Abstract
Acanthamoeba polyphaga mimivirus is the largest known virus in both particle size and genome complexity. Its 1.2-Mb genome encodes 911 proteins, among which only 298 have predicted functions. The composition of purified isolated virions was analyzed by using a combined electrophoresis/mass spectrometry approach allowing the identification of 114 proteins. Besides the expected major structural components, the viral particle packages 12 proteins unambiguously associated with transcriptional machinery, 3 proteins associated with DNA repair, and 2 topoisomerases. Other main functional categories represented in the virion include oxidative pathways and protein modification. More than half of the identified virion-associated proteins correspond to anonymous genes of unknown function, including 45 “ORFans.” As demonstrated by both Western blotting and immunogold staining, some of these “ORFans,” which lack any convincing similarity in the sequence databases, are endowed with antigenic properties. Thus, anonymous and unique genes constituting the majority of the mimivirus gene complement encode bona fide proteins that are likely to participate in well-integrated processes.
Acanthamoeba polyphaga mimivirus (mimivirus) is the largest virus isolated so far (23). Based on its highly specific characteristics, this double-stranded-DNA icosahedral virus (47) is the first member of the new Mimiviridae family (33, 43). Computational annotation of its 1.2-Mb genome (33) revealed many atypical features, including the presence of key translation enzymes, a full complement of DNA repair pathway components, and the unique presence of three different topoisomerases (of types IA, IB, and II) (2, 33). Another unique characteristic of mimivirus is the presence of nearly identical promoter sequence motifs upstream of half of its 911 protein-encoding genes (42), which are presumably associated with proteins expressed during the early or late-early phase. Only 23% of the predicted coding genes exhibit convincing homology to proteins of known function, and 39% of them do not exhibit a clear (E values, <10−5) sequence database match (33). Such coding regions without sequence similarity to other genes in databases are considered orphan open reading frames (ORFs) and termed “ORFans” (12). The origin and function of ORFan genes are still a matter of controversy, with opinions ranging from considering them pieces of junk DNA (1, 8, 40, 44) to seeing them as quickly evolving sequences encoding normally expressed functional proteins (38, 39). Recent clinical evidence raised the possibility that mimivirus might be a human pathogen causing pneumonia (4, 24, 34), as suspected when it was first isolated from a cooling tower following an outbreak of pneumonia (23).
Mass spectrometry-based analysis has recently emerged as a technique of choice to identify more comprehensively the set of viral proteins associated with viral particles (19, 29, 49). We now present the application of this technique to the largest known, and presumably most complex, viral particle, Acanthamoeba polyphaga mimivirus.
MATERIALS AND METHODS
Sample preparation for bidimensional (2D) gel electrophoresis.
Acanthamoeba polyphaga mimivirus (23) was purified through a sucrose gradient (25%) and washed twice with phosphate-buffered saline (PBS) in the presence of protease inhibitors (Complete; Roche, Mannheim, Germany). The resulting pellet was solubilized in 40 mM Tris-HCl, pH 7.5, supplemented with 2% (wt/vol) sodium dodecyl sulfate (SDS; Sigma-Aldrich) and 60 mM dithiothreitol (DTT), followed by 5 min of heating at 95°C. The insoluble fraction was removed by centrifugation (12,000 × g, 4°C, 10 min), and soluble proteins were precipitated using a PlusOne 2-D cleanup kit (Amersham Biosciences) to remove SDS. The final pellet was resuspended in solubilization buffer {7 M urea, 2 M thiourea, 4% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)} and stored at −80°C until isoelectric focusing (IEF) was performed.
2D gel electrophoresis and silver staining.
Immobiline DryStrips (18 cm, pH 4 to 7 or 6 to 11; Amersham) were rehydrated overnight using 350 μl rehydration buffer (8 M urea, 2% [wt/vol] CHAPS, 60 mM DTT, 2% [vol/vol] Immobiline pH gradient (IPG) buffer [Amersham]) containing 200 μg of solubilized mimivirus proteins, and IEF was carried out according to the manufacturer's protocol (Multiphor II; Amersham). Before the second-dimension electrophoresis was performed, strips were equilibrated twice in 10 ml equilibration buffer (30% [vol/vol] glycerol, 2% [wt/vol] SDS, 6 M urea, 50 mM Tris-HCl, bromophenol blue, pH 8.8) for 15 min. This buffer was supplemented with 65 mM DTT for the first equilibration and with 100 mM iodoacetamide for the second one. The strips were then embedded in 0.5% agarose, and the proteins were resolved by 10% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (Ettan DALT; Amersham) at 5 W/gel for 30 min, followed by 4 to 5 h at 17 W/gel. Following migration, gels were stained by a method compatible with mass spectrometry (36). Spots excised from the gel were stored at −20°C until identification. For accuracy, the spectra of at least two separate samples of each protein were analyzed and compared.
In-gel digestion and MALDI-TOF mass spectrometry (MS).
Spots excised from silver-stained gels were destained and subjected to in-gel digestion with trypsin (sequencing-grade modified porcine trypsin; Promega, Madison, WI) (36). Tryptic peptides were then extracted from the gel by successive treatments with 5% formic acid and 50% acetonitrile-5% formic acid. Extracts were pooled and dried in a Speedvac evaporator. Peptides resuspended in an α-cyano-4-hydroxycinnamic acid matrix solution (prepared by diluting 3 times a saturated solution in 50% acetonitrile-0.3% trifluoroacetic acid) were then spotted on the matrix-assisted laser desorption ionization (MALDI) target. Mass analyses were performed on a MALDI-time of flight (MALDI-TOF) Brüker Ultraflex spectrometer (Brüker Daltonique, Wissembourg, France). Mass spectra were internally calibrated using autolytic peptides from trypsin. Tryptic peptide mass lists were used to identify the proteins, using Mascot software. Searches were performed against all available sequences in public databases, including those for eukaryotes (http://www.matrixscience.com).
Immunization and Western blotting.
Acanthamoeba polyphaga mimivirus proteins resolved by 2D gel electrophoresis were transferred onto nitrocellulose membranes (Semi-Phor unit; Hoefer Scientific, San Francisco, CA). Membranes were then blocked in PBS supplemented with 0.2% Tween 20 and 5% nonfat dry milk (PBS-Tween-milk) for 1.5 h before incubation with anti-Acanthamoeba polyphaga mimivirus sera. The sera were obtained from BALB/c mice immunized by three intraperitoneal injections (with 15-day intervals between injections) of 5 μg of purified viral particles resuspended in CpG as an adjuvant. After 1 h of incubation (1:6,400 dilution in PBS-Tween-milk), membranes were washed three times with PBS-Tween and probed with horseradish peroxidase-conjugated goat anti-mouse secondary antibodies (1:1,000; Amersham). Detection was achieved by chemiluminescence (ECL; Amersham).
SDS-PAGE coupled with electrospray ionization-ion-trap MS/MS analysis.
An Acanthamoeba polyphaga mimivirus sample (175 μg) prepared as described above was separated by SDS-PAGE on an 11% acrylamide gel (22), and the proteins were revealed afterwards by Coomassie blue R-250 staining. Bands of 1-mm thickness were systematically cut from the gel, resulting in 49 pieces to be analyzed. These bands were reduced and alkylated before trypsin digestion, and the resulting peptides were analyzed through liquid chromatography (LC) (Finnigan Surveyor HPLC system; Thermo Electron, San Jose, CA) coupled to an electrospray ionization-ion-trap mass spectrometer (LCQ-Deca XP; Thermofinnigan). Tryptic peptides were resuspended in 20 μl formic acid at 5% in water, and 1/20 of the whole extract obtained from each piece of gel was desalted on a C18 trapping column (Zorbax 300SB-C18 [5-μm inner diameter, 5 by 0.3 mm]; Agilent) by a 20-min washing step with 0.1% formic acid. Peptide separation was achieved by a 60-min linear gradient of acetonitrile (0 to 65%) in 0.2% formic acid on a C18 reverse-phase column (PicoFrit column [5-μm BioBasic C18, 300-Å pore size, 75-μm inner diameter, 100-mm long, 15-μm tip]). The peptides were then ionized at a capillary temperature of 160°C with a 2-kV spray voltage. A collision energy of 35% was applied for the MS/MS scan events. The MS/MS spectrum of the three most intense peaks was obtained after each full MS scan. The dynamic exclusion features were set at a repeat count of 2 within 0.5 min, with an exclusion duration of 3 min.
Protein identification was performed using the TurboSequest algorithm (11) in the Bioworks 3.2 software package (Thermo Electron) and the Acanthamoeba polyphaga mimivirus database (33). TurboSequest automatically identifies proteins by matching a peptide's experimental MS/MS spectrum to a predicted mass spectrum for amino acid sequences within databases. Identified peptides were evaluated using the cross-correlation number (Xcorr) versus the charge state. Criteria for positive identification of peptides were Xcorr values of >1.8 for singly charged ions, >2.5 for doubly charged ions, and >3.5 for triply charged ions. Only the best match was considered, with two or more unique peptides identifying a protein. Four proteins with masses below 19 kDa were assigned by a single peptide after visual inspection of the MS/MS spectrum.
Protein glycosylation analysis.
A purified pellet of Acanthamoeba polyphaga mimivirus was resuspended in β-mercaptoethanol (Sigma-Aldrich) and incubated overnight at 4°C. The insoluble fraction was removed by centrifugation (12,000 × g, 4°C, 10 min). Proteins present in the supernatant were separated by SDS-PAGE on a 15% acrylamide gel (22) and visualized by Coomassie R-250 staining. Glycoproteins were revealed using a GlycoProfile III kit (Sigma-Aldrich), which allows specific and sensitive in-gel fluorescence detection of glycoproteins.
Sequence annotation and ORF functional predictions.
ORF sequences were annotated using homology searches against all available databases, as previously described (33). In addition, anonymous (homologous to proteins of unknown function) and orphan sequences were reanalyzed using the 3D structure-based homology program FUGUE (37), generating more tentative structural and/or functional assignments (indicated within parentheses in Table 1). Only assignments scored as “likely” or “certain” were reported.
TABLE 1.
Mimivirus proteins identified in this study
| Protein name and category | ORFa | 2D spot no.b |
|---|---|---|
| DNA topology and repair | ||
| Topoisomerase IB (pox-like) | MIMI_R194 | NP |
| Topoisomerase IA (bacterial type—untwisting) | MIMI_L221 | NP |
| DNA polymerase family X | MIMI_L318 | NP |
| Regulator of chromosome condensation RCC1 | MIMI_R345 | 1, 2, 3, 4, 5 |
| DNA UV damage repair endonuclease | MIMI_L687 | NP |
| Transcription | ||
| DNA-directed RNA polymerase subunit 5 | MIMI_L235 | 93 |
| DNA-directed RNA polymerase subunit 2 | MIMI_L244 | NP |
| VV D6R-like helicase | MIMI_R350 | NP |
| DNA-directed RNA Pol2 G subunit | MIMI_L376 | 55 |
| D6R-like putative early transcription factor | MIMI_L377 | NP |
| African swine fever virus (ASFV) NP868R-like mRNA capping enzyme | MIMI_R382 | NP |
| DNA-directed RNA polymerase subunit L | MIMI_R470 | 37 |
| DNA-directed RNA polymerase subunit 1 | MIMI_R501 | NP |
| VV I8-like helicase | MIMI_L540 | NP |
| NPH-1-like transcription termination factor | MIMI_L538 | NP |
| TFIIB-like transcription initiation factor | MIMI_L544 | NP |
| NPH-1-like transcription termination factor | MIMI_R563 | NP |
| Oxidative pathways | ||
| Choline dehydrogenase | MIMI_R135 | 69, 70, 71, 72 |
| Amine oxidoreductase | MIMI_R188 | 10 |
| Glutaredoxin (ESV128 type) | MIMI_R195 | NP |
| Thioredoxine domain | MIMI_R362 | 96 |
| Thioredoxine domain | MIMI_R443 | 67 |
| Zn-dependent alcohol dehydrogenase | MIMI_L498 | NP |
| Putative oxidoreductase (C terminus) | MIMI_L893 | NP |
| Putative oxidoreductase (N terminus) | MIMI_L894 | NP |
| Thiol oxidoreductase E10R (C-C bonding) | MIMI_R596 | 87 |
| Protein/lipid modification | ||
| Ubiquitin-specific protease | MIMI_L293 | NP |
| Serine/threonine protein phosphatase | MIMI_R307 | NP |
| Phosphoesterase | MIMI_R398 | NP |
| F10L-like S/T protein kinase | MIMI_R400 | NP |
| Putative S/T protein kinase | MIMI_L516 | NP |
| Putative triacylglycerol lipase | MIMI_R526 | 45, 46 |
| Prolyl 4-hydroxylase | MIMI_L593 | NP |
| Particle structure | ||
| A16L-like virion-associated membrane protein | MIMI_L65 | NP |
| Major core protein | MIMI_L410 | 11, 12 |
| Capsid protein D13L | MIMI_L425 | 6, 7, 8, 9 |
| Putative spore coat assembly factor | MIMI_L454 | NP |
| Lipocalin-like outer membrane lipoprotein | MIMI_R877 | NP |
| Other | ||
| Cytoskeletal protein | MIMI_L294 | 21, 22 |
| RNA methyl transferase | MIMI_R383 | NP |
| tRNA (uracil-5-)methyltransferase | MIMI_R407* | 88, 89 |
| Putative exoribonuclease | MIMI_R528 | NP |
| Cytochrome P450 | MIMI_L532* | 75 |
| Putative elongation factor EF-Tu homolog | MIMI_R553 | 73, 74 |
| Proline-rich protein | MIMI_R610 | 59, 60 |
| Mannose-6-phosphate isomerase | MIMI_L612 | 68 |
| Phosphatidyl-ethanolamine-binding protein | MIMI_R644 | NP |
| Chemotaxis | MIMI_R706 | NP |
| Chemotaxis | MIMI_R721 | 44 |
| ORFan | MIMI_L48* | 65, 66 |
| ORFan | MIMI_L208 | NP |
| Unknown (WD repeat) | MIMI_L264 | NP |
| ORFan | MIMI_L274 | NP |
| Chilo iridescent virus 380R-like anonymous domain | MIMI_R301 | NP |
| ORFan | MIMI_L309 | NP |
| ORFan | MIMI_L330 | 83 |
| ORFan | MIMI_L352 | NP |
| ORFan | MIMI_L389 | NP |
| ORFan | MIMI_L399 | NP |
| Unknown (PBCV-1 A488R) | MIMI_L417 | NP |
| ORFan | MIMI_L442 | 23, 24, 25, 26, 30, 31, 32, 33, 34, 35, 36 |
| ORFan | MIMI_L452 | NP |
| Unknown (ankyrin repeat) | MIMI_L484 | NP |
| ORFan (biotinyl carrier) | MIMI_L485* | 95 |
| ORFan | MIMI_L488 | NP |
| ORFan (coenzyme A binding) | MIMI_L492 | NP |
| Unknown | MIMI_L515 | NP |
| ORFan (nuclease) | MIMI_L533 | NP |
| ORFan (leucine zipper) | MIMI_L550 | 97 |
| Unknown | MIMI_L567 | 78, 79, 80, 81, 82 |
| ORFan | MIMI_L585 | NP |
| ORFan | MIMI_L591 | 77 |
| ORFan (leucine zipper) | MIMI_L647 | 94 |
| ORFan | MIMI_L688 | 52, 53 |
| Unknown (WD repeat) | MIMI_L690 | 27, 28, 29 |
| Unknown (COG 3979) | MIMI_L701 | NP |
| ORFan (fusion protein [Nipah virus]) | MIMI_L724 | 85, 86 |
| ORFan | MIMI_L725 | 47, 48, 49, 50, 51 |
| ORFan | MIMI_L778 | NP |
| Unknown | MIMI_L829 | 13, 14, 15, 16, 17, 18, 54, 62 |
| ORFan | MIMI_L851 | NP |
| ORFan (aromatic amino acid hydrolase) | MIMI_L872 | 43 |
| ORFan | MIMI_L899 | NP |
| ORFan | MIMI_R160 | NP |
| Unknown | MIMI_R161 | NP |
| Unknown | MIMI_R253 | 38, 39, 40, 76 |
| ORFan | MIMI_R326 | NP |
| Unknown | MIMI_R327 | NP |
| Unknown (ASFV C475L) | MIMI_R341 | 41 |
| ORFan | MIMI_R347 | NP |
| Unknown (ASFV S273R) | MIMI_R355 | NP |
| ORFan (apolipophorin III) | MIMI_R387 | 42 |
| Unknown | MIMI_R402 | 19 |
| ORFan (kinase) | MIMI_R403 | NP |
| ORFan | MIMI_R457 | NP |
| ORFan | MIMI_R459 | 91, 92 |
| ORFan | MIMI_R463 | NP |
| Unknown | MIMI_R472 | NP |
| Unknown | MIMI_R486 | NP |
| Unknown (PBCV-1 A352L) | MIMI_R489 | 56, 57, 58 |
| ORFan | MIMI_R557 | NP |
| ORFan (acetyltransferase) | MIMI_R584 | 63, 64, 84 |
| ORFan | MIMI_R646 | NP |
| Unknown (WD repeat) | MIMI_R648 | 20 |
| ORFan | MIMI_R653 | 61 |
| ORFan | MIMI_R658 | NP |
| ORFan | MIMI_R679 | NP |
| ORFan | MIMI_R691 | NP |
| ORFan | MIMI_R692 | 90 |
| ORFan | MIMI_R695 | NP |
| ORFan | MIMI_R705 | NP |
| ORFan (alpha-synuclein) | MIMI_R710 | NP |
| Unknown | MIMI_R722 | NP |
| ORFan | MIMI_R727 | NP |
*, ORF products identified solely through 2D MS.
NP, proteins not positioned on 2D gels.
Immunogold labeling.
Immunolabeling of ultrathin sections for electron microscopy was performed as previously described (24). Briefly, ultrathin sections (70 nm) were cut using an ultramicrotome (Ultracut E; Leica), collected on 300-mesh nickel grids without Formvar (Electron Microscopy Sciences), and then incubated in PBS supplemented with 0.2% bovine serum albumin (Roche Diagnostics). The sections were then treated with 0.05 M lysine (Sigma-Aldrich) in order to neutralize putative residual aldehyde groups. After repeated washings with PBS, the sections were then incubated for 3 h at 37°C under a humidified atmosphere with a monoclonal anti-Mimi_L725 antibody (Mab6E2; 1:100) (23) diluted in PBS supplemented with 3% nonfat dry milk. The secondary reaction used goat anti-mouse antibodies conjugated to 25-nm colloidal gold particles (Aurion EM Reagents, The Netherlands), followed by washings with PBS and H2O. Gold particles were visualized using an R-GENT SE-EM silver enhancement kit (Aurion) according to the instructions of the manufacturer. After washings and aldehyde fixation, a treatment with 5% uranyl acetate (ICN Biomedicals) was done to produce intense electron-opaque staining, and samples were viewed with a Philips electron microscope (MORGAGNI 268D) at 80 kV. A negative control experiment was performed with preimmune serum.
RESULTS
Global proteomic analysis of mimivirus particle by LC-MS/MS.
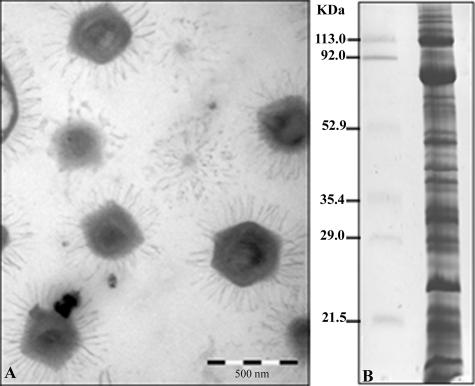
A critical parameter for every proteome project is the purity of the sample to be analyzed. This is particularly true when considering intracellular pathogens. For this study, the absence of major eukaryotic debris was assessed by negative-staining electron microscopy (Fig. 1A). Mimivirus proteins separated by one-dimensional SDS-PAGE (Fig. 1B) were fractionated in 49 pieces of gel, and corresponding tryptic digests were analyzed by capillary LC-MS/MS. Alternatively, mimivirus samples were resolved by 2D gel electrophoresis (Fig. 2), and protein identification was achieved by MALDI-TOF-MS. These complementary approaches allowed 114 proteins to be identified. These are listed in Table 1, together with their predicted functions (where applicable), and are classified into broad functional categories. The diversity of functions represented in the viral particle is quite large, with bona fide “structural” proteins amounting for a small fraction of the number of proteins. We considered only five candidates, whose sole role is to constitute the virion particle, as belonging to such a class. They are an A16L-like virion-associated membrane protein, the major core protein, the capsid protein D13L, a putative spore coat assembly factor, and a lipocalin-like outer membrane lipoprotein. Transcriptional enzymes and factors (12 gene products) constitute the largest functional category associated with the viral particle. This set includes all five DNA-directed RNA polymerase subunits as well as two helicases, one mRNA guanylyltransferase, and four transcription factors. Incidentally, one of the best identification scores was obtained with the β-subunit of the RNA polymerase (MIMI_R501), where the intron computationally identified within the gene was found to be excised as predicted (33). The next largest functional group is constituted of nine gene products, all of which are associated with oxidative pathways. Some of these enzymes might help the virus to cope with the oxidative stress generated by the host defense. The protein/lipid modification functional category is also well represented, including a phosphoesterase and a lipase, which are eventually used to digest the cell membrane, two protein kinases, and a protein phosphatase. Finally, five proteins associated with DNA topology and damage repair were identified, including the topoisomerase IA (MIMI_L221) and a DNA UV damage repair endonuclease (MIMI_L687), which were never described before as viral proteins. Other identified proteins belonging to this category are a tRNA methyltransferase and a putative mannose-6-phosphate isomerase (see Table S1 in the supplemental material). Yet the most abundant class of proteins associated with the mimivirus particle corresponds to those with unknown function (65 proteins), among which are 45 ORFans exhibiting no convincing sequence match in the databases. We noticed that the fraction of ORFan gene products found to be associated with the virion (45/114 [39.47%]) is equivalent to their proportion within the whole mimivirus genome (39%), a finding consistent with the notion that these genes are in no way different from other mimivirus genes and probably all encode bona fide proteins of importance to virus physiology. In a landmark study, Iyer et al. (16) identified a set of nucleocytoplasmic large DNA virus core genes that they further classified into four classes according to their decreasing levels of conservation across the main nucleocytoplasmic large DNA virus clades. We found that mimivirus particles incorporated four of the nine class I core gene products (including the major capsid protein), none of the class II core gene products, five of the class III gene products, and three of the class IV gene products (see Table S1 in the supplemental material). Finally, a small fraction (13/114) of the products originated from genes preceded by the highly conserved predicted mimivirus promoter (found in front of about half of all mimivirus genes) (42), confirming its putative role in governing the transcription of early or late-early genes (see Table S1 in the supplemental material).
FIG. 1.
Electron micrograph and SDS-PAGE of mimivirus. (A) Purified mimivirus particles (bar, 500 nm) separated by 11% SDS-PAGE and stained with Coomassie blue R250. (B) Bands in gels were excised for subsequent LC-MS/MS analysis after in-gel trypsin digestion. Identified proteins are listed in Table 1 (also see details in Table S1 in the supplemental material).
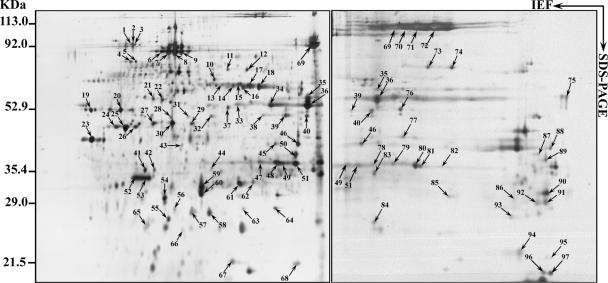
FIG. 2.
2D gel electrophoresis patterns of proteins. Acanthamoeba polyphaga mimivirus was separated using 18-cm pI 4 to 7 (left) or 6 to 11 (right) IPG strips for the first dimension (IEF) followed by 10% linear SDS-PAGE for the second dimension. Spots revealed by silver staining were cut out from the gel and subjected to trypsin digestion followed by MALDI-TOF-MS analysis. Molecular sizes and pI ranges are indicated. Identified proteins are listed in Table 1 (also see details in Table S1 in the supplemental material).
Posttranslational modifications.
As shown on 2D gels, most of the mimivirus proteins were not resolved into single spots but rather as a train of spots (Fig. 2). This was the case, for instance, for the capsid protein D13L (spots 6 to 9), which is glycosylated (Fig. 3). We observed that 66% of ORFan-encoded proteins exhibited molecular weights corresponding to their in silico predictions. Others, such as the MIMI_L442 ORFan gene product, were not located at their expected locations on the gel. Extensive analysis of the 11 corresponding isoforms showed that tryptic peptides from spot 23 matched exclusively with the central part of the protein, while spots 24 to 26 and 30 to 36 matched the N- and C-terminal extremities of the full-length 140-kDa precursor, respectively. In other cases, both the mature protein and cleavage products were identified. Other cases of discrepancy might correspond to computational errors in the identification of the true N-terminal boundary of the protein (i.e., the bona fide initiation codon). The same problem was noticed for proteins with functional attributes. According to both the areas and intensities of silver-stained spots, we can assume that some of the ORFan-encoded proteins are strongly expressed (spots 47 to 51, MIMI_L725; spots 52 to 53, MIMI_L688; and spot 61, MIMI_R653). Only four proteins not detected by LC-MS/MS were identified through 2D electrophoresis coupled with MALDI-TOF-MS.
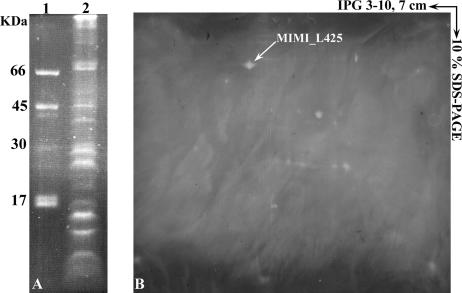
FIG. 3.
Glycosylation pattern of Acanthamoeba polyphaga mimivirus proteins detected by fluorescence assay. Mimivirus proteins were separated by 10% SDS-PAGE (A) or by 2D electrophoresis using 7-cm IPG strips (pI 3 to 10) (B), and glycosylated proteins were revealed with GlycoProfile III. Lane 1, standard glycosylation markers; lane 2, mimivirus.
Antigenic properties of ORFan-encoded mimivirus proteins.
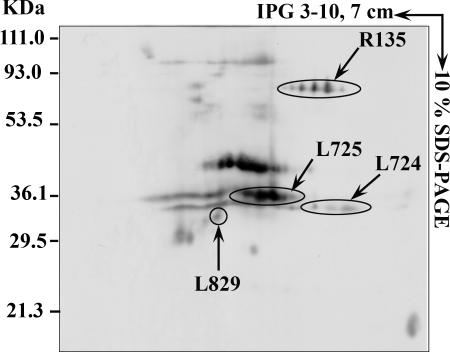
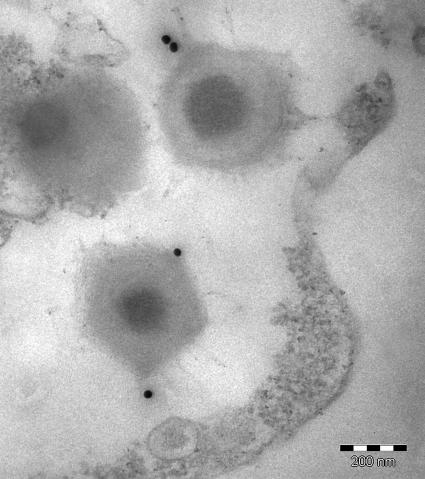
To investigate the antigenicity of mimivirus proteins, immunoblots were performed on samples resolved by 2D-PAGE, using sera from mice immunized with the whole particle (Fig. 4). Interestingly, two ORFan-encoded proteins, namely, MIMI_L724 (218 amino acids) and MIMI_L725 (224 amino acids), were specifically recognized, while no protein was recognized by preimmune sera. Both proteins are rich in cysteine residues and might be involved in a cross-linked structure accessible at the periphery of the particle. Immunogold labeling using an anti-MIMI_L725 monoclonal antibody further revealed gold particles surrounding the capsid core structure of mimivirus, confirming the exposure of the corresponding epitope to the surface (Fig. 5).
FIG. 4.
Western blot analysis of mimivirus proteins. Mimivirus proteins were separated by 2D electrophoresis using 7-cm IPG strips (pI 3 to 10) and transferred to nitrocellulose membranes before incubation with serum from an immunized mouse as described in Materials and Methods.
FIG. 5.
Localization of Mimi_L725-encoded epitope. Ultrathin sections of mimivirus-infected amoebae were immunogold labeled with a monoclonal antibody raised against the MIMI_L725-encoded protein followed by goat anti-mouse antibodies conjugated to 25-nm colloidal gold particles before immunoelectron microscopy analysis. Bar, 200 nm.
DISCUSSION
The proteomic analysis of mimivirus allowed us to identify 114 distinct proteins. Most of these proteins were identified through 1D electrophoresis coupled with LC-MS/MS, as this approach eliminated the methodological deficiencies encountered in separating low-abundance and hydrophobic proteins that are difficult to resolve by 2D electrophoresis (31). Completion of 2D gels coupled to MALDI-TOF-MS therefore provided additional and complementary information, as concluded from two recently published vaccinia virion proteome analyses (6, 48). First, for the majority of proteins identified from these gels, a good correlation between observed and theoretical pI and Mw values was observed. Analysis of 2D gels also revealed the existence of several isoforms due, in part, to protein glycosylation. Mimivirus contains six ORFs encoding putative glycosyltransferases and is thus, like Paramecium bursaria Chlorella virus (PBCV-1) (14, 46), likely to provide its own glycosylation machinery (33). The 2D gel profiles revealed cleaved proteins, a phenomenon found in many viruses and associated with synthesis and maturation steps (18, 27, 45). However, possible partial degradation of our samples due to contaminating proteases (although antiproteases were used) cannot be formally excluded, despite the reproducibility of our gel profiles.
This study also provided direct evidence that a large number of ORFan gene products contribute to the making of the mimivirus particle, with some of them acting as major components (based on the areas and intensities of the corresponding spots on 2D gels). Moreover, we demonstrated that some of these ORFan proteins are endowed with antigenic properties. Thus, proteins encoded by MIMI_L724 and MIMI_L725 were recognized by sera of mice experimentally infected with mimivirus. The antigenicity of MIMI_L724 as well as that of other ORFan proteins, encoded by MIMI_L330, MIMI_L442, and MIMI_L591, was also observed in a case of human infection (34). The surface exposure of the MIMI_L725 epitopes was further evidenced by immunogold staining with a monoclonal antibody raised against this protein. In the future, complementary experiments will be planned to localize other identified antigens. It is noteworthy that no eukaryotic proteins were identified in our samples, confirming their purity and demonstrating that eukaryotic proteins are not encapsidated within mimivirus particles. In contrast, proteomic analyses of vaccinia virus carried out by two distinct laboratories identified a significant proportion (>20%) of host proteins associated with purified intracellular mature virions (6, 17). Virus-associated host proteins include highly conserved proteins such as β-actin (6). This observation, associated with the fact that two amoebal genomes are available in the databases (10, 25), led us to discard a possible failure of identification. Genome replication and assembly of viruses often take place in specific cytoplasmic compartments, termed the viral factory (28), that may be instrumental in limiting the packaging of eukaryotic components within the viral particle. This hypothesis fits well with our own observation of mimivirus factories in infected amoebae (unpublished data).
Pending future studies of mimivirus physiology and future characterization of the virus-host interaction, the functions predicted for the proteins associated with the virion allow us to speculate on a possible molecular scenario for the early stage of the virus cycle. For instance, it is likely that the encapsidated lipolytic and proteolytic enzymes are part of the mechanism allowing the virus to gain access to the amoeba cytoplasm from its initial vacuolar location. The two topoisomerases (IA and IB) might then be involved in the ejection of the viral DNA from the particle and its injection into the cytoplasm, as well as allowing transcription to take place. These enzymes, which regulate the superhelical density of DNA (7), could indeed provide a driving force for DNA ejection into the bacterial cytoplasm by using the energy gained from changes of DNA topology (5). The DNA repair machinery components might then turn to action, allowing the viral DNA to be repaired prior to starting its transcription. Finally, the completeness of the transcription machinery components found in the viral particle is highly reminiscent of what has been described for poxviruses (15, 49), strongly suggesting that early transcription could take place in the host cytoplasm immediately upon mimivirus infection.
Altogether, these results contribute to the ongoing debate on the evolutionary origin of the gene content of large DNA viruses epitomized by mimivirus. Part of the community considers large viral genomes a “bag of genes” randomly accumulating unused laterally transferred genes (26). However, the recent release of two whole amoeba genomes (10, 25) does not favor this hypothesis (30). The fact that some of the mimivirus ORFans match unknown DNA sequences obtained from the Sargasso Sea suggests that mimivirus's uniqueness might well be related to a poor knowledge on viruses from the environment (13), which could partly explain the huge proportions of unmatched sequences in metagenomic studies (9). According to this view, the sequences of these genes might quickly lose any resemblance to their homologues under no functional constraint. Our finding of multiple ORFan products in viral particles rather suggests that ORFan proteins participate in a highly coordinated multimolecular process normally associated with tight evolutionary constraints. The unprecedented conservation of mimivirus promoters (42), as well as evidence for gene duplication generating well-conserved paralogues (20, 41), is also in favor of a well-ordered rather than chaotic model of genome evolution.
In conclusion, we believe that this work fuels the debate on the nature of viruses. Mimivirus was thought to be as complex and large as microorganisms of comparable genome size, such as rickettsiae or mycoplasmas (21). As a matter of fact, this proteomic analysis confirms that the virus's protein complexity is comparable to that of Rickettsia conorii (35). This observation, in association with the apparently common origin of the capsid protein of mimivirus and those of viruses infecting different hosts in the three domains of life, i.e., Eukarya, Bacteria, and Archaea (3), makes the question of the origin of viruses more relevant than ever (32). Finally, recent metagenomic studies indicated that up to 70% of randomly sampled DNA sequences from various environments have no database match, while 10% have similarities with viruses. Our data, with the finding of mimivirus relatives in the Sargasso Sea, suggest that environmental ORFans may indeed correspond to bona fide viral proteins whose functions and origins remain to be discovered.
Supplementary Material
Acknowledgments
We thank Jean-Yves Patrice for his help with Acanthamoeba polyphaga mimivirus culture, Claude Nappez for production of antibodies, and Bernard Campagna for immunolabeling and electron microscopy analysis. We gladly acknowledge the use of the bioinformatics platform of Marseille-Nice Génopole.
Footnotes
Published ahead of print on 13 September 2006.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Amiri, H., W. Davids, and S. G. Andersson. 2003. Birth and death of orphan genes in Rickettsia. Mol. Biol. Evol. 20:1575-1587. [DOI] [PubMed] [Google Scholar]
- 2.Benarroch, D., J.-M. Claverie, D. Raoult, and S. Shuman. 2006. Characterization of mimivirus DNA topoisomerase IB suggests horizontal gene transfer between eukaryal viruses and bacteria. J. Virol. 80:314-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson, S. D., J. K. H. Bamford, D. H. Bamford, and R. M. Burnett. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell. 16:673-685. [DOI] [PubMed] [Google Scholar]
- 4.Berger, P., L. Papazian, M. Drancourt, B. La Scola, J. P. Auffray, and D. Raoult. 2006. Ameba-associated microorganisms and diagnosis of nosocomial pneumonia. Emerg. Infect. Dis. 12:248-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bustamante, C., Z. Bryant, and S. B. Smith. 2003. Ten years of tension: single-molecule DNA mechanics. Nature 421:423-427. [DOI] [PubMed] [Google Scholar]
- 6.Chung, C. S., C. H. Chen, M. Y. Ho, C. Y. Huang, C. L. Liao, and W. Chang. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 80:2127-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbett, K. D., and J. M. Berger. 2004. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 33:95-118. [DOI] [PubMed] [Google Scholar]
- 8.Dujon, B. 1996. The yeast genome project: what did we learn? Trends Genet. 12:263-270. [DOI] [PubMed] [Google Scholar]
- 9.Edwards, R. A., and F. Rohwer. 2005. Viral metagenomics. Nat. Rev. Microbiol. 3:504-510. [DOI] [PubMed] [Google Scholar]
- 10.Eichinger, L., J. A. Pachebat, G. Glockner, M. A. Rajandream, R. Sucgang, M. Berriman, J. Song, R. Olsen, K. Szafranski, Q. Xu, B. Tunggal, S. Kummerfeld, M. Madera, B. A. Konfortov, F. Rivero, A. T. Bankier, R. Lehmann, N. Hamlin, R. Davies, P. Gaudet, P. Fey, K. Pilcher, G. Chen, D. Saunders, E. Sodergren, P. Davis, A. Kerhornou, X. Nie, N. Hall, C. Anjard, L. Hemphill, N. Bason, P. Farbrother, B. Desany, E. Just, T. Morio, R. Rost, C. Churcher, J. Cooper, S. Haydock, N. van Driessche, A. Cronin, I. Goodhead, D. Muzny, T. Mourier, A. Pain, M. Lu, D. Harper, R. Lindsay, H. Hauser, K. James, M. Quiles, M. Madan Babu, T. Saito, C. Buchrieser, A. Wardroper, M. Felder, M. Thangavelu, D. Johnson, A. Knights, H. Loulseged, K. Mungall, K. Oliver, C. Price, M. A. Quail, H. Urushihara, J. Hernandez, E. Rabbinowitsch, D. Steffen, M. Sanders, J. Ma, Y. Kohara, S. Sharp, M. Simmonds, S. Spiegler, A. Tivey, S. Sugano, B. White, D. Walker, J. Woodward, T. Winckler, Y. Tanaka, G. Shaulsky, M. Schleicher, G. Weinstock, A. Rosenthal, E. C. Cox, R. L. Chisholm, R. Gibbs, W. F. Loomis, M. Platzer, R. R. Kay, J. Williams, P. H. Dear, A. A. Noegel, B. Barrell, and A. Kuspa. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435:43-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eng, J. K., A. J. McCormak, and J. R. I. Yates. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass. Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 12.Fischer, D., and D. Eisenberg. 1999. Finding families for genomic ORFans. Bioinformatics 15:759-762. [DOI] [PubMed] [Google Scholar]
- 13.Ghedin, E., and J.-M. Claverie. 2005. Mimivirus relatives in the Sargasso Sea. Virol. J. 2:62. [Online.] http://www.virologyj.com/content/2/1/62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graves, M. V., C. T. Bernadt, R. Cerny, and J. L. Van Etten. 2001. Molecular and genetic evidence for a virus-encoded glycosyltransferase involved in protein glycosylation. Virology 285:332-345. [DOI] [PubMed] [Google Scholar]
- 15.Gross, C. H., and S. Shuman. 1996. Vaccinia virions lacking the RNA helicase nucleoside triphosphate phosphohydrolase II are defective in early transcription. J. Virol. 70:8549-8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer, L. M., L. Aravind, and E. V. Koonin. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720-11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen, O. N., T. Houthaeve, A. Shevchenko, S. Cudmore, T. Ashford, M. Mann, G. Griffiths, and J. K. Locker. 1996. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J. Virol. 70:7485-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, J. E. 1996. Functional implications of protein-protein interactions in icosahedral viruses. Proc. Natl. Acad. Sci. USA 93:27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kattenhorn, L. M., R. Mills, M. Wagner, A. Lomsadze, V. Makeev, M. Borodovsky, H. L. Ploegh, and B. M. Kessler. 2004. Identification of proteins associated with murine cytomegalovirus virions. J. Virol. 78:11187-11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin, E. V. 2005. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39:309-338. [DOI] [PubMed] [Google Scholar]
- 21.Koonin, E. V. 2005. Virology: Gulliver among the Lilliputians. Curr. Biol. 15:R167-R169. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.La Scola, B., S. Audic, C. Robert, L. Jungang, X. de Lamballerie, M. Drancourt, R. Birtles, J.-M. Claverie, and D. Raoult. 2003. A giant virus in amoebae. Science 299:2033. [DOI] [PubMed] [Google Scholar]
- 24.La Scola, B., T. J. Marrie, J. P. Auffray, and D. Raoult. 2005. Mimivirus in pneumonia patients. Emerg. Infect. Dis. 11:449-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loftus, B., I. Anderson, R. Davies, U. C. Alsmark, J. Samuelson, P. Amedeo, P. Roncaglia, M. Berriman, R. P. Hirt, B. J. Mann, T. Nozaki, B. Suh, M. Pop, M. Duchene, J. Ackers, E. Tannich, M. Leippe, M. Hofer, I. Bruchhaus, U. Willhoeft, A. Bhattacharya, T. Chillingworth, C. Churcher, Z. Hance, B. Harris, D. Harris, K. Jagels, S. Moule, K. Mungall, D. Ormond, R. Squares, S. Whitehead, M. A. Quail, E. Rabbinowitsch, H. Norbertczak, C. Price, Z. Wang, N. Guillen, C. Gilchrist, S. E. Stroup, S. Bhattachary, A. Lohia, P. G. Foster, T. Sicheritz-Ponten, C. Weber, U. Singh, C. Mukherjee, N. M. El-Sayed, W. A. Petri, Jr., C. G. Clark, T. M. Embley, B. Barrell, C. M. Fraser, and N. Hall. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433:865-868. [DOI] [PubMed] [Google Scholar]
- 26.Moreira, D., and P. Lopez-Garcia. 2005. Comment on “the 1.2-megabase genome sequence of mimivirus”. Science 308:1114. [DOI] [PubMed] [Google Scholar]
- 27.Moulard, M., and E. Decroly. 2000. Maturation of HIV envelope glycoprotein precursors by cellular endoproteases. Biochim. Biophys. Acta 1469:121-132. [DOI] [PubMed] [Google Scholar]
- 28.Novoa, R. R., G. Calderita, R. Arranz, J. Fontana, H. Granzow, and C. Risco. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97:147-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Connor, C. M., and D. H. Kedes. 2006. Mass spectrometric analyses of purified rhesus monkey rhadinovirus reveal 33 virion-associated proteins. J. Virol. 80:1574-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogata, H., C. Abergel, D. Raoult, and J.-M. Claverie. 2005. Response to comment on the “1.2-megabase genome sequence of mimivirus”. Science 308:1114b. [DOI] [PubMed] [Google Scholar]
- 31.Rabilloud, T. 2002. Two-dimensional gel electrophoresis in proteomics: old, old fashioned, but it still climbs up the mountains. Proteomics 2:3-10. [PubMed] [Google Scholar]
- 32.Raoult, D. 2006. The journey from Rickettsia to mimivirus. ASM News 71:278-284. [Google Scholar]
- 33.Raoult, D., S. Audic, C. Robert, C. Abergel, P. Renesto, H. Ogata, B. La Scola, M. Suzan, and J.-M. Claverie. 2004. The 1.2-megabase genome sequence of mimivirus. Science 306:1344-1350. [DOI] [PubMed] [Google Scholar]
- 34.Raoult, D., P. Renesto, and P. Brouqui. 2006. Laboratory infection of a technician by giant mimivirus. Ann. Intern. Med. 144:702-703. [DOI] [PubMed] [Google Scholar]
- 35.Renesto, P., S. Azza, A. Dolla, P. Fourquet, G. Vestris, J. P. Gorvel, and D. Raoult. 2005. Proteome analysis of Rickettsia conorii by two-dimensional gel electrophoresis coupled with mass spectrometry. FEMS Microbiol. Lett. 245:231-238. [DOI] [PubMed] [Google Scholar]
- 36.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 37.Shi, J., T. L. Blundell, and K. Mizuguchi. 2001. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310:243-257. [DOI] [PubMed] [Google Scholar]
- 38.Siew, N., and D. Fischer. 2004. Structural biology sheds light on the puzzle of genomic ORFans. J. Mol. Biol. 342:369-373. [DOI] [PubMed] [Google Scholar]
- 39.Siew, N., H. K. Saini, and D. Fischer. 2005. A putative novel alpha/beta hydrolase ORFan family in Bacillus. FEBS Lett. 579:3175-3182. [DOI] [PubMed] [Google Scholar]
- 40.Skovgaard, M., L. J. Jensen, S. Brunak, D. Ussery, and A. Krogh. 2001. On the total number of genes and their length distribution in complete microbial genomes. Trends Genet. 17:425-428. [DOI] [PubMed] [Google Scholar]
- 41.Suhre, K. 2005. Gene and genome duplication in Acanthamoeba polyphaga Mimivirus. J. Virol. 79:14095-14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suhre, K., S. Audic, and J.-M. Claverie. 2005. Mimivirus gene promoters exhibit an unprecedented conservation among all eukaryotes. Proc. Natl. Acad. Sci. USA 102:14689-14693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzan-Monti, M., B. La Scola, and D. Raoult. 2005. Genomic and evolutionary aspects of Mimivirus. Virus Res. 117:145-155. [DOI] [PubMed] [Google Scholar]
- 44.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2001. The COG Database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.VanSlyke, J. K., C. A. Franke, and D. E. Hruby. 1991. The multistep proteolytic maturation pathway utilized by vaccinia virus P4a protein: a degenerate conserved cleavage motif within core proteins. Virology 183:467-478. [DOI] [PubMed] [Google Scholar]
- 46.Wang, I. N., Y. Li, Q. Que, M. Bhattacharya, L. C. Lane, W. G. Chaney, and J. L. Van Etten. 1993. Evidence for virus-encoded glycosylation specificity. Proc. Natl. Acad. Sci. USA 90:3840-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao, C., P. R. Chipman, A. Battisti, V. D. Bowman, P. Renesto, D. Raoult, and M. G. Rossmann. 2005. Cryo-electron microscopy of the giant mimivirus. J. Mol. Biol. 353:493-496. [DOI] [PubMed] [Google Scholar]
- 48.Yoder, J. D., T. S. Chen, C. R. Gagnier, S. Vemulapalli, C. S. Maier, and D. E. Hruby. 2006. Pox proteomics: mass spectrometry analysis and identification of vaccinia virion proteins. Virol. J. 3:10 [Online.] http://www.virologyj.com/content/3/1/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zachertowska, A., D. Brewer, and D. H. Evans. 2005. Characterization of the major capsid proteins of myxoma virus particles using MALDI-TOF mass spectrometry. J. Virol. Methods 132:1-12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.