Abstract
Recombinant antibody cloning and phage display technologies were used to produce single-chain antibodies (scFv) against Clostridium difficile toxin B. The starting material was the mouse B cell hybridoma line 5A8, which generates a monoclonal antibody against the toxin. The integrated cloning, screening, and phage display system of Krebber et al. (J. Immunol. Methods 201:35-55, 1997) allowed us to rapidly obtain toxin B-binding scFv sequences derived from the hybridoma cell line. The best candidate scFv sequences, based on preliminary enzyme-linked immunosorbent assay (ELISA) screening data were then subcloned into the compatible expression vector. Recombinant single-chain antibodies were expressed in Escherichia coli. A 29-kDa band was observed on polyacrylamide gel electrophoresis as predicted. The expressed product was characterized by immunoblotting and detection with an anti-FLAG antibody. The toxin B-binding function of the single-chain antibody was shown by a sandwich ELISA. The antibody was highly specific for toxin B and did not cross-react with material isolated from a toxin B-negative C. difficile strain. The sensitivity of the soluble single-chain antibody is significantly higher than the original monoclonal antibody based on ELISA data and could detect a minimum of 10 ng of toxin B/well. Competitive ELISAs established that the affinity of the 5A8 parent antibody and the best representative (clone 10) of the single-chain antibodies were similar and in the range of 10−8 M. We propose that recombinant antibody technology is a rapid and effective approach to the development of the next generation of immunodiagnostic reagents.
Clostridium difficile, a gram-positive, spore-forming anaerobic bacillus, was first described in 1935 (14), but it was not associated with antibiotic-related diarrhea until the late 1970s (3, 4, 30). C. difficile infection can lead to severe complications and currently is the most common cause of nosocomial diarrhea, often adding up to 2 weeks to the length of the hospitalization, at an additional cost of $6,000 to $10,000 per case (1, 10, 22, 43, 44, 47, 55).
The organism produces two exotoxins responsible for the pathogenesis of this diarrhea, toxins A and B (12, 39). The most sensitive and specific test available for diagnosis is a tissue culture assay for the cytotoxicity of toxin B, which uses preincubation with a neutralizing antibody to demonstrate specificity (21, 40). This test can detect as little as 10 pg of toxin in stool specimens and has a high sensitivity (94 to 100%) and specificity (99%) (21, 22). However, the test takes 1 to 3 days to complete and requires specialized tissue culture facilities. More recently, enzyme-linked immunosorbent assays (ELISAs) have been developed to detect toxin A and/or toxin B in stool specimens and they have a sensitivity of 71 to 94% and a specificity of 92.5 to 98% (40, 54). Because of the rapidity of testing and ease of performance, ELISAs for toxins A and B are now used most frequently by clinical laboratories for the diagnosis of C. difficile infection, but the anti-toxin B antibody employed in the ELISA is difficult to produce and a prime target for genetic manipulation.
During the past decade, advances in antibody cloning technology have greatly facilitated the genetic manipulation of antibody fragments (6, 25). These innovations have permitted the development of a large variety of engineered antibody molecules for research, diagnosis, and therapy with specificities out of reach of conventional antibody technology. Once cloned, it is possible to improve the affinity and specificity of antigen binding by mimicking somatic hypermutation during an immune response (11).
Due to these considerations regarding traditional polyclonal and monoclonal antibody technology for the diagnosis of C. difficile infection, we have exploited recombinant antibody and phage display technologies to produce an optimized reagent. We started from the mouse B-cell hybridoma cell line 5A8 (8) which generates a monoclonal antibody against C. difficile toxin B. The cloning and phage display system employed was developed by Krebber et al. (27). Using this approach, we have successfully produced highly specific single-chain antibodies directed against C. difficile toxin B.
MATERIALS AND METHODS
We employed the antibody cloning and phage display system of Krebber et al. (27) with the following modifications.
Preparation of RNA.
The hybridoma cell line 5A8 was obtained from Meridian Biosciences. Total RNA was extracted from 5 × 106 5A8 hybridoma cells by using the Trizol total RNA extraction protocol (Gibco BRL) (5).
First-strand cDNA synthesis.
Five micrograms of total RNA was reversed transcribed in a reaction volume of 33 μl by using separate reactions for light chains and heavy chains with the primers specified by Krebber et al. (27) according to the manufacturer's protocol (first-strand cDNA synthesis kit, catalog no. 27-9261-01; Pharmacia, Piscataway, N.J.).
Cloning of variable fragment of 5A8 antibody gene.
We amplified the entire first-strand reaction mixture (33 μl) to include all of the cDNA of interest in the final mixture and added only the PCR primers and Taq DNA polymerase (catalog no. M1661; Promega, San Luis Obispo, Calif.). A cool start protocol was used. We added 40 pmol of LB and LF primer mixes for amplification of the light chain variable domain gene (VL) or 40 pmol of HB and HF primer mixes for amplification of the heavy chain variable domain gene (VH) (27). Taq polymerase (2.5 U) was added to 33 μl of cDNA mixed with water to a final reaction volume of 100 μl. The mixes were retained on ice before running the PCR program. The PCR program was as follows: denaturation at 92°C for 5 min followed by 7 cycles of 1 min at 92°C, 30 s at 63°C, 50 s at 58°C, and 1 min at 72°C and 23 cycles of 1 min at 92°C, 30 s at 63°C, and 1 min at 72°C. Subsequently, the reaction products were analyzed by running a 5-μl sample on a 1% agarose gel.
Assembly PCR.
The purified PCR products of the VL and VH sequences were prepared with a QIAquick PCR purification kit (catalog no. 28104; QIAGEN, Inc., Valencia, Calif.). Approximately 30 ng of each VL and VH DNA was combined by SOE-PCR (splicing by overlap extension) (31). Again, the cool start method was used. Taq polymerase (2.5 U) was added as before, and the solution was kept on ice. The following program was used: denaturation at 95°C for 5 min followed by 5 cycles of 1 min at 95°C, 30 s at 63°C, 50 s at 58°C, and 3 min at 72°C without primers. After adding the outer primers scback and scfor, we repeated the denaturation at 95°C for 5 min followed by 5 cycles of 1 min at 95°C, 30s at 63°C, 50 s at 58°C, and 3 min at 72°C and 35 cycles of 1 min at 95°C, 1 min at 55°C, 3 min at 72°C, and a final extension at 72°C for 7 min was employed. Five microliters of PCR mixture was analyzed by 1% agarose gel electrophoresis.
Isolation of immunoglobulin sequences from the 5A8 hybridoma.
We cloned and assembled sequences from the 5A8 hybridoma. In order to obtain enough scFv product, five assembly SOE-PCR runs were performed as above, pooling the PCR products, with ethanol precipitation and washing as usual (34, 45). We then purified the DNA on a 1% gel, and approximately 1 μg of the gel-purified scFv fragment was digested with 30 U of SfiI in an 85-μl reaction mixture at 50°C for 4 h. We purified the digested PCR insert (750 bp) on a 1% agarose gel, and the concentration was determined by running an aliquot on a 1% gel and quantitated by comparison with the known amounts of DNA on the ladder. The phage display vector pAK100 was prepared with a Wizard miniprep kit (lot no. 145302; Promega, Inc.) according to the manufacturer's instructions. Ten micrograms of pAK100 was digested with 60 U of SfiI in a 100-μl reaction volume at 50°C for 4 h. DNA was purified on a 0.5% gel as described above. After SfiI digestion, 200 ng of the vector was ligated with 20 ng of scFv (molar ratio of vector to insert, 1:1.5) and transformed into Escherichiacoli strain XL1-Blue (Stratagene). For the isolation of light and heavy chain sequences, all the transformants were spread on nonexpression medium (NE; 2× yeast-tryptone [YT] containing 1% glucose and 25 μg of chloramphenicol/ml) (27) plated in 5 dishes (100 by 15 mm, catalog no. 08-757-12; Fisher) and incubated overnight at room temperature. We then scraped off all the colonies into 5 ml of NE. Cultures were stored at −80°C after adding 15% glycerol.
Phage rescue.
Titers of phage were determined by plating. Fifty microliters of the phage culture was inoculated into 25 ml of NE medium and grown at 37°C with shaking. When an optical density at 600 nm (OD600) between 0.5 and 1.0 was reached, 500 μl of M13K07 (catalog no. 27-1524-01; Pharmacia Biotech) helper phage (about 1012 PFU), 37.5 μl of 1 M isopropyl-β-d-thiogalactopyranoside (IPTG) (final IPTG concentration, 0.5 mM), and 50 ml of NE were added and then the culture was shaken at 225 rpm at 26°C overnight for phage production.
Two hours after IPTG induction, kanamycin was added to the culture to reach a final concentration of 30 μg/ml. The next day we collected the culture medium and centrifuged it at 4,000 × g at 4°C for 10 min to eliminate bacterial cells. To precipitate the phage particles, we added to the supernatant (about 70 ml) 20 ml of 5× polyethylene glycol (PEG)-NaCl (20% PEG 8000, 2.5 M NaCl) (20) on ice for 30 min and performed centrifugation at 10,000× g at 4°C for 15 min. We then poured off the supernatant and resuspended the pellet in 30 ml of phosphate-buffered saline (PBS). The phage solution was once again mixed with 5.5 ml of 5× PEG-NaCl on ice for 20 min and centrifuged again at 10,000 × g at 4°C for 15 min. We discarded the supernatant and resuspended the phage pellet in 2 ml of PBS (containing 10% glycerol) for storage at 4°C. The phage was either subjected to further panning after titers were determined or used in a phage ELISA to determine phage antibody activity. Only freshly prepared phage was employed for panning in order to optimize binding ability.
Biopanning.
Nunc immunotubes were coated overnight with 1 ml of toxin B solution (10 μg of toxin B/ml in PBS solution, pH 7.4) at 4°C. The next day the tubes were washed 3 times with PBS by rinsing and immediately emptying the tube. For blocking, the tubes were filled with PBS plus 2% dry milk and incubated at 37°C for 2 h. The tubes were washed 3 times with PBS and 1012 PFU of phage in 1 ml of 2% dry milk in PBS was added. The tubes were left in a stationary position at 37°C for 1 h, and the unbound phage in the supernatant was discarded. We washed the tubes 10 times with PBS containing 0.1% Tween 20, and then we washed them 10 times with PBS to remove the detergent. Each washing step was performed by pouring buffer in and immediately discarding it. The excess PBS was discarded, and the phage was eluted by adding 1 ml of 100 mM triethylamine (700 μl of triethylamine in 50 ml of water, diluted on the day of use), with the tube kept stationary at room temperature for 10 min during the incubation. Elution by competition or by pH change have both proven satisfactory in our hands, as per Krebber et al. (27).
The eluted 1 ml of phage was added to another microcentrifuge tube prepared with 0.5 ml of 1 M Tris (pH 7.4) ready for quick neutralization. Half of this volume (750 μl) was added to 5 ml of mid-log phase (OD600 of 0.6 to 0.8) cultures of E. coli XL1-Blue. The cells were shaken at 250 rpm at 37°C for 1 h and centrifuged at 3,000 × g for 5 min. The supernatant was carefully discarded, and the cells were resuspended in 500 μl of 2× YT. The entire contents were spread onto 5 NE plates (100 μl per plate) and grown overnight at 26°C. After overnight growth, 1.5 ml of 2 × YT medium per plate was added. Next, we scraped off all the colonies and pooled all the cells into Eppendorf tubes. If a further round of selection was required, we then inoculated 25 ml of NE medium with 50 μl of these cells and grew them to an OD600 of 1.0. We then proceeded with the phagemid rescue as above. The remainder of the scraped cells were stored at −70°C.
Screening for binders by ELISA.
We picked 20 colonies before panning, 6 colonies after the second panning, and 5 colonies after the third panning for the binding test against toxin B. Purified toxin B and cross-reacting protein (CRP; a protein digest isolated from a toxin B-negative strain of C. difficile) were isolated by Meridian Bioscience. For isolation of the CRP, toxin B-negative bacterial strains were grown in brain heart infusion for 3 days. The cultures were harvested by centrifugation. The supernatant was clarified by filtration and passed over a G50 gel filtration column. The first peak is a mixture of proteins containing epitopes which cross-react with unabsorbed polyclonal antibodies raised against toxin B (K. Kozak, personal communication). For isolation of toxin B, positive strains were grown in brain heart infusion for 3 days. Cells were harvested by centrifugation, and the pellet was discarded. The supernatant was clarified by filtration, and a 60% saturated ammonium sulfate cut was made and passed over a DEAE column. The bound toxin was eluted with a salt gradient and further purified by passage through an S300 column. The CRP and the toxin were stored in glycerol buffer at −20°C.
ELISA plates were coated with 10 μg of toxin B/ml (100 μl/well) in PBS at 4°C, and after coating overnight, the wells were rinsed three times with PBS and blocked with 300 μl of 5% dry milk in PBS per well for 2 h at 37°C. For the preparation of phage antibody supernatant to be used as the first antibody in the ELISA, colonies of XL-1 Blue were grown separately in 2 ml of NE at 37°C. After an OD600 of 0.5 was reached, we added 2 ml of NE, 1 mM IPTG, and 109 PFU of M13K07 helper phage and grew the culture overnight at 26°C in a shaking incubator at 225 rpm. We then collected phage supernatants by centrifuging the cultures at 3,000 × g at 4°C for 10 min. We added an equal volume of 4% dry milk in PBS (MPBS) to the phage supernatants and used them directly in the assay. After blocking with 5% MPBS, wells were rinsed three times with PBS and loaded with 100 μl of phage supernatant prepared as above and incubated for 1.5 h at room temperature. The wells were washed three times for 2 min each with PBS-0.1% Tween 20. We diluted the horseradish peroxidase (HRP)-conjugated anti-M13 antibody (catalog no. 27-9411-01; Amersham Pharmacia Biotech AB) in 100 μl of 2% dry milk in PBS (1:1,000) per well and incubating the mixture for 1 h at room temperature. The plates were washed as described above. We prepared the substrate by mixing 20 ml of citric acid buffer, 8 mg of O-phenylenediamine, and 80 μl of 3% H2O2, and we incubated the plates for at least 10 min at room temperature in the dark with 150 μl of substrate per well. The reaction was stopped by adding 50 μl of 1 M H2SO4 per well. The plates were read at 492 nm in the 312e ELISA autoreader (Bio-Tek Instruments, Inc., Winooski, Vt.).
Soluble expression of single-chain antibodies.
The clones giving the highest signals against toxin B were chosen for further analysis. The scFv in pAK100 was subcloned into the expression vector pAK400 (27), which contains the same SfiI restriction sites as pAK100. The E. coli strain JM83 (which lacks an amber suppressor allowing soluble single-chain antibody expression) was transformed with pAK400 harboring the appropriate scFv candidate. The transformant was plated out on an NE plate, incubated overnight at 26°C, and then stored at 4°C. We then inoculated 50 ml of super broth medium (45) with a single bacterial colony at 26°C overnight with shaking at 250 rpm. We next inoculated 1 liter of super broth medium with this 50-ml preculture and shook the culture at 26°C at 200 rpm. To induce antibody expression, we grew the cultures to an OD600 of 0.5, added 0.25 ml of 1 M IPTG (final concentration, 0.5 mM), and continued shaking until the culture reached the stationary phase. Usually, this was within 4 to 6 h after induction, when the final OD600 reached 3 to 6. We then harvested the cells by centrifugation (8,000 × g, 10 min, 4°C). To prepare the whole-cell extract, we resuspended the cell pellet in 20 ml of HEPES extraction buffer (20 mM HEPES and 0.5 M NaCl adjusted to pH with NaOH) (53), disrupted the cells in a French press, added DNase I to a final concentration of 10 μg/ml, centrifuged the suspension at 27,000 rpm (4,820 × g) in a 70.1 T1 rotor at 4°C for 30 min, and carefully collected the supernatant containing the single-chain antibody. The solution was filtered through a 0.22-μm-pore-size filter and stored at −70°C. To prepare the periplasm extract, we resuspended the cells in 10 ml of ice-cold 1× TES (0.2 M Tris-HCl [pH 8.0], 0.5 mM EDTA. 0.5 M sucrose). To prepare 1/5× TES, we added 1 volume of 1× TES buffer to 4 volumes of distilled water. We added 15 ml of ice cold 1/5× TES and vortexed it to resuspend the bacteria (this step induces a mild osmotic shock). We incubated the mixture on ice for 30 min and then centrifuged it at 10,000 × g for 10 min, carefully transferring the supernatant, which contains the soluble antibodies from the periplasm (33). The scFv fragments were purified over a Ni2+ column (His trap kit, catalog no. 17-1880-01; Amersham Pharmacia Biotech AB) and a cation-exchange column (His trap SP HP, catalog no. 17-115-01; Amersham Pharmacia Biotech AB) (23, 27, 37).
Electrophoresis and immunoblotting.
Bacterial extracts were mixed with sample buffer with sodium dodecyl sulfate (SDS) and dithiothreitol and electrophoresed by SDS-polyacrylamide gel electrophoresis (PAGE) (29) on a 10% polyacrylamide gel. Proteins were electroblotted onto nitrocellulose to be assayed by Western blot analysis (53). For immunostaining, blots were blocked in Tris-buffered saline (TBS) containing 5% nonfat dry milk and incubated with an anti-FLAG antibody diluted 1:2,000 in TBS with 1 mM CaCl2 at room temperature for 30 min. Blots were washed three times with TBS containing 1 mM CaCl2 and incubated in HRP-conjugated goat anti-mouse antibodies at a ratio of 1:3,000 at room temperature for 30 min (catalog no. A106PS; American Qualex, San Clemente, Calif.). The last washings were performed three times in TBS containing CaCl2 for 15 min each time. The blots were developed with a chemiluminescence kit (pro no. 34080; Pierce, Rockford, Ill.) and visualized on Kodak film.
Comparison of the single-chain antibody activity with the 5A8 monoclonal antibody.
The sandwich ELISA to detect antibody activity against toxin B was modified as follows. The antibody to be tested was coated on a 96-well ELISA plate at a 1:32 dilution (15 ng/μl) and incubated at 4°C overnight. Antibody binding was determined by serially diluting the toxin B beginning at 100 ng/well. A 1:100-diluted anti-toxin B chicken immunoglobulin Y (IgY) (prepared in our laboratory) was used as the second antibody, and 1:2,000 HRP-conjugated goat anti-IgY (catalog no. A176PN; American Qualex) was used as a third antibody. The developed plates were read at 490 nm in the ELISA autoreader.
Affinity determination.
The binding constants of the soluble antibody fragments were determined by competitive ELISA (40). Briefly, wells of a microtiter plate were coated at 4°C overnight with 100 μl of toxin B (200 ng/well) in PBS. The wells were washed three times and blocked with 1% bovine serum albumin in PBS at 37°C for 3 hours. The scFv supernatants were mixed with toxin B at different concentrations in PBS-bovine serum albumin and incubated in the wells at 37°C for 2 h. The plates were washed with PBS-Tween (0.1%), anti-FLAG antibody was added, and the plates were incubated at 37°C for 30 min. The plates were washed as described, goat anti-mouse IgG conjugated to HRP (catalog no. 27-9411-01; Amersham Pharmacia Biotech AB) was added at a ratio of 1:1,000, and the plates were incubated at 37°C for 30 min. After incubation, the substrate was added and the plate was read as described. Apparent affinities were determined as the reciprocal of the toxin B concentration required to inhibit 50% maximal binding in the competitive ELISA. This is a close approximation to the true affinity and permitted the ranking of the binding activities.
RESULTS
Cloning of variable fragment of antibody genes to form the 5A8 single-chain antibody phage display cassette.
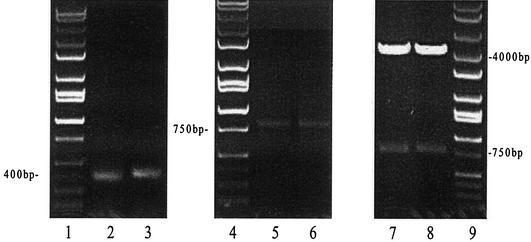
Total RNA preparation from the hybridoma line 5A8 and cDNA synthesis were performed as described above. The use of the whole-cDNA reaction mixture for PCR amplification of the variable light and heavy chain genes allowed us to obtain sufficient amounts of products for cloning. The light and heavy chains both generated a sharp band at around 400 bp as predicted previously (27). The antibody heavy and light chain DNA were subsequently joined into a single-chain fragment (scFv) by a (Gly4 Ser)3 linker sequence. The scFv could be seen on the agarose gel as a ca. 750-bp band. After purification, the scFv was then digested by SfiI and ligated into the phage display vector pAK100 to form the cassette (Fig. 1).
FIG. 1.
Agarose gels showing the sequence of cloning of the variable heavy and light chain antibody genes from the hybridoma line 5A8. Lanes 1, 4, and 9 are the DNA ladders. Lanes 2 and 3 are the light and heavy chain DNAs, respectively. Lanes 5 and 6 are assembly PCR products (the complete scFv DNA). Lanes 7 and 8 are two different samples of the recombinant expression cassette after SfiI digestion. After amplification from cDNA, the VH and VL DNA appeared on the 1% agarose gel as sharp bands at ca. 400 bp. The VH and VL were then combined into a single-chain fragment through the following assembly PCR, which is shown in lanes 5 and 6 as a 750-bp band. This scFv product was further cloned into the phage display vector pAK100 to form the single-chain antibody phage display cassette. As shown in lanes 7 and 8, the display cassette is comprised of two fragments after SfiI digestion, one is pAK100 DNA, which is about a 4-kb band on the gel, and another is the scFv insert, which is a 750-kb band on the gel as predicted.
Panning to select the phages displaying toxin B-positive recombinant antibodies.
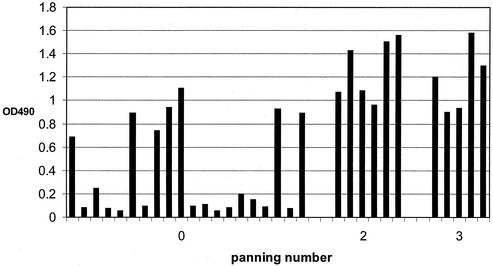
The panning procedure was used to selectively capture toxin B-positive recombinant phage with toxin B bound to an immunotube. The recombinant vector pAK100 was transformed into XL1-Blue and rescued by the M13K07 helper phage. Then the single-chain antibody was expressed as an anchored protein on the head of the phage particle. Those phage antibodies were subjected to biopanning. Three panning cycles have been carried out. The phage antibody pools prepared after 0, 1, 2, and 3 rounds of panning were tested for toxin B-specific binding in a phage ELISA. The phage ELISA results are shown in Fig. 2, in which the ELISA signal increases with each round of panning. The phage antibody gave a very weak signal against toxin B before panning with an OD490 of only 0.372. The OD490 increased as a function of the number of rounds of panning: 0.68 after the first panning, 1.41 after the second panning, and 1.78 after the third panning. The bacteriophage containing the nucleotide sequences encoding a functional scFv sequence were enriched by repeating the biopanning.
FIG. 2.
ELISA signal from phage pools as a function of the round of the biopanning cycle. Phage antibodies after 0, 1, 2, and 3 rounds of panning were tested in parallel for their antibody activity against 50/50 serially diluted toxin B or CRP, starting from 1 μg/well. The bound phage was detected with an anti-m13 antibody. The CRP antigen was coated on the plates in the same way as toxin B. Binding levels to wells coated with toxin B or CRP for pooled phage after rounds 0 (rows 1 and 2), 1 (rows 3 and 4), 2 (rows 5 and 6), and 3 (rows 7 and 8) are shown.
Based on these findings, we chose those individual clones which produce toxin B-binding phage antibody and used them to express soluble scFv antibody. For this purpose, 20 individual colonies were chosen prior to panning, 6 colonies were picked after the second panning, and 5 colonies were chosen after the third panning. They were grown and infected by M13K07 helper phage. The sequences expressed in the phage head from those single colonies were then screened for binding with toxin B by the phage ELISA (Fig. 3). The percentage of clones producing phage with specific toxin B-binding sequences increases as a function of the number of rounds of biopanning. It is 35% (7 of 20) before panning, 100% (6 of 6) after second panning, and 100% (5of 5) after the third panning. The strongest binders were chosen for further analysis: clone 10 from round 0, clone 6 from round 2, and clone 4 from round 3. These three clones were then processed to generate soluble, single-chain antibodies.
FIG. 3.
Performance of single clones screened for binding with toxin B by phage ELISA. Each column represents the ELISA signal from an individual clone isolated after the indicated number of pannings. The columns are arranged in the order in which the clones were isolated: clones 1 to 20, before panning; clones 21 to 26, after the second panning; clones 27 to 31, after the third panning. Three clones, the highest representative from each panning, were set aside for further study.
Expression of soluble antibody from cloned scFv fragments.
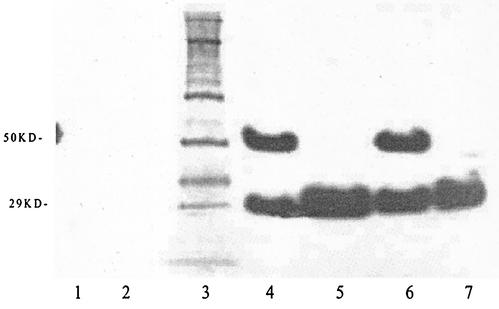
Three scFv sequences which give the strongest ELISA signals against toxin B were subcloned into the expression vector pAK400. After IPTG induction, both periplasm and whole-cell fractions were prepared to determine the location of the recombinant proteins. For further characterization, the culture extracts were subjected to reducing SDS-PAGE, immunoblotted, and probed with the anti-FLAG antibody, which recognizes the N-terminal FLAG tag fused to the antibody sequence. Only recombinant proteins in the whole-cell and periplasmic fractions demonstrated reactivity with the anti-FLAG antibody at the predicted 29-kDa molecular mass, the monomer form of the scFv antibody (Fig. 4, lanes 4 and 7). This species appears in the periplasm. Possibly due to the oligo-histidine tags in the C-terminal end of the scFv fragment, dimerized molecules also appeared in the immunoblot, at double the size of the scFv antibody (Fig. 4, lanes 4 and 6). The dimerized form was seen mainly in the whole-cell extract. The FLAG-tagged recombinant proteins were not observed in the uninduced culture or in the vector without the insert. Periplasmic fractions containing the soluble FLAG-tagged recombinant antibodies were purified for all further experiments, including antibody evaluation through ELISA.
FIG. 4.
Immunoblot of scFv antibody detected with an anti-FLAG antibody. Inserts were grown in the expression vector pAK400. Cells were harvested and samples were extracted, separated by PAGE, and transferred to nitrocellulose membranes. They were blocked, reacted with peroxidase-coupled antibody, and visualized with a bioluminescence kit. Lane 1, uninduced with IPTG. Lane 2, blank vector. Lane 3, molecular mass marker (value indicated at the left, in kilodaltons). Lane 4, whole-cell extract from clone 10. Lane 5, periplasm extract from clone 10. Lane 6, whole-cell extract from clone 6. Lane 7, periplasm extraction from clone 6.
Sandwich ELISA comparison with scFv antibodies and 5A8 hybridoma supernatant as the capture antibodies.
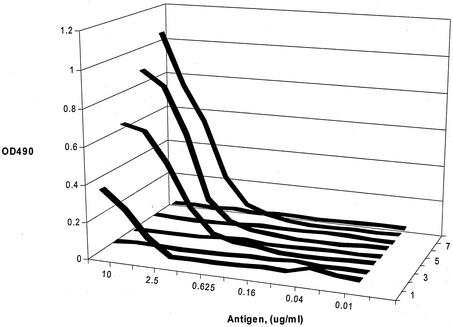
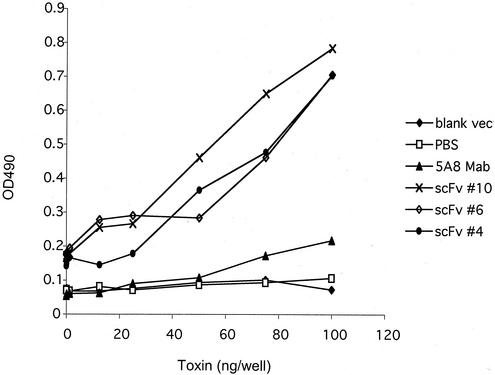
The purified soluble antibodies were used in a capture assay similar to that employed in the Meridian Bioscience diagnostic kits. The initial concentration of the antibody in the sandwich ELISA was determined, and the same amounts of all the antibodies were tested at different serial concentrations. At the 1:32 dilution (equivalent to 15 ng of IgG antibody/μl), the hybridoma supernatant monoclonal antibody no longer produced a measurable signal. However, the scFv antibody retained the ability to capture measurable antigen down to less than 10 ng of toxin B (Fig. 5). Under these conditions, all of the single-chain antibodies from different clones gave values that were substantially higher than the 5A8 monoclonal antibody. Low nonspecific reactivity was observed with the blank vector.
FIG. 5.
Sandwich ELISA of equivalent amounts of soluble scFv antibodies compared with 5A8 hybridoma supernatant. Lines are as indicated on the figure. Antibody concentrations were constant, as indicated in Methods and Materials. The toxin B concentration was serially diluted.
ELISAs for affinity measurements.
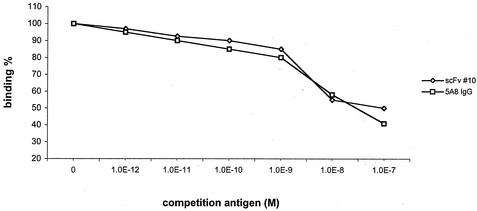
We evaluated the comparative affinities of the 5A8 parent hybridoma and the best representative of our scFv antibodies, based on the initial ELISA data. We found that the comparative affinity of the clone 10 scFv and the 5A8 parent hybridoma were the same within the range of experimental error, ca. 10−8 M (Fig. 6).
FIG. 6.
Competition ELISA to determine relative affinity for scFv clone 10 and 5A8 IgG. The relative affinities were determined by inhibition ELISA with toxin B as the competing antigen.
DISCUSSION
There is a growing need in clinical microbiology for diagnostic methods that do not rely upon culturing and cytotoxicity assays. Immunological methods are widely used, as they are rapid and relatively economical. In the last few years, a variety of molecular approaches to antibody development have become widely available. Generating polyclonal antibodies is time consuming, requires large animals, and may be subject to the vagaries of biological variability. While hybridoma technology represents a considerable advance, it is slow, expensive, and labor intensive and it may require many rounds of evaluation to achieve a satisfactory antibody. However, phage display technology when combined with cloning of antibody genes, can eliminate these drawbacks.
Recombinant antibody cloning coupled with phage display (7) is a relatively recent approach to the issue of developing better diagnostic reagents. Phage display was proposed in the mid-1980s by Smith (49). In contrast to the conventional hybridoma technology (24) for the production of monoclonal antibodies, phage display is not based on immortalization and cloning of the antibody-producing lymphocytes themselves. Rather, screening of antibody libraries can recreate the vertebrate immune system in vitro.
An enormous sample (up to 109 clones) can be isolated from a single immunized animal, and the affinity of these antibodies can be enhanced by in vitro affinity maturation (13). A prerequisite for the success of phage antibody display techniques is the reliable cloning of functional immunoglobulin genes. A DNA library of antibody sequences may be created from spleen cells from unimmunized animals or animals immunized with antigens or from hybridoma cells (if available) (32). In the present study, we cloned the variable light and heavy chain antibody fragments from the 5A8 hybridoma line, which generates a monoclonal antibody against toxin B. We also have available spleen cells taken directly from toxin B-immunized mice, which may provide a better representation of the possible antibody gene repertoire (16, 36, 52). But the most direct approach is to start from a hybridoma rather than by screening combinatorial libraries. The latter approach was reported to frequently result in the production of scFvs with low solubility and reduced binding affinities in the range 10−5 to 10−7 M, which are unsuitable for diagnostic use.
For this investigation, we employed the cloning and phage display system of Krebber et al. (27), which was quite satisfactory. The extended primer set can incorporate all of the mouse VH and VL sequences collected in the Kabat database (18) and can be modified to accommodate other species as well. In addition, the system included a FLAG peptide introduced at the N terminus of the VL chain for purification by commercial kits (19, 23) and SOE-PCR assembly from two rather than three pieces (31). Incorrect overlaps during assembly PCR were prevented by four (Gly3Ser) repeats, encoded by different codons (27, 50), and SfiI restriction sites that produce two different sticky ends at the same time (53). This is the only enzyme necessary for directional cloning of the scFv fragments into the phage display vector pAK100, preventing self-dimerization by either insert or vector (17). Other useful features of the system are described by Krebber et al. and in related papers (26, 27, 37, 48).
In our hands, the amount of light and heavy chain DNA was the critical factor for success of the assembly PCR. In order to obtain enough light and heavy chain DNA, we used the entire first-strand reaction for amplification. Both light and heavy chain DNA show up as two sharp bands of 400 bp on agarose gels. These are of the anticipated molecular weights, which were confirmed by DNA sequencing of scFv portions of the genes (Fig. 6). The concentration of both DNAs has to be determined very carefully in order to keep their amounts the same, which is critical for the success of the subsequent SOE-PCR. For us, the most accurate measure was obtained by running gel electrophoresis and correlating the intensity of bands with the known quantities on the DNA ladder. These amounts were assembled in the subsequent SOE-PCR, which yielded the single-chain variable fragment DNA of 750 bp. More than one assembly PCR was performed to obtain enough pure scFv DNA.
It appears that the function of the panning was to eliminate nonfunctional sequences from the original pool, since the ELISA performance of individual clones from later pannings did not improve. Rather, the average value of the pool increased due to the elimination of nonfunctional members. This bodes well for the future success of the many options that are available to us for improving our scFv performance.
The single chain antibody was expressed on the phage as a membrane-anchored protein. This allows us to perform antibody selection very efficiently and to isolate the best candidate scFv antibodies through biopanning. Once the best antibody candidate was chosen, the sequences were transferred into the appropriate expression plasmid for large-scale expression. Phage antibodies showed high specificities for toxin b, either as a component of the phage or as soluble antibodies, without cross-reaction with the CRP. We determined the best three candidates in our sample: from round 0, clone 10; from round 2, clone 6; from round 3, clone 4. These sequences were moved from the pAK100 vector to the pAK400 vector for expression.
For soluble scFv antibody expression, the nonsuppressed E. coli host JM83 was used. The soluble scFv antibody was visualized as a 29-kDa band by PAGE. During the process of generating the light chain and the heavy chain, the behavior of the scFv could be altered from that of the parental IgG, which may explain the superior performance of the scFv over the hybridoma supernatants. However, the binding of the single-chain antibody was determined by its performance in the sandwich ELISA, in order to duplicate the conditions employed in the Meridian Bioscience kit for toxin B diagnosis.
As shown by the sandwich ELISA data, antibody performance has been improved compared with the starting monoclonal antibody. The ELISA signal from clone 6 is much higher than the 5A8 monoclonal antibody, but the binding between the scFv and toxin is highly specific, as evidenced by the lack of cross-reactivity with the CRP. Furthermore, there is no cross-reaction between the toxin B and the bacterial extract containing the blank pAK400 vector without the scFv insert. These observations are especially striking in light of the fact that we have not yet mutagenized or shuffled the original 5A8 sequences. Clearly, if these first investigations have provided a satisfactory reagent, future manipulations potentially could produce a reagent with extremely high binding affinity.
The competitive ELISAs demonstrate conclusively that the scFvs are specific for toxin B and that the comparative affinities are similar to that of the parent monoclonal antibody (ca. 10−8). Our scFv should lend itself to favorable improvements through mutagenesis by using error-prone PCR, allowing even stronger binding affinities to be obtained.
It should be noted that we used our single-chain antibodies as the capture antibody in the ELISA procedure, although under these conditions the small scFv molecule can undergo partial denaturation (A. Plückthun, personal communication) which should lower its performance. We believe that much of the activity observed is due to spontaneous dimerization, as revealed by the immunoblot. The phenomenon of dimerization or even higher oligomerization of single-chain antibody Fv fragments has been reported previously (2). These observations are a manifestation of the phenomenon of domain swapping, which is defined as one domain of a monomeric protein being replaced by the same domain of a second, related monomeric protein chain. The scFv fragment was found to form a dimer after freezing and thawing. Additionally, the expression conditions can play a role. The difference in a dimeric fraction resulting from the expression method can also be best explained by a trapped dimer during periplasmic expression. Under these conditions, the dimeric form is favored, probably due to the at least 500-fold-higher concentration during folding in the periplasm. Other factors, such as ionic strength and pH, were shown to influence the rate of the dimer-to-monomer conversion (2). The noncovalent association of two single-chain Fv fragments leads to a considerable gain in binding ability, and thus, sensitivity is observed to be increased for all dimerization constructs compared to the monomeric scFv fragment. This is consistent with the simultaneous binding of two or even more binding sites to the same surface. It is also probably the most important reason why our antibody has better performance in ELISA than the original 5A8 antibody.
The amount of toxin B we employed in our detection techniques was 10 μg/ml. By using phage display, we are allowing for multiple interactions of the scFv on the surface of the bacteriophage with the toxin B. This allows for greater sensitivity for the antigen, so detection of the toxin within the patient stool in the presence of other bacteria and inhibitory factors can be enhanced. Using purified samples enabled us to develop the antibody, which can be altered for greater affinity if necessary for the detection of C. difficile within clinical samples.
We employed an extract from a toxin B-negative strain of C. difficile to determine if the phage scFv could cross-react with nontoxin epitopes. We recognize that other bacterial components such as other species of Clostridium or toxin B-negative C. difficile strains, which has had the toxin B rescued by plasmid expression, should be used to confirm 5A8 specificity.
It has been shown that C. difficile toxin B works in conjunction with toxin A to produce pathological effects on the host. Toxin B is also capable of producing virulent effects alone as well. Therefore, ensuring the specificity of 5A8 can enable us to target C. difficile present in the gut, even if the strain is only producing toxin B and not toxin A, which is capable of conferring pathological effects to the patient.
We are currently designing diabodies and other structural modifications of our original starting material, which we anticipate will further augment the performance of our recombinant antibody toxin B detection system (2, 19).
These data establish a proof of principle demonstrating that the system is working optimally, and we now have available soluble, single-chain antibodies against the C. difficile toxin, which will serve as a starting point for further investigations. We believe this strategy will result in antibodies with greater levels of sensitivity, by using rational design and gene shuffling protocols (9, 13, 15, 20, 28, 35, 41, 44, 50).
Acknowledgments
This work was supported by a contract from Meridian Bioscience, Cincinnati, Ohio, and by the Howard Hughes Medical Research Institute Texas Tech Undergraduate Research Training program.
REFERENCES
- 1.Anand, A., B. Bashey, T. Mir, and A. E. Glatt. 1994. Epidemiology, clinical manifestations, and outcome of Clostridium difficile-associated diarrhea. Am. J. Gastroenterol. 89:519-523. [PubMed] [Google Scholar]
- 2.Arndt, K., K. M. Muller, and A. Pluckthun. 1998. Factors influencing the dimer to monomer transition of an antibody single-chain Fv fragment. Biochemistry 37:12918-12926. [DOI] [PubMed] [Google Scholar]
- 3.Bartlett, J. G., A B. Onderdonk, and R. L. Cinseros. 1977. Clindamycin-associated colitis due to a toxin-producing species of Clostridium in hamsters. J. Infect. Dis. 136:701-705. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett, J. G., T. W. Chang, M. Gurwith, S. L. Gorbach, and A. B. Onderdonk. 1978. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. N. Engl. J. Med. 298:531-534. [DOI] [PubMed] [Google Scholar]
- 5.Berger, S. L., and J. M. Chirgwin. 1989. Isolation of RNA. Methods Enzymol. 180:3-13. [DOI] [PubMed] [Google Scholar]
- 6.Boss, M. A., J. H. Kenth, C. R. Wood, and J. S. Emtage. 1984. Assembly of functional antibodies from immunoglobulin heavy and light chains synthesized in Escherichiacoli. Nucleic Acid Res. 12:3791-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clackson, T., H. P. Hoogenboom, A. D. Griffiths, and G. Winter. 1991. Making antibody fragments using phage display libraries. Nature 352:624-628. [DOI] [PubMed] [Google Scholar]
- 8.Coughlin, R. T., M. Annunziato, J. Roberson, and D. J. Marciani. 1994. Characterization of six murine monoclonal antibodies specific for toxin B of Clostridium difficile. Hybridoma 13:147-152. [DOI] [PubMed] [Google Scholar]
- 9.Crameri, R., and M. Suter. 1993. Display of biologically active proteins on the surface of filamentous phages: a cDNA cloning system for selection of functional gene products linked to the genetic information responsible for their production. Gene 137:69-75. [DOI] [PubMed] [Google Scholar]
- 10.Djuretic, T., M. J. Ryan, D. M. Fleming, and P. G. Wall. 1996. Infectious intestinal disease in elderly people. Commun. Dis. Rep. Wkly. 19:R107-R112. [PubMed] [Google Scholar]
- 11.Gram, H., L. A. Marconi, and C. F. Barbas, T. A. Collet, R. A. Lerner, and A. S. Kang. 1992. In vitro selection and affinity maturation of antibodies from a naïve combinatorial immunoglobulin library. Proc. Natl. Acad. Sci. USA 89:3576-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grossmann, E. M., W. E. Longo, D. L. Kaminski, G. S. Smith, C. E. Murphy, R. L. Durham, M. J. Shapiro, J. G. Norman, and J. E. Mazuski. 2000. Clostridium difficile toxin: cytoskeletal changes and lactate dehydrogenase release in hepatocytes. J. Surg. Res. 88:165-172. [DOI] [PubMed] [Google Scholar]
- 13.Gunneriusson, E., K. Nord, M. Uhlen, and P. Nygren. 1999. Affinity maturation of a Taq DNA polymerase specific affibody by helix shuffling. Protein Eng. 12:873-878. [DOI] [PubMed] [Google Scholar]
- 14.Hall, I. C., and E. O'Toole. 1935. Intestinal flora in newborn infants with description of a new pathogenic anaerobe, Bacillus difficilis. Am. J. Dis. Child. 49:390-402. [Google Scholar]
- 15.Hoogenboom, H. R., A. D. Griffiths, K. S. Johnson, D. J. Chiswell, P. Hudson, and G. Winter. 1991. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 19:4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huse, W. D., L. Sastry, S. A. Iverson, A. S. Kang, M. Alting-Mees, D. R. Burton, S. J. Benkovic, and R. A. Lerner. 1989. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science 246:1275-1281. [DOI] [PubMed] [Google Scholar]
- 17.Jung, S., A. Honegger, and A. Plückthun. 1999. Selection for improved protein stability by phage display. J. Mol. Biol. 294:163-180. [DOI] [PubMed] [Google Scholar]
- 18.Kabat, E. A., T. T. Wu, M. Reid-Miller, H. M. Perry, K. S. Gottesman, and C. Foeller. 1991. Sequences of protein of immunological interest, 5th ed. National Institutes of Health, Public Health Service, U.S. Department of Health and Human Services, Washington, D.C.
- 19.Kalinke, U., A. Krebber, C. Krebber, E. Bucher, A. Plückthun, R. M. Zinkernagel, and H. Hengartner. 1996. Nucleotide monovalent single-chain Fv fragments and bivalent miniantibodies bound to vesicular stomatitis virus protect against lethal infection. Eur. J. Immunol. 26:2801-2806. [DOI] [PubMed] [Google Scholar]
- 20.Kang, A. S., C. F. Barbas, K. D. A. Janda, S. J. Benkovic, and R. A. Lerner. 1991. Linkage of recognition and replication functions by assembling combinatorial antibody Fab libraries along phage surfaces. Proc. Natl. Acad. Sci. USA 88:4363-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, C. P., C. Pothoulakis, and J. T. LaMont. 1994. Clostridium difficile colitis. N. Engl. J. Med. 330:257-262. [DOI] [PubMed] [Google Scholar]
- 22.Kelly, C. P., and J. T. LaMont. 1998. Clostridium difficile infection. Annu. Rev. Med. 49:375-390. [DOI] [PubMed] [Google Scholar]
- 23.Knappik, A., and A. Plückthun. 1994. An improved affinity tag based on the FLAG peptide for the detection and purification of recombinant antibody fragments. BioTechniques 17:754-761. [PubMed] [Google Scholar]
- 24.Kohler, G., and C. Milstein. 1975. Continuous cultures of fused cells secreting antibodies of predefined specificity. Nature 256:495-497. [DOI] [PubMed] [Google Scholar]
- 25.Kontermann, R. E., and R. Muller. 1999. Intracellular and cell surface displayed single chain diabodies. J. Immunol. Methods 226:179-188. [DOI] [PubMed] [Google Scholar]
- 26.Krebber, A., J. Burmester, and A. Plückthun. 1996. Inclusion of an upstream transcriptional terminator in phage display vectors abolishes background expression of toxic fusions with coat protein g3p. Gene 178:71-74. [DOI] [PubMed] [Google Scholar]
- 27.Krebber, A., S. Bornhauser, J. Burmester, A. Honegger, J. Willuda, H. R. Bosshard, and A. Pluckthun. 1997. Reliable cloning of functional antibody variable domains from hybridoma and spleen cell repertoires employing a reengineered phage display system. J. Immunol. Methods 201:35-55. [DOI] [PubMed] [Google Scholar]
- 28.Krebber, C., S. Spada, D. Desplancq, and A. Plückthun. 1995. Co-selection of cognate antibody-antigen pairs by selectively-infective phages. FEBS Lett. 377:227-231. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Larson, H. E., J. V. Parry, A. B. Price, D. R. Davies, and D. A. Tyrrell. 1977. Undescribed toxin in pseudomembranous colitis. Br. Med. J. 1:1246-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lefebvre, B., P. Formstecher, and P. Lefebvre. 1995. Improvement of the gene splicing overlap (SOE) method. BioTechniques 19:186-188. [PubMed] [Google Scholar]
- 32.Lowman, H. B., S. H. Bass, N. Simpson, and J. A. Wells. 1991. Selecting high-affinity binding proteins by monovalent phage display. Biochemistry 30:10832-10834. [DOI] [PubMed] [Google Scholar]
- 33.Neu, H. C., and L. A. Heppel. 1965. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J. Biol. Chem. 240:3685-3892. [PubMed] [Google Scholar]
- 34.Nohno, T., T. Saito, and J. S. Hong. 1986. Cloning and complete nucleotide sequence of the Escherichia coli glutamine permease operon (glnHPQ). Mol. Gen. Genet. 205:260-269. [DOI] [PubMed] [Google Scholar]
- 35.Ohlin, M., H. Owman, M. Mach, and C. A. Borrebaeck. 1996. Light chain shuffling of a high affinity antibody results in a drift in epitope recognition. Mol. Immunol. 33:47-56. [DOI] [PubMed] [Google Scholar]
- 36.Orlandi, R., D. H. Gussow, P. T. Jones, and G. Winter. 1989. Cloning immunoglobulin variable domains for expression by the polymerase chain reaction. Proc. Natl. Acad. Sci. USA 86:3833-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plückthun, A., A. Krebber, C. Krebber, U. Horn, U. Knüpfer, R. Wenderoth, L. Nieba, K. Proba, and D. Riensenberg. 1996. Antibody engineering, p. 203-249. Oxford University Press, Oxford, United Kingdom.
- 38.Pothoulakis, C. 2000. Effects of Clostridium difficile toxins on epithelial cell barrier. Ann. N. Y. Acad. Sci. 915:347-356. [DOI] [PubMed] [Google Scholar]
- 39.Pothoulakis, C., and J. T. LaMont. 1993. Clostridium difficile colitis and diarrhea. Gastroenterol. Clin. N. Am. 22:623-637. [PubMed] [Google Scholar]
- 40.Rath, S., C. M. Stanley, and M. W. Steward. 1988. An inhibition enzyme immunoassay for estimating relative antibody affinity and affinity heterogeneity. J. Immunol. Methods 106:245-249. [DOI] [PubMed] [Google Scholar]
- 41.Riechmann, L., and G. Winter. 2000. Novel folded protein domains generated by combinatorial shuffling of polypeptide segments. Proc. Natl. Acad. Sci. USA 97:10068-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riley, T. V., M. Cooper, B. Bell, and C. L. Golledge. 1995. Community-acquired Clostridium difficile-associated diarrhea. Clin. Infect. Dis. 20(Suppl. 2):S263-S265. [DOI] [PubMed] [Google Scholar]
- 43.Rubin, M. S., L. E. Bodenstein, and K. C. Kent. 1995. Severe Clostridium difficile colitis. Dis. Colon Rectum 38:350-354. [DOI] [PubMed] [Google Scholar]
- 44.Ryu, D. D., and D. H. Nam. 2000. Recent progress in biomolecular engineering. Biotechnol. Prog. 16:2-16. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Samore, M. H., L. Venkataraman, P. C. DeGirolami, R. D. Arbeit, and A. W. Karchmer. 1996. Clinical and molecular epidemiology of sporadic and clustered cases of nosocomial Clostridium difficile diarrhea. Am. J. Med. 100:32-40. [DOI] [PubMed] [Google Scholar]
- 47.Sastry, L., M. Alting-Mees, W. D. Huse, J. M. Short, J. A. Sorge, B. N. Hay, K. D. A. Janda, S. J. Benkovic, and R. A. Lerner. 1989. Cloning of the immunological repertoire in Escherichia coli for generation of monoclonal catalytic antibodies: construction of a heavy chain variable region-specific cDNA library. Proc. Natl. Acad. Sci. USA 86:5728-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skerra, A., and A. Plückthun. 1988. Assembly of a functional immunoglobulin Fv fragment in Escherichia coli. Science 240:1038-1041. [DOI] [PubMed] [Google Scholar]
- 49.Smith, G. P. 1985. Filamentous fusion phage: novel expression vectors that display clone antigen on the virion surface. Science 228:1315-1317. [DOI] [PubMed] [Google Scholar]
- 50.Stoop, A. A., L. Jespers, I. Lasters, E. Eldering, and H. Pannekoek. 2000. High-density mutagenesis by combined DNA shuffling and phage display to assign essential amino acid residues in protein-protein interactions: application to study structure-function of plasminogen activation inhibitor 1 (PAI-I). J. Mol. Biol. 301:1135-1147. [DOI] [PubMed] [Google Scholar]
- 51.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaughan, T. J., A. J. Williams, K. Pritchard, J. K. Osbourn, A. R. Pope, J. C. Earnshaw, J. McCafferty, R. A. Hodits, J. Wilton, and K. S. Johnson. 1996. Human antibodies with sub-nanomolar affinities isolated from a large nonimmunized phage display library. Nat. Biotechnol. 14:309-314. [DOI] [PubMed] [Google Scholar]
- 53.Wentzell, L. M., T. J. Nobbs, and S. E. Halford. 1995. The SfiI restriction endonuclease makes a four-strand DNA break at two copies of its recognition sequence. J. Mol. Biol. 248:581-595. [DOI] [PubMed] [Google Scholar]
- 54.Whittier, S., D. S. Shapiro, and W. F. Kelly. 1993. Evaluation of four commercially available enzyme immunoassays for laboratory diagnosis of Clostridium difficile-associated diseases. J. Clin. Microbiol. 31:2861-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilcox, M. H., J. G. Cunniffe, C. Trundle, and C. Redpath. 1996. Financial burden of hospital-acquired Clostridium difficile infection. J. Hosp. Infect. 34:23-30. [DOI] [PubMed] [Google Scholar]