scholar]
- 18.Magliery, T. J., C. G. Wilson, W. Pan, D. Mishler, I. Ghosh, A. D. Hamilton, and L. Regan. 2005. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J. Am. Chem. Soc. 127:146-157. [DOI] [PubMed] [Google Scholar]
- 19.McGeoch, D. J., S. Cook, A. Dolan, F. E. Jamieson, and E. A. Telford. 1995. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 247:443-458. [DOI] [PubMed] [Google Scholar]
- 20.Menard, C., M. Wagner, Z. Ruzsics, K. Holak, W. Brune, A. E. Campbell, and U. H. Koszinowski. 2003. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 77:5557-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michnick, S. W. 2003. Protein fragment complementation strategies for biochemical network mapping. Curr. Opin. Biotechnol. 14:610-617. [DOI] [PubMed] [Google Scholar]
- 23.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Hagerstown, Md.
- 24.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 25.Neubauer, A., J. Rudolph, C. Brandmuller, F. T. Just, and N. Osterrieder. 2002. The equine herpesvirus 1 UL34 gene product is involved in an early step in virus egress and can be efficiently replaced by a UL34-GFP fusion protein. Virology 300:189-204. [DOI] [PubMed] [Google Scholar]
- 26.Nord, O., A. Gustrin, and P. A. Nygren. 2005. Fluorescent detection of beta-lactamase activity in living Escherichia coli cells via esterase supplementation. FEMS Microbiol. Lett. 242:73-79. [DOI] [PubMed] [Google Scholar]
- 27.Nyfeler, B., S. W. Michnick, and H. P. Hauri. 2005. Capturing protein interactions in the secretory pathway of living cells. Proc. Natl. Acad. Sci. USA 102:6350-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Callaghan, C. H., A. Morris, S. M. Kirby, and A. H. Shingler. 1972. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob. Agents Chemother. 1:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddehase, M. J., F. Weiland, K. Munch, S. Jonjic, A. Luske, and U. H. Koszinowski. 1985. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J. Virol. 55:264-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Remy, I., and S. W. Michnick. 1999. Clonal selection and in vivo quantitation of protein interactions with protein-fragment complementation assays. Proc. Natl. Acad. Sci. USA 96:5394-5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Remy, I., I. A. Wilson, and S. W. Michnick. 1999. Erythropoietin receptor activation by a ligand-induced conformation change. Science 283:990-993. [DOI] [PubMed] [Google Scholar]
- 32.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossi, F., C. A. Charlton, and H. M. Blau. 1997. Monitoring protein-protein interactions in intact eukaryotic cells by beta-galactosidase complementation. Proc. Natl. Acad. Sci. USA 94:8405-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rupp, B., Z. Ruzsics, T. Sacher, and U. H. Koszinowski. 2005. Conditional cytomegalovirus replication in vitro and in vivo. J. Virol. 79:486-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Spector, F. C., L. Liang, H. Giordano, M. Sivaraja, and M. G. Peterson. 1998. Inhibition of herpes simplex virus replication by a 2-amino thiazole via interactions with the helicase component of the UL5-UL8-UL52 complex. J. Virol. 72:6979-6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramaniam, R., D. Desveaux, C. Spickler, S. W. Michnick, and N. Brisson. 2001. Direct visualization of protein interactions in plant cells. Nat. Biotechnol. 19:769-772. [DOI] [PubMed] [Google Scholar]
- 38.Uetz, P., Y. A. Dong, C. Zeretzke, C. Atzler, A. Baiker, B. Berger, S. V. Rajagopala, M. Roupelieva, D. Rose, E. Fossum, and J. Haas. 2006. Herpesviral protein networks and their interaction with the human proteome. Science 311:239-242. [DOI] [PubMed] [Google Scholar]
- 39.Wagner, M., D. Michel, P. Schaarschmidt, B. Vaida, S. Jonjic, M. Messerle, T. Mertens, and U. Koszinowski. 2000. Comparison between human cytomegalovirus pUL97 and murine cytomegalovirus (MCMV) pM97 expressed by MCMV and vaccinia virus: pM97 does not confer ganciclovir sensitivity. J. Virol. 74:10729-10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wehrman, T., B. Kleaveland, J. H. Her, R. F. Balint, and H. M. Blau. 2002. Protein-protein interactions monitored in mammalian cells via complementation of beta-lactamase enzyme fragments. Proc. Natl. Acad. Sci. USA 99:3469-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitley, R. J. 2001. Herpes simplex viruses, p. 2301-2326. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Hagerstown, Md.
- 42.Yamauchi, Y., C. Shiba, F. Goshima, A. Nawa, T. Murata, and Y. Nishiyama. 2001. Herpes simplex virus type 2 UL34 protein requires UL31 protein for its relocation to the internal nuclear membrane in transfected cells. J. Gen. Virol. 82:1423-1428. [DOI] [PubMed] [Google Scholar]
- 43.Zlokarnik, G., P. A. Negulescu, T. E. Knapp, L. Mere, N. Burres, L. Feng, M. Whitney, K. Roemer, and R. Y. Tsien. 1998. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science 279:84-88. [DOI] [PubMed] [Google Scholar]