Abstract
The type 2 iodothyronine deiodinase (D2) is an integral membrane ER-resident selenoenzyme that activates the pro-hormone thyroxine (T4) and supplies most of the 3,5,3′-triiodothyronine (T3) that is essential for brain development. D2 is inactivated by selective conjugation to ubiquitin, a process accelerated by T4 catalysis and essential for the maintenance of T3 homeostasis. A yeast two-hybrid screen of a human-brain library with D2 as bait identified von Hippel–Lindau protein–interacting deubiquitinating enzyme-1 (VDU1). D2 interaction with VDU1 and VDU2, a closely related deubiquitinase, was confirmed in mammalian cells. Both VDU proteins colocalize with D2 in the ER, and their coexpression prolongs D2 half-life and activity by D2 deubiquitination. VDU1, but not VDU2, is markedly increased in brown adipocytes by norepinephrine or cold exposure, further amplifying the increase in D2 activity that results from catecholamine-stimulated de novo synthesis. Thus, deubiquitination regulates the supply of active thyroid hormone to brown adipocytes and other D2-expressing cells.
Introduction
Thyroxine (T4), the main product of thyroid secretion, is a pro-hormone that must be activated by deiodination to 3,5,3′-triiodothyronine (T3) by one of two selenoproteins, the types 1 and 2 iodothyronine deiodinases (D1 and D2). The T3 produced by the ER-resident D2 rapidly enters the cell nucleus, contributing more than 50% of the nuclear T3 in the brain, pituitary, and brown adipose tissue (BAT) (for review, see ref. 1).
Conversion of T4 to T3 by D2 accelerates the ubiquitination of D2, inactivating it and thus serving as an important posttranslational feedback mechanism for maintaining T3 homeostasis (2–5). While the inactive ubiquitinated D2 (Ub-D2) is subject to ER-associated degradation (ERAD), our present studies demonstrate that Ub-D2 is the first recognized substrate for the recently discovered von Hippel–Lindau protein–interacting (pVHL-interacting) deubiquitinating enzyme-1 (VDU1) (6). VDU1 was isolated from human brain using the COOH-terminal portion of human D2 as bait in a yeast two-hybrid system. The interaction of D2 with VDU1, and the closely related VDU2 (7), was confirmed in mammalian cells. We also show in this report that VDU1 expression is regulated by catecholamines in BAT. This adaptive reactivation of D2 by deubiquitination constitutes a previously unrecognized pathway for amplifying the rapid cold-induced increase in D2 synthesis and consequent thyroid hormone activation in this tissue (8, 9). The expression of these ER proteins in the brain supports the concept that the ubiquitination-deubiquitination cycle plays a major role in determining intracellular T3 in D2-expressing tissues.
Methods
Yeast two-hybrid screening.
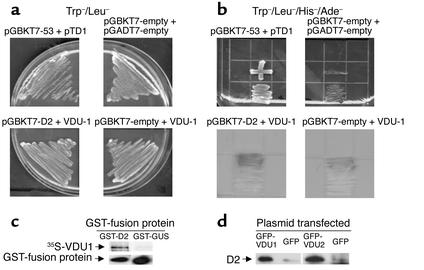
The yeast two-hybrid Matchmaker 3 system (CLONTECH Laboratories Inc., Palo Alto, California, USA) was used to screen for potential D2-interacting proteins. A human-brain cDNA library cloned into pACT2 such that it contains a GAL4 activation domain (GAL4 AD) and an hemaglutinin (HA) tag amino-terminal was pretransformed into the yeast strain Y187. This library was then screened by mating with AH109 yeast containing the D2 bait construct pGBKT7-D2. pGBKT7-D2 encodes a fusion protein that contains amino acids 1–147 of the GAL4 DNA binding domain (GAL4 DBD), a myc tag, and then amino acids 166–273 of human D2, with the selenocysteine at 266 mutated to cysteine. The D2 fragment was prepared by PCR amplification of a clone of human D2. This PCR product was digested and cloned in NcoI/EcoRI sites of pGBKT7, and the resulting plasmid was confirmed by sequencing. Western blot analysis using anti-myc antibody (CLONTECH Laboratories Inc.) further confirmed pGBKT7-D2 expression in AH109. According to the manufacturer’s instructions, yeast were initially selected on triple-dropout media that lacked the vector-selective markers leucine and tryptophan, as well as histidine; then they were re-streaked on the most stringent selection media, i.e., quadruple-dropout (QDO) media that lacked leucine, tryptophan, histidine, and adenine and contained 20 μg/ml X-α-gal, allowing for blue/white selection. Single colonies were picked and then re-streaked on QDO media containing X-α-gal two more times. Yeast colonies were then grown up, and DNA was extracted by lyticase digestion followed by detergent lysis. Purified DNA was transformed back into Escherichia coli and isolated by Qiaprep Spin Miniprep Kit (Qiagen, Valencia, California, USA). Each clone was amplified by PCR using PCR Platinum SuperMix (Invitrogen Life Technologies, Carlsbad, California, USA) and pACT2-specific primers flanking the region of the library insert to confirm a single PCR product, hence one unique clone. This DNA was reintroduced into the yeast strain Y187 and mated with yeast AH109 that contained either pGBKT7-D2 or, as a control for nonspecific interactions, empty pGBKT7 expressing only the GAL4 DBD. Clones that grew appropriately on QDO media containing X-α-gal and were determined not to interact with the GAL4 DBD alone were sequenced and blasted against the GenBank database. Since these clones are expressed with the GAL4 AD fused amino-terminally, only clones determined to be in the same reading frame as the GAL4 AD were pursued. The interaction between D2 and VDU1 was confirmed by mating of yeast that contained pGBKT7-D2 with yeast that contained pGADT7-VDU1. The resulting yeast cells grew on QDO media, whereas the negative and positive controls behaved as expected (Figure 1b).
Figure 1.
Yeast two-hybrid mating for pGBKT7-D2 and VDU1-expressing clone. (a and b) Positive control interaction for the mating: AH109 (pGBKT7-53) plus Y187 (pTD1); negative control for the mating: AH109 (pGBKT7-empty) plus Y187 (pGADT7-empty); mating: AH109 (pGBKT7-D2) plus Y187 (pGADT7-VDU1); negative control for the VDU1 mating: AH109 (pGBKT7-empty) plus Y187 (pGADT7-VDU1). (a) Mating grown on Trp–Leu– double-dropout (DDO) media. (b) Mating grown on Trp–Leu–His–Ade– QDO media plus X-α-Gal substrate. While all yeast strains grew on DDO media, indicating the presence of both expression plasmids, D2 did not interact with the GAL4 AD alone (data not shown), and, on QDO media (b), VDU1 did not interact with the GAL4 DBD alone, while pGBKT7-D2 interacted with the pGADT7-VDU1 clone. The top left panel indicates the growth of the positive control, while the negative control is indicated in the top right panel. The growth of PGBKT7-D2 + VDU-1 on QDO media plus X-α-Gal is shown on the lower left panel, while pGBKT7-empty + VDU-1 is shown on the lower right panel, indicating specificity of VDU-1 interaction with D2. (c) In vitro coimmunoprecipitation of GST-D2 and 35S-VDU1. Crude bacterial lysates expressing GST-D2 or GST-GUS were incubated with 35S-VDU1 followed by GST pulldown. Pellets were resolved by SDS-PAGE, and 35S-VDU1 is indicated. Levels of GST-fusion proteins (GST-D2 and GST-GUS) were determined by Western analysis using anti-GST antibody. Approximately 1% of the input 35S-VDU1 is specifically pulled down. This experiment was performed twice. (d) In vivo coimmunoprecipitation of D2 and VDU1 or VDU2. HEK-293 cells were cotransfected with FLAG-CysD2 and GFP-VDU1 or GFP-VDU2. Anti-GFP antibody was used for immunoprecipitation, and the pellets were resolved by SDS-PAGE and probed with anti-FLAG antibody by Western analysis. GFP refers to a vector containing only GFP, not fused to any other protein. This experiment was performed twice with similar results.
Plasmids, transfections, and D2 activity.
HEK-293 cells were transfected by the CaPO4 method (3) with plasmids containing human wild-type D2 (wtD2) (10), FLAG-CysD2 in which the Sec133 was replaced by Cys (4), FLAG-wtD2 (11), GFP-VDU1 (6), GFP-VDU2 (7), wild-type D1 (wtD1) (12), or empty vector (D10). In all experiments, human growth hormone (thymidine kinase growth hormone) was used as a control for the transfection efficiency, as described previously (13). For the VDU-D2 interaction studies (Figure 1, b and c), immunofluorescence (Figure 2b), and identification of HA-Ub-D2 (Figure 3c), a catalytically active CysD2 was used, because Sec-encoding mRNAs are translated at a much lower rate (20- to 400-fold) than non–Sec-encoding mRNAs (14). We have previously demonstrated that both wild-type CysD2 and transiently expressed CysD2 in HEK-293 cells are targeted by the ubiquitin-proteasome system (3). For all other experiments, either wtD2 or FLAG-wtD2 was transiently expressed in HEK-293 cells. Because of the small amounts of plasmid DNA that were used for transfection (1–5 μg), D2 activity in these cells was not higher than in a human mesothelioma cell line that endogenously expresses D2 (15). The fusion with the FLAG epitope does not alter D2 properties (11). In experiments using glutatione-S-transferase (GST) pulldown of in vitro translated proteins, the plasmid pET28C-VDU1 (6) or pET28C-VDU2 (7) was used. D2 activity was measured in the presence of outer ring–labeled [125I]-T4 (NEN Life Science Products Inc., Boston, Massachusetts, USA) and 1 mM propylthiouracyl, as described previously (2). In some experiments, 100 μM cycloheximide (Sigma-Aldrich, St. Louis, Missouri, USA) was added to cells transiently expressing D2 and VDU1 or VDU2 as described above. All activity values were corrected by protein concentration in each sample. In transfected HEK-293 cells, activity was also corrected by human growth hormone transfection values and expressed as fold stimulation based on control activity for each group. Each experiment was performed with n = 2–4 cell plates for each condition.
Figure 2.
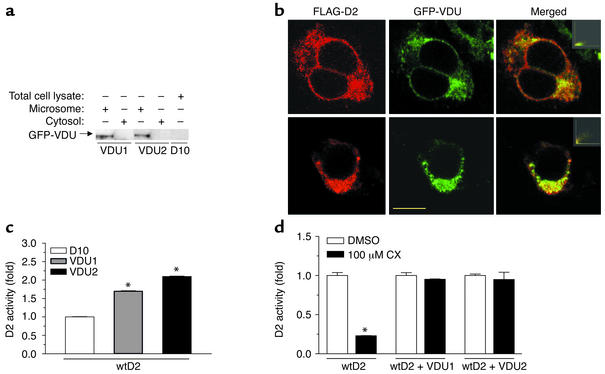
VDU1 and VDU2 are ER-resident integral membrane proteins and colocalize with D2. (a) Subcellular fractionation of cell lysates transiently expressing GFP-VDU1, GFP-VDU2, or D10. Fractions of total cell lysate, particulate fraction, and cytosol were isolated and analyzed by Western blot using anti-GFP antibody. This experiment was performed twice. (b) Colocalization of VDU1 (top panels) and VDU2 (bottom panels) with D2. Cells transiently expressing the indicated pairs of proteins were permeabilized with Triton X-100 and processed for immunocytochemistry with anti-FLAG antibody. Visualization was achieved with goat anti-mouse Texas red F(ab′)2 fragment. D2 is shown in red, while the VDUs are shown in green. The yellow bar indicates 10 μm. The distribution spectrum of image pixels is shown in the insets. (c) D2 activity in HEK-293 cells transiently coexpressing D2 and/or VDU1 or VDU2. Cells were harvested 48 hours after transfection. *P < 0.05 vs. D10-transfected cells. (d) D2 activity, as in c except that cells were treated with 0.2% DMSO (vehicle) or 100 μM cycloheximide (CX) and harvested 2 hours later. *P < 0.05 vs. DMSO-treated cells. Values in c and d are the mean ± SD of two to four cell plates; experiments were performed two to three times.
Figure 3.
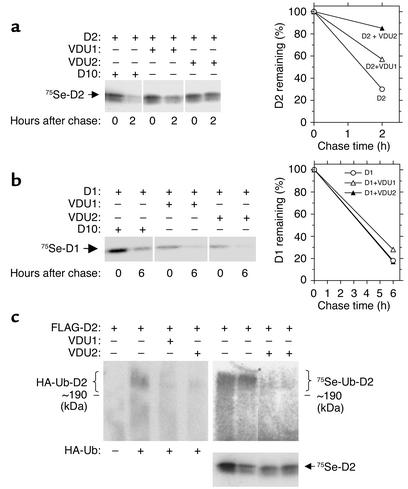
Stabilization of 75Se-D2 is due to decreased Ub-D2. (a) HEK-293 cells transiently coexpressing FLAG-wtD2 and/or VDU1 or VDU2 were labeled with Na2[75Se]O3 for 16 hours and pulse-chased with 100 nM Na2SeO3; cells were harvested at 0 or 2 hours. (b) Same as in a except that wtD1 was coexpressed with VDU1 or VDU2, with a chase time of 6 hours. Cell lysates in a and b were processed for immunoprecipitation with anti-FLAG antibody, and pellets were resolved by SDS-PAGE. The densitometric analysis is presented for each gel. (c) HEK-293 cells transiently expressing FLAG-CysD2, HA-Ub, and VDU1 or VDU2 were lysed and processed for immunoprecipitation with anti-FLAG antibody. The pellets were resolved by SDS-PAGE and probed with anti-HA antibody by Western analysis. HEK-293 cells transiently coexpressing FLAG-wtD2 and/or VDU1 or VDU2 were labeled with 75Se for 16 hours, and the cell lysates were processed as described in a. The corresponding immunoprecipitated nonubiquitinated 75Se-D2 is shown below each lane of 75Se-Ub-D2. Each experiment in a–c was performed twice.
75Se incorporation and pulse chase of D1 and D2 selenoproteins.
75Se incorporation was performed as previously described (2). Transiently expressing D2 or D1 cells were labeled with 4–6 μCi Na2[75Se]O3 per dish, kindly provided by the University of Missouri Research Reactor (St. Louis, Missouri, USA) courtesy of Marla Berry and Dolph L. Hatfield. After 16 hours of labeling, cells were washed twice with DMEM 10% FBS medium. Some plates were immediately harvested (0 hours), and some plates were kept in DMEM 10% FBS medium plus 100 nM Na2SeO3 for 2 hours (D2) or 6 hours (D1). Immunoprecipitation (IP) with anti-FLAG antibody was performed as previously described (15).
In vitro and in vivo coimmunoprecipitation.
An in vitro coupled transcription/translation system (Promega Corp., Madison, Wisconsin, USA) and L-[35S]-methionine/cysteine EasyTag (Perkin-Elmer Life Sciences, Boston, Massachusetts, USA) were used to generate recombinant proteins. Ten microliters of the indicated in vitro translated pET28C-VDU1 or pET28C-VDU2 was added to the GST-D2 or GST-GUS (Arabidopsis thaliana β-glucoronidase) previously bound to GST resin, and the GST pulldown was performed as previously described (11). Pellets were resolved in a 10% SDS-PAGE gel, dried, and exposed to autoradiography for 35S-labeled band detection. For the in vivo coimmunoprecipitation experiments, HEK-293 cells transiently expressing 5 μg GFP-VDU1 or 5 μg GFP-VDU2 and 5 μg FLAG-CysD2 were resuspended in lysis buffer containing 1× PBS, 5 mM EDTA, 0.5% Triton X-100 (Sigma-Aldrich), type I protease inhibitor cocktail (Calbiochem-Novabiochem Corp., San Diego, California, USA), and 1 mM PMSF. Cell lysates with each transfection combination were incubated with Living Colors Full-Length Av polyclonal antibody (anti-GFP antibody; BD Biosciences Clontech, Palo Alto, California, USA) at a dilution of 1:300. Forty microliters of Protein G PLUS/Protein A–Agarose bead suspension (Oncogene Research Products, San Diego, California, USA) was added, and IP pellets were washed five times with 1× PBS, resuspended in sample loading buffer, boiled for 5 minutes, resolved in a 10% SDS-PAGE, and used for Western blot analysis for FLAG-CysD2 detection. FLAG-CysD2 detection, using anti–FLAG M2 mAb (Sigma-Aldrich) at a dilution of 1:2,000 (4), was performed using the BM chemiluminescence Western blotting kit (Roche, Indianapolis, Indiana, USA) according to the manufacturer’s instructions.
In vivo D2 deubiquitination.
HEK-293 cells transiently expressing 5 μg FLAG-CysD2 and 5 μg HA-Ub, with or without 5 μg VDU1 or 5 μg VDU2, were processed for IP as described above using anti-FLAG antibody at 1:250. The IP pellets were resolved in a 7.5% SDS-PAGE gel and used for Western blot analysis with anti-HA polyclonal antibody (CLONTECH Laboratories Inc.).
Immunofluorescence confocal microscopy.
Immunofluorescence confocal microscopy was done as described previously (16) with minor modifications. HEK-293 cells were transfected with GFP-VDU1 or GFP-VDU2 and FLAG-CysD2. After paraformaldehyde fixation, cells were permeabilized with 0.25% Triton X-100 and incubated with anti-FLAG antibody at a dilution of 1:500. This was followed by 30 minutes’ incubation with affinity-purified goat anti-mouse Texas red F(ab′)2 fragment (Molecular Probes Inc., Eugene, Oregon, USA) at a dilution of 1:1,000. Specimens were examined by conventional confocal microscopy on a Bio-Rad (Hercules, California, USA) MRC-1024/2P system interfaced with a Zeiss Axiovert microscope (Carl Zeiss MicroImaging Inc., Thornwood, New York, USA).
Subcellular fractionation.
Lysates of HEK-293 cells transiently expressing GFP-VDU1, GFP-VDU2, or D10 were processed for isolation of the particulate fraction (microsomes). The total cell lysate, the particulate pellet (obtained by 100,000 g centrifugation for 90 minutes), and the cytosol (supernatant of the centrifugation) were isolated and analyzed by Western blot using anti-GFP antibody at a dilution of 1:1,000.
Mice.
The Standing Committee on Animal Research at Harvard Medical School approved all aspects of animal care and experimentation in this study. Wild-type male C57BL/6 mice (The Jackson Laboratory, Bar Harbor, Maine, USA) weighing 20–30 g were kept in individual cages on a 12-hour/12-hour dark/light cycle with free access to chow and water. Animals were maintained at room temperature or exposed to 4°C for up to 48 hours, as indicated in the figures. As indicated, animals were injected with 0.75 μg norepinephrine (NE) per 1 g body weight. Animals were euthanized, and the tissues were immediately isolated and frozen in liquid nitrogen for RNA isolation and/or cell lysis for subsequent deiodinase assay.
Real-time PCR.
Total RNA was extracted using TRIzol reagent (Invitrogen Life Technologies) and used to synthesize cDNA using SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen Life Technologies). The generated cDNAs were used in a real-time PCR using the QuantiTect SYBR Green PCR kit in I-Cycler (Bio-Rad). Standard curves representing five-point serial dilution of mixed cDNA of the experimental and control groups were analyzed in each assay and used as calibrators to the relative quantification of product generated in the exponential phase of the amplification curve. The r2 was greater than 0.99 for all standard curves, and the amplification efficiency varied between 80% and 100%. Sample quantification was calculated by the standard curve and corrected by the internal control β-actin mRNA in all experiments. Specific oligonucleotides for mouse VDU1, VDU2, D2, and β-actin were designed using Beacon Designer 2.06 (PREMIER Biosoft International, Palo Alto, California, USA).
Results
VDU1 and VDU2 interact specifically with D2 in vivo and colocalize in the ER.
We screened a human-brain library with a C-terminal fragment of human D2 (residues 166–273) using a yeast two-hybrid system and identified a specific interacting protein containing the amino acids 585–786 of the recently identified VDU1 (GenBank accession no. AF383173) (Figure 1, a and b). Using GST pulldown, we verified that the interaction between GST-D2 and 35S-VDU1 occurs in vitro (Figure 1c). Moreover, in HEK-293 cells, we demonstrated that D2 interacts not only with VDU1 but also with the closely related VDU2, as FLAG-CysD2 is coimmunoprecipitated with either GFP-VDU1 or GFP-VDU2, but not in vector-transfected cells (Figure 1d).
Subcellular fractionation of HEK-293 cells transiently expressing VDU1 and VDU2 revealed that both proteins are found in the particulate pellet (microsomes) but not in the cytosolic fraction (Figure 2a). Analysis of the protein sequences suggests a low probability of a single transmembrane domain (17), suggesting that VDUs are membrane-associated, rather than integral membrane, proteins. Immunofluorescence confocal microscopy studies of HEK-293 cells transiently coexpressing FLAG-CysD2 and GFP-VDU1 or FLAG-CysD2 and GFP-VDU2 indicate that both VDU1 and VDU2 colocalize with D2 in the perinuclear region (Figure 2b). Furthermore, VDU1 and VDU2 also colocalize with BIP, an ER-resident chaperone (data not shown).
D2 is a substrate for VDU1 and VDU2.
Coexpression of VDU1 or VDU2 with D2 in HEK-293 cells increased basal D2 activity 1.7- and 2.1-fold, respectively (Figure 2c). Protein synthesis was blocked with cycloheximide to ascertain whether coexpression with VDU1 or VDU2 stabilizes D2 activity. Blockade of protein synthesis for 2 hours reduced D2 activity to about 25% of that in vehicle-treated cells, which is compatible with previous observations (3). However, D2 activity remained stable after protein synthesis was blocked in cells coexpressing either VDU1 or VDU2, indicating a marked increase in the half-life of active D2 (Figure 2d).
Deubiquitination of D2 would be predicted to decrease its uptake by proteasomes, which is known to be rate limiting in D2 degradation (2, 3). This was demonstrated in HEK-293 cells coexpressing FLAG-D2 and VDU1 or VDU2 labeled with Na2[75Se]O3 and pulse-chased with 100 nM Na2SeO3. Pulse-chase studies revealed that 75Se-D2 protein decreases about 70% within 2 hours (Figure 3a), which is compatible with a 30-minute half-life. When coexpressed with VDU1 or VDU2 in a large series of experiments, the rate of decrease in 75Se-D2 is reduced to 15–40% within 2 hours, confirming that both VDUs stabilize D2 protein. This effect is specific, since the levels of 75Se-D1, an isoenzyme that is not ubiquitinated, were not affected by coexpression with VDU1 or VDU2 (Figure 3b).
To confirm that it is deubiquitination by VDU1 and VDU2 that stabilizes D2, lysates of HEK-293 cells transiently expressing FLAG-CysD2, HA-Ub, and VDU1 or VDU2 were immunoprecipitated with anti-FLAG antibody, and the pellets were resolved by SDS-PAGE and probed with anti-HA antibody. The accumulation of HA-Ub-D2 containing high–molecular weight bands was blocked by coexpression of VDU1 and VDU2 (Figure 3c). Similar findings were observed when Ub-D2 bands were visualized by labeling of wild-type FLAG-D2 with 75Se (Figure 3c).
Adrenergic coinduction of D2 and VDU1 mRNA levels in brown adipocytes.
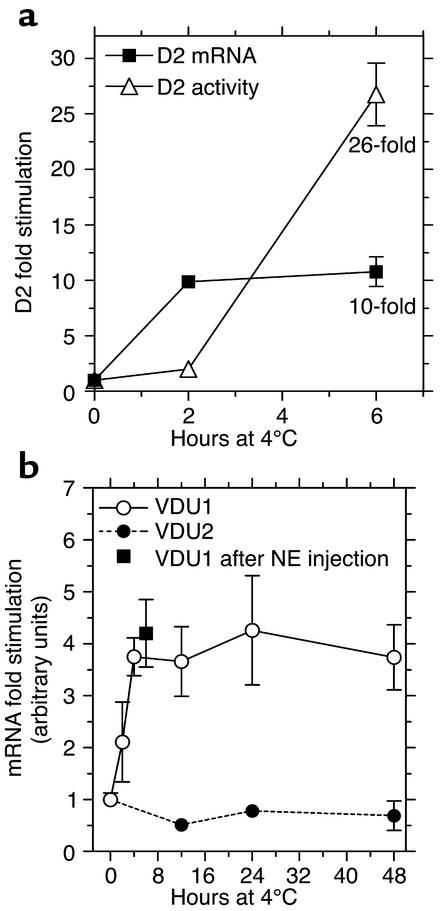
The increase in heat loss after birth is a substantial stress to thermal homeostasis in small mammals such as the human newborn. Acute exposure of rodents to 4°C, a typical experimental paradigm for this event, requires an adaptive increase in T3 via D2-catalyzed T4-to-T3 conversion in brown adipocytes. Within a few hours of cold exposure, D2 activity in brown adipocytes is substantially increased due to stimulation by the sympathetic nervous system (8). To test whether VDU1 or VDU2 in these cells could modulate D2 expression, mice were placed at 4°C for 1–48 hours. After 6 hours, the expected approximately tenfold increase in D2 mRNA levels occurred (Figure 4a), but D2 activity was increased to a much greater extent, approximately 26-fold. Remarkably, VDU1 mRNA was increased approximately fourfold in BAT within 4 hours of cold exposure, an effect that is mimicked by NE injection in mice at room temperature (Figure 4b). Interestingly, VDU2 mRNA levels were not increased in brown adipocytes of cold-exposed mice, indicating that this autonomic response is VDU subtype–specific.
Figure 4.
Sympathetic stimulation markedly induces VDU1 expression in brown adipocytes. (a) Mice were exposed to cold for the indicated times. D2 mRNA levels in the BAT were measured by real-time PCR. β-Actin mRNA was used as internal control, and the results are expressed as fold change in the mRNA ratio compared with that of controls. D2 activity is expressed as fold change versus controls. (b) VDU1 and VDU2 mRNA levels were measured as in a. Experimental animals were treated with 0.75 μg NE per 1 g body weight, and control animals received saline. Values are the mean ± SD of two to four animals. The entire experiment was performed twice.
Discussion
The present studies identify D2 as the only known specific substrate of VDU1 and VDU2, which in turn are the first ubiquitin-specific processing proteases (UBPs) known to specifically deubiquitinate an ERAD substrate. These results show that protein recognition is involved not only in the E3-mediated ubiquitination process but also in the deubiquitination pathway catalyzed by UBPs. Both VDUs are downstream targets for ubiquitination by pVHL E3 ligase, and VHL mutations that disrupt the interaction between the VDUs and pVHL abrogate their ubiquitination (6, 7). Although hundreds of UBP enzymes have been cloned, only a few examples of substrate recognition by UBP enzymes have been reported, and, to our knowledge, none are ER-resident proteins (18–22).
VDU1 specifically binds to D2 in vitro (Figure 1c), as well as in yeast (Figure 1, a and b) and mammalian cells (Figure 1d). Since only the COOH-terminal portion of D2 (amino acids 166–273) was used as bait in the yeast two-hybrid screening, the VDU1 binding site is localized to this region of D2, which is predicted to be in the cytosol (16). There is no apparent homology between D2 and pVHL (23, 24), to which VDU1 binds specifically and of which it is a substrate, suggesting that different VDU1 binding sites are involved when it interacts with a substrate or pVHL. Our confocal studies (Figure 2b) indicate that both VDUs colocalize with D2, itself an integral membrane ER-resident protein. Although they are present in the particulate fraction and not in the cytosol (Figure 2a), it is not clear based on their hydrophobic profile whether VDU1 and VDU2 are integral membrane proteins. Their physical colocalization with D2, however, provides the opportunity for catalysis and D2 deubiquitination (Figure 2b).
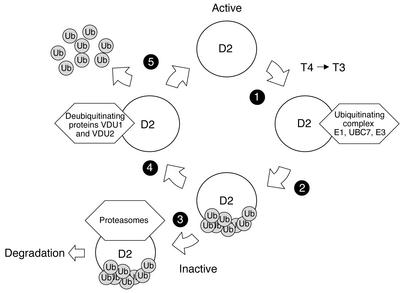
D2 contains the rare amino acid selenocysteine in its active center. Decoding of the selenocysteine-encoding UGA codon to signal for cotranslational insertion of selenocysteine, not termination, in selenoproteins such as D2 decreases translation efficiency 20- to 400-fold (14). The transient expression of wtD2 in HEK cells results in D2 activities that are similar to those in a human mesothelioma cell line, MSTO211H (15). This argues for the physiological relevance of the observation that transient coexpression of either UBP in D2-expressing HEK-293 cells increases D2 activity, as would be expected if D2 is a specific VDU substrate. This observation is based on stabilization of both D2 activity (Figure 2, c and d) and 75Se-labeled protein levels (Figure 3a), in agreement with the finding of decreased levels of Ub-D2 during coexpression with VDU1 or VDU2 (Figure 3c). These data indicate that VDU1- or VDU2-catalyzed deubiquitination recycles inactive Ub-D2 to its active deubiquitinated form, bypassing the proteasomal degradation pathway. Thus, Ub-D2 can be either reactivated or degraded, with the balance between these two processes determined, at least in part, by the VDUs’ activities (Figure 5).
Figure 5.
Proposed model for reversible D2 ubiquitination. D2 is an ER-resident selenoprotein that is inactivated by selective ubiquitination via a process that is accelerated by catalysis of T4 deiodination (step 1). D2 ubiquitination involves a ubiquitin-activating enzyme (E1) (4), a ubiquitin conjugase (UBC7) (5, 31), and an as-yet unidentified ubiquitin ligase (E3) (step 2). Ub-D2 can either be taken up by the proteasome and irreversibly degraded (step 3) or interact with (step 4) and be reactivated by (step 5) VDU1- or VDU2-mediated deubiquitination.
The present study also provides evidence that VDU1-catalyzed D2 deubiquitination is an important part of the adaptive mechanism that regulates thyroid hormone action. In stimulated brown adipocytes, normal D2 induction increases intracellular T3 production, resulting in isolated tissue thyrotoxicosis (9). This is an important mechanism for cold acclimatization, in that mice with targeted inactivation of the D2 gene develop hypothermia and marked weight loss during cold exposure due to impaired BAT thermogenesis (25). Our results demonstrate that increased VDU1-catalyzed deubiquitination of a pool of Ub-D2 and its rescue from proteasomal degradation are an integral part of this mechanism. VDU1 mRNA levels are markedly upregulated by cold exposure or NE (Figure 4), amplifying the transcriptional increase in D2 activity and hence T3 production approximately 2.5-fold (Figure 4a). Even though UBP induction is known to play a physiological role in a number of cellular processes (26–30), to our knowledge this is the first example of enzyme reactivation due to deubiquitination.
Thus, due to the intrinsic inefficiency of the selenoprotein synthesis, the availability of a reversible ubiquitination-dependent mechanism to control the activity of D2 constitutes a biochemical and physiological advantage that allows for rapid control of thyroid hormone activation (Figure 5). The finding that VDU1 and VDU2 are coexpressed with D2 in many human tissues, including brain, heart, and skeletal muscle (1, 6), indicates that the importance of this mechanism may extend well beyond thermal homeostasis to include brain development, cardiac performance, glucose utilization, and energy expenditure.
Acknowledgments
The authors are grateful to P. Reed Larsen for valuable discussions and review of this manuscript. This work was supported by NIH grant DK-58538. Balazs Gereben is a Magyary Zoltán postdoctoral fellow of the Hungarian Education Ministry and is supported by a Felsõoktatási Kutatási és Fejlesztési Pályázat grant (FKFP0036/2001) and the Fifth European Community Framework Program (QLG3 2000-00844).
Footnotes
See the related Commentary beginning on page 145.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: thyroxine (T4); 3,5,3′-triiodothyronine (T3); types 1 and 2 iodothyronine deiodinase (D1 and D2); brown adipose tissue (BAT); ubiquitinated D2 (Ub-D2); ER-associated degradation (ERAD); von Hippel–Lindau protein (pVHL); VHL-interacting deubiquitinating enzyme (VDU); GAL4 activation domain (GAL4 AD); hemaglutinin (HA); GAL4 DNA binding domain (GAL4 DBD); quadruple dropout (QDO); wild-type D2 (wtD2); glutatione-S-transferase (GST); immunoprecipitation (IP); norepinephrine (NE); ubiquitin-specific processing protease (UBP).
References
- 1.Bianco AC, Salvatore D, Gereben B, Berry MJ, Larsen PR. Biochemistry, cellular and molecular biology and physiological roles of the iodothyronine selenodeiodinases. Endocr. Rev. 2002;23:38–89. doi: 10.1210/edrv.23.1.0455. [DOI] [PubMed] [Google Scholar]
- 2.Steinsapir J, Harney J, Larsen PR. Type 2 iodothyronine deiodinase in rat pituitary tumor cells is inactivated in proteasomes. J. Clin. Invest. 1998;102:1895–1899. doi: 10.1172/JCI4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinsapir J, Bianco AC, Buettner C, Harney J, Larsen PR. Substrate-induced down-regulation of human type 2 deiodinase (hD2) is mediated through proteasomal degradation and requires interaction with the enzyme’s active center. Endocrinology. 2000;141:1127–1135. doi: 10.1210/endo.141.3.7355. [DOI] [PubMed] [Google Scholar]
- 4.Gereben B, Goncalves C, Harney JW, Larsen PR, Bianco AC. Selective proteolysis of human type 2 deiodinase: a novel ubiquitin-proteasomal mediated mechanism for regulation of hormone activation. Mol. Endocrinol. 2000;14:1697–1708. doi: 10.1210/mend.14.11.0558. [DOI] [PubMed] [Google Scholar]
- 5.Botero D, et al. Ubc6p and Ubc7p are required for normal and substrate-induced endoplasmic reticulum-associated degradation of the human selenoprotein type 2 iodothyronine monodeiodinase. Mol. Endocrinol. 2002;16:1999–2007. doi: 10.1210/me.2002-0135. [DOI] [PubMed] [Google Scholar]
- 6.Li Z, et al. Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 2002;277:4656–4662. doi: 10.1074/jbc.M108269200. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, et al. Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel-Lindau tumor suppressor. Biochem. Biophys. Res. Commun. 2002;294:700–709. doi: 10.1016/S0006-291X(02)00534-X. [DOI] [PubMed] [Google Scholar]
- 8.Silva JE, Larsen PR. Adrenergic activation of triiodothyronine production in brown adipose tissue. Nature. 1983;305:712–713. doi: 10.1038/305712a0. [DOI] [PubMed] [Google Scholar]
- 9.Bianco AC, Silva JE. Cold exposure rapidly induces virtual saturation of brown adipose tissue nuclear T3 receptors. Am. J. Physiol. 1988;255:E496–E503. doi: 10.1152/ajpendo.1988.255.4.E496. [DOI] [PubMed] [Google Scholar]
- 10.Salvatore D, Tu H, Harney JW, Larsen PR. Type 2 iodothyronine deiodinase is highly expressed in human thyroid. J. Clin. Invest. 1996;98:962–968. doi: 10.1172/JCI118880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curcio-Morelli C, et al. In vivo dimerization of types 1, 2, and 3 iodothyronine selenodeiodinases. Endocrinology. 2003;144:3438–3443. doi: 10.1210/en.2002-220960. [DOI] [PubMed] [Google Scholar]
- 12.Berry MJ, Banu L, Larsen PR. Type I iodothyronine deiodinase is a selenocysteine-containing enzyme. Nature. 1991;349:438–440. doi: 10.1038/349438a0. [DOI] [PubMed] [Google Scholar]
- 13.Toyoda N, Zavacki AM, Maia AL, Harney JW, Larsen PR. A novel retinoid X receptor-independent thyroid hormone response element is present in the human type 1 deiodinase gene. Mol. Cell. Biol. 1995;15:5100–5112. doi: 10.1128/mcb.15.9.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berry MJ, Maia AL, Kieffer JD, Harney JW, Larsen PR. Substitution of cysteine for selenocysteine in type I iodothyronine deiodinase reduces the catalytic efficiency of the protein but enhances its translation. Endocrinology. 1992;131:1848–1852. doi: 10.1210/endo.131.4.1396330. [DOI] [PubMed] [Google Scholar]
- 15.Curcio C, et al. The human type 2 iodothyronine deiodinase is a selenoprotein highly expressed in a mesothelioma cell line. J. Biol. Chem. 2001;276:30183–30187. doi: 10.1074/jbc.C100325200. [DOI] [PubMed] [Google Scholar]
- 16.Baqui MM, Gereben B, Harney JW, Larsen PR, Bianco AC. Distinct subcellular localization of transiently expressed types 1 and 2 iodothyronine deiodinases as determined by immunofluorescence confocal microscopy. Endocrinology. 2000;141:4309–4312. doi: 10.1210/endo.141.11.7872. [DOI] [PubMed] [Google Scholar]
- 17.Moller S, Croning MDR, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 18.Taya S, Yamamoto T, Kanai-Azuma M, Wood SA, Kaibuchi K. The deubiquitinating enzyme Fam interacts with and stabilizes beta-catenin. Genes Cells. 1999;4:757–767. doi: 10.1046/j.1365-2443.1999.00297.x. [DOI] [PubMed] [Google Scholar]
- 19.Gnesutta N, et al. Cloning and characterization of mouse UBPy, a deubiquitinating enzyme that interacts with the ras guanine nucleotide exchange factor CDC25(Mm)/Ras-GRF1. J. Biol. Chem. 2001;276:39448–39454. doi: 10.1074/jbc.M103454200. [DOI] [PubMed] [Google Scholar]
- 20.Taya S, et al. The Ras target AF-6 is a substrate of the fam deubiquitinating enzyme. J. Cell Biol. 1998;142:1053–1062. doi: 10.1083/jcb.142.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Zhang B, Fischer JA. A specific protein substrate for a deubiquitinating enzyme: liquid facets is the substrate of Fat facets. Genes Dev. 2002;16:289–294. doi: 10.1101/gad.961502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ideguchi H, et al. Structural and functional characterization of the USP11 deubiquitinating enzyme, which interacts with the RanGTP-associated protein RanBPM. Biochem. J. 2002;367:87–95. doi: 10.1042/BJ20011851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gnarra JR, et al. Molecular cloning of the von Hippel-Lindau tumor suppressor gene and its role in renal carcinoma. . Biochim. Biophys. Acta. 1996; 1242:201–210. doi: 10.1016/0304-419x(95)00012-5. [DOI] [PubMed] [Google Scholar]
- 24.Croteau W, Davey JC, Galton VA, St. Germain DL. Cloning of the mammalian type II iodothyronine deiodinase. A selenoprotein differentially expressed and regulated in human and rat brain and other tissues. J. Clin. Invest. 1996;98:405–417. doi: 10.1172/JCI118806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Jesus LA, et al. The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J. Clin. Invest. 2001;108:1379–1385. doi:10.1172/JCI200113803. doi: 10.1172/JCI13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Baker RT, Fischer-Vize JA. Control of cell fate by a deubiquitinating enzyme encoded by the fat facets gene. Science. 1995;270:1828–1831. doi: 10.1126/science.270.5243.1828. [DOI] [PubMed] [Google Scholar]
- 27.Moazed D, Johnson D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y, Carroll M, Papa FR, Hochstrasser M, D’Andrea AD. DUB-1, a deubiquitinating enzyme with growth-suppressing activity. Proc. Natl. Acad. Sci. U. S. A. 1996;93:3275–3279. doi: 10.1073/pnas.93.8.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naviglio S, et al. UBPY: a growth-regulated human ubiquitin isopeptidase. EMBO J. 1998;17:3241–3250. doi: 10.1093/emboj/17.12.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park KC, et al. Antagonistic regulation of myogenesis by two deubiquitinating enzymes, UBP45 and UBP69. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9733–9738. doi: 10.1073/pnas.152011799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, B.W., Zavacki, A.M., Harney, J.W., Larsen, P.R., and Bianco, A.C. 2002. The human type 2 iodothyronine selenodeiodinase (D2) is ubiquitinated via interaction with the mammalian ubiquitin conjugases MmUBC7 and MmUBC6. Program and abstracts of the 74th Annual Meeting of the American Thyroid Association, program number 110. Los Angeles, California, USA. 165.