Abstract
Crenarchaeota are ubiquitous and abundant microbial constituents of soils, sediments, lakes, and ocean waters. To further describe the cosmopolitan nonthermophilic Crenarchaeota, we analyzed the genome sequence of one representative, the uncultivated sponge symbiont Cenarchaeum symbiosum. C. symbiosum genotypes coinhabiting the same host partitioned into two dominant populations, corresponding to previously described a- and b-type ribosomal RNA variants. Although they were syntenic, overlapping a- and b-type ribotype genomes harbored significant variability. A single tiling path comprising the dominant a-type genotype was assembled and used to explore the genomic properties of C. symbiosum and its planktonic relatives. Of 2,066 ORFs, 55.6% matched genes with predicted function from previously sequenced genomes. The remaining genes partitioned between functional RNAs (2.4%) and hypotheticals (42%) with limited homology to known functional genes. The latter category included some genes likely involved in the archaeal–sponge symbiotic association. Conversely, 525 C. symbiosum ORFs were most highly similar to sequences from marine environmental genomic surveys, and they apparently represent orthologous genes from free-living planktonic Crenarchaeota. In total, the C. symbiosum genome was remarkably distinct from those of other known Archaea and shared many core metabolic features in common with its free-living planktonic relatives.
Keywords: Archaea, Crenarchaea, environmental genomics, marine microbiology, population genomics
Planktonic Crenarchaeota of the domain Archaea (1, 2) now are recognized to comprise a significant component of marine microbial biomass, ≈1028 cells in today's oceans (3–5). Although marine Crenarchaeota span the depth continuum (6), their numbers are greatest in waters just below the photic zone (3, 7). Isotopic analyses of lipids suggest that marine Crenarchaeota have the capacity for autotrophic carbon assimilation (8–12). The recent isolation of Nitrosopumilus maritimus, the first cultivated nonthermophilic crenarchaeon, demonstrated conclusively that bicarbonate and ammonia can serve as sole carbon and energy sources for at least some members of this lineage (13). The presence and distribution of gene fragments in environmental samples, bearing weak homology to one of the subunits of ammonia monooxygenase (amoA), also has been recently reported (14–18). Comparative environmental genomic studies have also identified the presence of a multiple genes in metabolic pathways potentially associated with crenarchaeotal ammonia oxidization and CO2 fixation (19).
Cenarchaeum symbiosum, the sole archaeal symbiont of the marine sponge Axinella mexicana (20), falls well within the lineage of ubiquitous and abundant planktonic marine Crenarchaeota (18, 20, 21). Although yet uncultivated, C. symbiosum can be harvested in significant quantities from host tissues, where it comprises up to 65% of the total microbial biomass (20, 21). These enriched uniarchaeal preparations of C. symbiosum have facilitated DNA analyses (20, 22, 23), as well as the identification and structural elucidation of nonthermophilic crenarchaeotal core lipids (8, 24, 25).
Fosmid libraries enriched in C. symbiosum genomic DNA previously were constructed and screened for phylogenetic and functionally informative gene sequences (19, 22). In a directed effort to genetically characterize C. symbiosum, we systematically selected overlapping fosmid clones to assemble the full genome complement. The composite full genome sequence of one ribotype of C. symbiosum, along with overlapping regions from related, sympatric genetic variants, provides new perspective on the biological properties of nonthermophilic Crenarchaeota, their predicted metabolic pathways, population biology, and gene representation in environmental samples.
Results
Genome Assembly and Population Structure.
The C. symbiosum genome was assembled from a set of 155 completed fosmid sequences selected from an environmental library enriched for C. symbiosum genomic DNA (see Supporting Text, Tables 2 and 3, and Fig. 4, which are published as supporting information on the PNAS web site) (19, 22). The fosmids AF083071 and AF083072 corresponding to previously described a- and b-type ribosomal variants (22) served as nucleation points for the separation of sequence variants into discrete genomic bins (Tables 2 and 3). Because only a relatively small number of clones comprise this library, each fosmid insert is expected to originate from an independent donor genome. Therefore, any assembly derived from this sample must represent a composite of related, but potentially nonidentical, genotypes; for the purposes of this study, a population genome equivalent. Remarkably, a single tiling path containing the complete genomic complement of C. symbiosum could be assembled from this complex data set, which corresponded to the a-type population of sequence variants (Table 4, which is published as supporting information on the PNAS web site).
C. symbiosum population structure was evaluated by analyzing fosmid sequence variation over the length of the assembled tiling path (Fig. 1). Overlapping fosmid sequences ranged between ≈80% and 100% nucleotide identity, with the a- and b-type variants dominating at the extremes. Overlapping a- and b-type fosmids, although virtually indistinguishable at the level of gene content and organization, differed in average nucleotide identity by ≈15% (Fig. 1). Average nucleotide identity within each set of overlapping a- or b-type fosmids was ≈98%, although the range of variation within the b-type population was considerably higher (Fig. 1). To facilitate analyses, fosmid sequences were partitioned by using a 93% identity cut-off, roughly corresponding to a standard demarcation of bacterial species based on whole-genome analysis (26, 27). To estimate the representation of a- and b-type donors in the fosmid library, a- and b-type sequences were queried against the set of fosmid end sequence reads ≥200 bp in length (see Materials and Methods). This approach identified on average 11 matches per a-type fosmid, and 8 matches per b-type fosmid, consistent with 60% representation of the a-type donor genomes in the DNA library (see supporting information for further information).
Fig. 1.
C. symbiosum fosmid population structure. (Upper) Fosmids partition into two distinct population bins corresponding to a-type and b-type ribosomal variants. Average nucleotide identity of each fully sequenced fosmid is plotted against the position of each fosmid in the assembled a-type scaffold. Blue lines represent the set of fosmids falling within the a-type population, and red lines indicate the set of fosmids falling within the b-type population. (Insets) Histograms represent the overall sequence divergence among overlapping fosmids. The distribution of observed sequence similarity (percentage identity) in high-scoring segment pairs for alignments between fosmid clones assigned to population “a” (Left), between “a + b” populations (Center), and between fosmid clones assigned to population “b” (Right). (Lower) Number of nucleotide polymorphisms per 1 kb of orthologous sequence shared between overlapping fosmids within the a-type population exhibiting >95% nucleotide identity to the genomic scaffold. Gaps in the distribution represent genomic intervals covered by a single fosmid clone (see Supporting Text). ⋀, Dashed line represents identity cut-off (≈92% average nucleotide identity) for type-a fosmids used in tiling path construction *, Sequence divergence within type-a fosmids based on comparison of fosmids sharing >95% average nucleotide identity.
To explore the coherence and diversity of donor genotypes within the a-type population, overlapping fosmid inserts with >95% nucleotide identity were evaluated for nucleotide polymorphisms (see Supporting Text). On average, these overlapping sequences exhibited 25–30 nucleotide polymorphisms per 1 kbp. The majority of these cases involved variation within intergenic regions or synonymous changes within ORFs (77% synonymous compared with 23% nonsynonymous changes). However, “hot-spots” of nucleotide variation were detected in some orthologous genes (>50–80 polymorphisms per kbp). These changes often were associated with the presence of a variant allele within one or more of the expanded gene families (see below), typically originating from a donor genotype not covered by sequenced fosmids. To gain insight into processes shaping allelic diversity between populations, the ratio of nonsynonymous to synonymous substitutions for 836 orthologous C. symbiosum genes common to a- and b-type populations was determined (Fig. 5, which is published as supporting information on the PNAS web site).
Genome Features.
The assembled C. symbiosum genome sequence is represented by a 2,045,086-bp single circular chromosome, with a 57.74% average G + C content (Table 1). No clear origin of replication could be identified using standard criteria (28, 29). A total of 2,017 protein-encoding genes were predicted in the genome sequence, as well as a single copy of a linked small subunit–large subunit ribosomal RNA (rRNA) operon, 1 copy of a 5S rRNA, 45 predicted transfer RNAs (tRNA) (Table 5, which is published as supporting information on the PNAS web site). Approximately 56% of all predicted protein-encoding genes could be assigned to functional or conserved roles based on homology searches (see Materials and Methods). The distribution of tRNAs was uneven, with the clear majority mapping to two distinct regions of the genome (Fig. 2).
Table 1.
C. symbiosum genome features
| Specifications* | |
| Size, bp | 2,045,086 |
| Average G + C content, % | 57.74 |
| Predicted ORFs | 2,066 |
| ORF density, gene/kb | 0.986 |
| Average ORF length (bp) | 924 |
| Coding percentage, % | 91.2 |
| ORF content | |
| Predicted functional | 1,067 |
| Predicted functional in COGs | 1,024 |
| Conserved hypothetical | 88 |
| Hypothetical | 863 |
| RNA genes | |
| 16S-23S rRNA operon | 1 |
| 5S rRNA | 1 |
| tRNAs | 45 |
| Expanded gene families† | |
| Number of families | 79 |
| Number of genes in families | 263 |
| Coding percentage, % | 26.78 |
*See Materials and Methods for fosmid assembly parameters.
†Based on following cutoffs: expection ≥ 1e−20, bitscore ≥ 100, identity ≥ 40%, and overlap ≥ 100 aa.
Fig. 2.
The C. symbiosum genome. Nested circles from outermost to innermost represent the following information. (i) Gene content predicted on the forward strand. (ii) Gene content predicted on the reverse strand. The color of predicted ORFs is based on COG functional categories (see key for color designations). (iii) Conservation of predicted C. symbiosum genes in the unassembled set of WGS data from the SAR. (iv) Conservation of predicted C. symbiosum genes in the set of published and completed microbial genomes (see Supporting Text). The height of the bars in circles 3 and 4 indicates the BSR for the set of predicted C. symbiosum proteins, queried against the public genomes and SAR, respectively, spanning a range of BSR values between ≈30% and 100% amino acid identity. (v) The extent of polymorphisms within the type-a population shown in Fig. 1 mapped on the genome (see Fig. 1 for details). (vi) Expanded gene families (discussed in the text; see key for color designations and Table 6 for additional information); note that high numbers of polymorphisms (circle 5) frequently coincide with the expanded protein families. (vii) tRNA and rRNA gene positions. (viii) G + C content deviation from the mean (57.5%) in 1,000-bp windows.
Expanded Gene Families.
The C. symbiosum genome contained an estimated 79 expanded gene families accounting for over 25% of its coding potential (see Materials and Methods and Table 6, which is published as supporting information on the PNAS web site). The majority of families were predicted to encode hypothetical proteins with no more than three representatives. However, 15 families contained at least four representatives (Table 6). Many families, including the two largest (containing 34 and 15 members, respectively), were predicted to encode hypothetical proteins with limited homology to surface-layer or extracellular matrix proteins. Representatives of these families often contained high levels of nucleotide polymorphism, corresponding to hot-spots of allelic diversity (Fig. 4, which is published as supporting information on the PNAS web site).
The largest family contained genes ranging between 2 and 35 kbp in size, predicted to encode numerous big archaeal proteins (bap) of unknown function. The bap genes represent ≈15% of the entire genome and 56% of all expanded gene families. Sequence analysis identified putative signal peptide cleavage sequences in just over 70% (24/34) of all predicted bap family members. Membrane-spanning domains in bap family members were not identified. All but three family members contained one or more WD40 or β propeller domain repeats, suggesting potentially shared protein-binding domains or interactions (30).
Central Metabolism.
Multiple components of a putative CO2 assimilation pathway based on a modified 3-hydroxypropionate cycle, and enzymes involved in the oxidative tricarboxylic acid (TCA) cycle, were present, as predicted (19), in the fully assembled C. symbiosum genome. A single operon encoding oxoacid:ferredoxin oxidoreductase subunits suggested that C. symbiosum utilizes at a minimum an incomplete branched TCA cycle for production of intermediates in cofactor and amino acid biosynthesis. Because succinate dehydrogenase and fumarase also are involved in the 3-hydroxypropionate cycle, it is possible they may serve dual roles as both hydroxypropionate and TCA cycle components in these Crenarchaeota. With the exception of glucokinase (EC 2.7.1.2) and pyruvate kinase (EC 2.7.1.40), C. symbiosum appears to contain an intact form of the Embden–Meyerhof–Parnas (EMP) pathway for the metabolism of hexose sugars. The lack of glucokinase and pyruvate kinase may indicate that the EMP functions in the gluconeogenic direction rather than the glycolytic pathway, as has been proposed for other Archaea (31). Several alternatives to glucokinase, however, including two genes predicted to encode carbohydrate kinases of unknown specificity and one gene predicted to encode a ROK-family ribokinase, were identified. Similarly, a gene predicted to encode phosphoenolpyruvate synthase also was present. The absence of glucose 1-dehydrogenase (EC 1.1.1.47), gluconolactonase (EC 3.1.1.17), and 2-keto-3-deoxy gluconate aldolase (EC 4.1.2.–) homologues suggested that C. symbiosum does not use the Entner–Duodoroff (ED) pathway in the catabolism of hexose sugars. In addition to the EMP pathway, an intact nonoxidative pentose phosphate pathway was identified, providing a mechanism for production of NADPH and ribose sugars for nucleotide biosynthesis.
Energy Metabolism.
Homologues of genes potentially associated with chemolithotrophic ammonia oxidation, including ammonia monooxygenase, ammonia permease, urease, a urea transport system, putative nitrite reductase, and nitric oxide reductase accessory protein (19) were represented in the C. symbiosum genome (Table 6). Several loci, including the ammonia permease and ammonia monooxygenase subunit C, were expanded gene-family members (Table 6). Homologues for (bacterial) hydroxylamine oxidoreductase (EC 1.7.3.4) and cytochromes c554 and c552 were not identified. If C. symbiosum does indeed derive energy directly from ammonia oxidation, it appears to employ different mechanisms than typical nitrifying bacteria for oxidizing hydroxylamine. Consistent with this hypothesis, 14 genes predicted to encode domains related of the plastocyanin/azurin family of blue type (I) copper proteins were identified with potential to substitute for cytochromes as mobile electron carriers (32, 33).
Other Genomic and Metabolic Features.
The complement of core genes expected for Archaea in general were mostly present in the genome of C. symbiosum. (More detailed discussion of specific metabolic features of can be found in Supporting Text). Genes required to synthesize all 20 aa, with the exception of proline, were present in the C. symbiosum genome. Nearly complete sets of genes required for the de novo synthesis of biotin, vitamin B12, riboflavin, thiamine, and pyridoxine all were identified. In the case of folic acid biosynthesis, genes encoding all steps for the conversion of the C1 carrier tetrahydrofolate (THF) to methyl-THF were identified. The C. symbiosum genome contains the full repertoire of genes necessary for chromosomal replication fork assembly and function, including components of the origin recognition complex (cdc6), two topoisomerases, single- and double-stranded helicases, three copies of a predicted bacterial/archaeal-type DNA primase, a two-subunit eukaryal/archaeal DNA primase system, RNase H, sliding clamp, and DNA ligase (cdc9). Genes encoding two distinct DNA polymerases were identified, including a single B family DNA polymerase I elongation subunit related to those of thermophilic Crenarchaeota (23) and a second euryarchaeotal-like DNA polymerase II including both large and small subunits. As predicted (34), a eukaryal-like, single copy histone H3–H4, the first to be found in any crenarchaeote, was present in the assembled C. symbiosum genome. Ten of the 45 predicted tRNAs in C. symbiosum contain putative introns (Table 5) (35). Most of the exon–intron boundaries form the conserved bulge–helix–bulge motif (BHB), although several appear to adopt structurally divergent forms (36). Such divergent features previously have been correlated with the presence of two distinct copies of the splicing endonuclease (endA) (37–40). Consistent with these observations, the C. symbiosum genome encodes two copies of endA.
Phylogeny and Comparative Environmental Genomics.
We phylogenetically compared the newly available C. symbiosum ORFs (including ribosomal proteins, elongation factors, SecY, and DNA repair proteins), to orthologues from other lineages (unpublished data; Fig. 6, which is published as supporting information on the PNAS web site). The lack of close relatives, and paucity of crenarchaeotal genomes in current databases, complicated these analyses because of unbalanced taxon representation. In aggregate, phylogenetic analyses of the r-protein alignments and of conserved nonribosomal proteins did not resolve the phylogenetic placement of C. symbiosum beyond previous results of rRNA analyses.
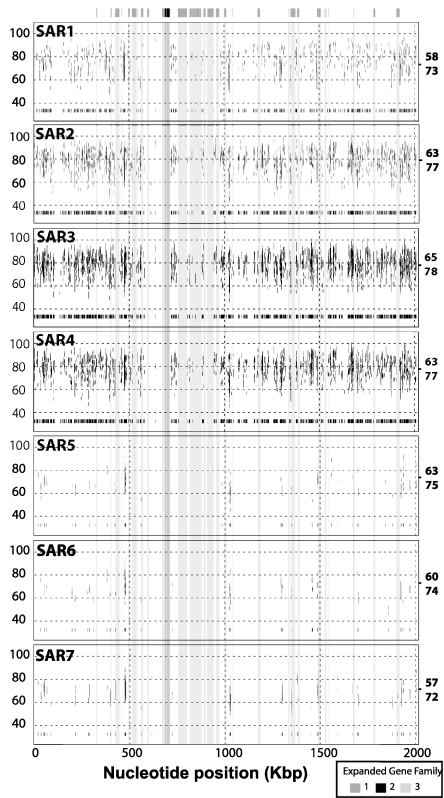
To explore the shared coding potential between C. symbiosum and its planktonic relatives, marine metagenomic data (15) were aligned to the assembled C. symbiosum genome (Fig. 2; see Materials and Methods). The distribution of planktonic crenarchaeotal homologues over the length of the C. symbiosum genome varied considerably between different marine samples. Coverage was greatest in the Sargasso Sea sample 3 (SAR3) with over 4,000 unique reads averaging 65% amino acid identity and 78% amino acid similarity over the length of the aligning read. This finding represents ≈1.25% of the total sequence population in the SAR3 sample. The depth of sequence coverage was uneven, varying between 1- and >20-fold between homologous intervals. More than 20% of the aligning sequences were derived from mate pairs mapping within the average range of insert sizes (3–6 kb), suggesting gene order is conserved between C. symbiosum and its planktonic relatives over short syntenic intervals. Numerous gaps in sequence coverage also were identified, indicating a significant proportion of C. symbiosum genes are absent or not well conserved within planktonic Crenarchaeota (Fig. 3).
Fig. 3.
Comparative analysis of C. symbiosum and SAR sample bins. Coverage plots for individual SAR sample bins aligned to the C. symbiosum genomic scaffold (see Materials and Methods). Each vertical bar represents an individual WGS read. The y axis on each plot corresponds to the percentage amino acid sequence similarity for each aligning read. The average percentage amino acid identity (top) and similarity (bottom) for SAR WGS reads aligning to the C. symbiosum genome is shown to the far right of each coverage plot. For simplified visualization of gaps in the alignment, all matches are replotted near the base of the x axis to form a normalized 1D plot spanning the reference sequence. To illustrate the gene content of gaps within the distribution of aligning WGS reads, several regions corresponding to expanded gene families 1–3 (Table 4) unique to the C. symbiosum genome are highlighted. (For coverage plots for SAR sample 3 aligned to five archaeal reference genomes, see Fig. 7, which is published as supporting information on the PNAS web site.)
All protein-encoding sequences predicted in the C. symbiosum genome were queried against the SAR whole-genome shotgun (WGS) data as well as the set of public genomes (see Materials and Methods). The resulting alignments were compared by using BLAST score ratios (BSR) to identify highly conserved genes shared between C. symbiosum, SAR, and public genomes (Fig. 2). A total of 65 genes with a BSR ≥ 30 were more highly conserved between C. symbiosum and the public genomes (Table 7, which is published as supporting information on the PNAS web site). Of these, 43 fell into defined clusters of orthologous group (COG) categories, including 10 genes associated with DNA replication, recombination, and repair (L), 9 genes associated with amino acid transport and metabolism (E), and 6 genes associated with posttranslational modification, protein turnover, and chaperones (O). The distribution of genes within these three categories was far from random. For instance, within the first category, seven genes were most similar to bacterial associated DNA modification methyltransferases, and within the third category, five genes were homologous to serine protease inhibitors (serpins). A total of 525 genes with a BSR ≥ 30 were more highly conserved between C. symbiosum and the SAR data set, corresponding to ≈26% of all predicted protein-encoding genes in the C. symbiosum genome (Table 8, which is published as supporting information on the PNAS web site). This set of shared genes spanned the complete spectrum of COG categories, with highest representation in energy production and conversion (C), amino acid transport and metabolism (E), translation, ribosomal structure, and biogenesis (J), transcription (K), and DNA replication, recombination, and repair (L). The remaining gene predictions either were shared equally between the SAR and public genomes or were not well conserved at all (Fig. 2 and data not shown). The latter case, represented by gaps in both the circular genome map (Fig. 2) and coverage plots (Fig. 3), encompassed >800 genes with a BSR < 30, corresponding to ≈39% of all predicted protein-encoding genes in the C. symbiosum genome.
Discussion
Population Structure and Genomic Coherence.
The C. symbiosum genome represents a composite sequence assembled from individual, closely related sympatric donor genotypes. As such, the genetic plasticity and population structure of host-associated C. symbiosum cells is in part reflected in genome sequence variants. Rearrangements or transpositions between overlapping syntenic regions were seldom detected, and in only two instances were recent intragenic recombination events unambiguously detected. Although we sampled only a small fraction of C. symbiosum donor genomes within the host, the rarity of recombination events suggests that recombination and hybridization are less important than are clonal diversification and genetic drift in C. symbiosum populations. This dynamic is qualitatively different from recently described acid-mine drainage archaeal populations, where widespread recombination is proposed to generate multiple mosaic genotypes (41).
The separation of syntenic a- and b-type sequences during assembly potentially reflects periodic selection within the sponge host tissues, partitioning population variants into distinct sequence clusters (42, 43). Given that gene content, order, and orientation among overlapping a- and b-type genotypes are identical, selective forces are likely acting on individual genes or their expression. The frequency of highly variable alleles (indicated by large peaks in Fig. 1) indicates that selective pressures act with variable intensity on different regions of the C. symbiosum genome. Fine scale determination of a- and b-type genotype spatial distribution in host tissues, as well as deeper sampling of allelic variation in C. symbiosum populations, are necessary to further explore the nature, expression, and consequences of this intrapopulation genetic variability.
Functional and Metabolic Relationships.
The results presented here, in combination with previous studies (13, 16, 19), support the notion that C. symbiosum and its planktonic marine relatives may derive cellular energy directly from the oxidation of ammonia. C. symbiosum harbors a number of genes known to be relevant to ammonia metabolism. Whether ammonia is the sole source of energy for either C. symbiosum or planktonic Crenarchaeota is unclear, but this appears to be so for its recently cultivated crenarchaeotal relatives (13). Although carbon fixation remains to be formally demonstrated for C. symbiosum, the hydroxypropionate pathway appears present in this archaeal symbiont (Table 6) (19). Closely related homologues of virtually all genes associated with ammonia oxidation and carbon fixation in C. symbiosum also were present in environmental samples having known high crenarchaeotal representation (Fig. 3 and Table 8) (19). Combined with well documented high crenarchaeotal biomass in marine plankton (3, 6, 11, 44), these data are consistent with a major role for crenarchaeotal nitrification in carbon and nitrogen cycling in the sea.
Comparisons with the SAR WGS data set indicated that the majority of other core metabolic subsystems identified in C. symbiosum also were well conserved in planktonic Crenarchaeota. In addition to information processing systems, homologies of C. symbiosum biosynthetic and housekeeping subsystems (including glycolysis, gluconeogenesis, pentose phosphate conversion, TCA cycle, cofactor and vitamin metabolism, amino acid biosynthesis, oxidative phosphorylation, and ATP synthesis) were most frequently found in environmental samples known to contain high levels of free-living planktonic Crenarchaeota. These results suggest that the majority of core metabolic functions found in C. symbiosum also are present in its planktonic relatives. Conversely, a considerable number of C. symbiosum-unique genes also were found, which potentially are involved in the archaeal–sponge symbiotic association (see below).
Evolutionary Relationships.
The domain Archaea remains well defined by two major subkingdoms, the Crenarchaeota and Euryarchaeota (2). Cultivation-independent phylogenetic surveys (45) have revealed many new environmentally significant clades within the domain Archaea. These environmental clades now outnumber archaeal lineages having cultivated representatives (1, 18, 46, 47). Ribosomal rRNA-based phylogenetic analyses of predominant cultivated and uncultivated archaeal groups suggest that Crenarchaeota and Euryarchaeota are best represented by polytomies, “star radiations,” with poor intragroup resolution relative to their common ancestral node (1). As a consequence, groups previously thought to be more deeply branching, for example, the Korarchaeota (46), appear now to fall well within the Crenarchaeota based on trees with well supported nodes having broad taxon representation (1).
In aggregate, analyses of individual or concatenated protein alignments and phylogenetic analyses did not resolve the phylogenetic placement of C. symbiosum beyond previous rRNA analyses (1, 47). This finding is mostly attributable to the currently poor representation of Crenarchaeota in existing genomic databases. Despite the relatively low number of crenarchaeotal genomes available for comparison, our results generally supported previous rRNA phylogenetic analyses, placing C. symbiosum peripheral to the crenarchaeotal lineage of cultivated hyperthermophiles.
Symbiosis.
Little is known about specific functional relationships between C. symbiosum and its sponge host, Axinella mexicana. Close relatives of C. symbiosum, however, seem commonly associated with other Axinella or other sponge hosts (48, 49), so similar associations may be common in the marine environment. With respect to potential symbiotic metabolic interactions, carbon and nitrogen exchanges are common in many microbial–eukaryal symbioses, and sponge-associated nitrification also has been reported previously (50). One possible interaction, consistent with the C. symbiosum genome complement and the nitrifying phenotype of N. maritimus, is removal of nitrogenous host-waste products (e.g., ammonia, urea). This could simultaneously fuel the symbiont's respiratory energy metabolism and might even provide new carbon to the host, via archaeal chemolithotrophic CO2 fixation, and subsequent symbiont–host carbon exchange.
Viable and dividing populations of C. symbiosum have been observed to persist in a single sponge host individual for up to 5 years (21). As an apparently nonmotile extracellular symbiont, C. symbiosum likely has developed mechanisms to inhibit or evade host consumption and defend against viral predation. A significant number of predicted genes encode domains homologous to cell surface, regulatory, or defense mechanisms, including numerous restriction modification systems to protect against foreign DNA, autotransporter adhesins potentially involved in mediating cell–cell contact, proteases that possibly modify or degrade extracellular matrix proteins, glycosyltransferases involved in cell wall biogenesis, and secreted seine protease inhibitors with the potential to mediate evasion of innate host defense systems. Many of these genomic features are not found in the planktonic relatives of C. symbiosum, and therefore may be specifically associated with the symbiotic lifestyle of C. symbiosum. Given the few known archaeal–metazoan symbioses, the C. symbiosum genome sequence provides a unique opportunity to further explore the genetic features mediating archaeal–eukaryal host contact, communication, and trophic exchange. It also provides a reference point for interpreting the genomic inventory, metabolic features, and evolution of its free-living relatives, which are abundant components of microbial plankton and may exert significant influence on energy and matter cycling in the sea.
Materials and Methods
Library Construction, Specifications, and Sequencing.
C. symbiosum cell enrichment, DNA extraction from sponge tissue, and fosmid library construction and sequencing protocols have been previously described (19, 51). Complete genome annotation files are available through the Joint Genome Institute's Integrated Microbial Genomes system (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi) and through the National Center for Biotechnology Information's web portal (www.ncbi.nlm.nih.gov). Individual fosmid sequences can be obtained from GenBank under the accession nos. DQ397540–DQ397640 and DQ397827–DQ397878, corresponding to a- and b-type population bins, respectively. The complete a-type genome sequence can be obtained from GenBank under accession no. DP000238. Additional information can be found in the Supporting Text.
Nucleotide Polymorphism Determination.
To identify orthologous regions (defined here as the reciprocal best matches with the BLASTn algorithm (52) and a minimum cut-off of 50% identity over a minimal 700-bp interval), the complete genomic scaffold was divided into 1,000-bp-long (1-kbp-long) consecutive fragments and searched against the set of fosmids. The same analysis was performed at the gene level as well, by using the set of genes annotated on the genomic scaffold as reference sequences. When the total length of orthologous sequences (gene-level comparisons) between a fosmid and the genomic scaffold was longer than 5 kbp, the fosmid was considered to derive from C. symbiosum, and the average nucleotide identity between the fosmid and the genome was calculated directly from the resulting BLASTn output. Orthologous regions between C. symbiosum fosmids (1-kbp window comparisons) subsequently were aligned by using clustalw (53). The number of invariable and variable bases in the 1-kbp fragments, which are shown in Fig. 1, was calculated directly from the clustalw alignments for the fosmids that showed >95% average nucleotide identity to the genomic scaffold.
Comparative Analysis Between C. symbiosum, Public Genomes, and the SAR Data Set.
The SAR database contained the complete set of unassembled, vector-trimmed, WGS sequences (15), whereas the public genomes database included all whole-genome sequences accessible through National Center for Biotechnology Information's ftp site as of December 2006 (260 genomes in total). Because the SAR average read-length is only ≈818 bases, C. symbiosum proteins longer than 300 aa were split into 300-aa-long consecutive fragments, which then were queried against the SAR database. Evaluation of gene conservation was based on analysis of BSR among C. symbiosum, SAR, and public genomes (54) by using tBLASTn.
Coverage plots relating the set of WGS reads from individual SAR sample bins, SAR1–7 (www.venterinstitute.org/sargasso) (15), to the C. symbiosum genomic scaffold were generated by using the Promer program implemented in MUMmer 3.18 (55). See Supporting Text for further details.
Phylogenetic Analysis.
Phylogenetic analyses were performed by using maximum-likelihood methods implemented in PHYML (http://atgc.lirmm.fr/phyml) (56). See Supporting Text for further details.
Supplementary Material
Acknowledgments
We thank Celine Brochier, Asuncion Martinez, Tsultrim Palden, Tracy Mincer, Matthew Sullivan, Maureen Coleman, Jarod Chapman, Sam Pitluck, Chris Detter, Krishna Palaniappan, and the Joint Genome Institute staff for computational and technical assistance. J.S. and Y.-i.W. thank Nozomu Yachie, Masaru Tomita, and Akio Kanai at Keio University for tRNA analysis with SPLITS. We thank two anonymous reviewers and Norman Pace, whose advice and suggestions greatly improved the final manuscript. This study was supported by National Science Foundation Grants MCB0236541, MCB0509923, and MCB0348001 (to E.F.D.), a Gordon and Betty Moore Foundation Award (to E.F.D.), the U.S. Department of Energy's Office of Science, Biological, and Environmental Research Program and the University of California–Lawrence Livermore National Laboratory under Contract W-7405-ENG-48, Lawrence Berkeley National Laboratory Contract DE-AC03–765F00098, and Los Alamos National Laboratory Contract W-7405-ENG-36.
Abbreviations
- TCA
tricarboxylic acid
- BSR
BLAST score ratios
- SAR
Sargasso Sea
- COG
clusters of an orthologous group
- WGS
whole-genome shotgun.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DP000238).
References
- 1.Robertson CE, Harris JK, Spear JR, Pace NR. Curr Opin Microbiol. 2005;8:638–642. doi: 10.1016/j.mib.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Woese CR, Kandler O, Wheelis ML. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karner MB, DeLong EF, Karl DM. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 4.DeLong EF. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuhrman JA, McCallum K, Davis AA. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 6.Massana R, Murray AE, Preston CM, DeLong EF. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, Martinez A, Sullivan MB, Edwards R, Brito BR, et al. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 8.Damste JS, Schouten S, Hopmans EC, van Duin AC, Geenevasen JA. J Lipid Res. 2002;43:1641–1651. doi: 10.1194/jlr.m200148-jlr200. [DOI] [PubMed] [Google Scholar]
- 9.Pearson A, McNichol AP, Benitez-Nelson BC, Hayes JM, Eglinton TI. Geochem Cosmochim Acta. 2001;65:3123–3137. [Google Scholar]
- 10.Wuchter C, Schouten S, Boschker HT, Sinninghe Damste JS. FEMS Microbiol Lett. 2003;219:203–207. doi: 10.1016/S0378-1097(03)00060-0. [DOI] [PubMed] [Google Scholar]
- 11.Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A, Pernthaler J. Appl Environ Microbiol. 2005;71:2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingalls AE, Shah SR, Hansman RL, Aluwihare LI, Santos GM, Druffel ER, Pearson A. Proc Natl Acad Sci USA. 2006;103:6442–6447. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 14.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 16.Wuchter C, Abbas B, Coolen MJ, Herfort L, van Bleijswijk J, Timmers P, Strous M, Teira E, Herndl GJ, Middelburg JJ, et al. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Treusch AH, Leininger S, Kletzin A, Schuster SC, Klenk HP, Schleper C. Environ Microbiol. 2005;7:1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- 18.Schleper C, Jurgens G, Jonuscheit M. Nat Rev Microbiol. 2005;3:479–488. doi: 10.1038/nrmicro1159. [DOI] [PubMed] [Google Scholar]
- 19.Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, Richardson PM, DeLong EF. PLoS Biol. 2006;4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Preston CM, Wu KY, Molinski TF, DeLong EF. Proc Natl Acad Sci USA. 1996;93:6241–6246. doi: 10.1073/pnas.93.13.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preston CM. PhD thesis. Santa Barbara: Univ of California; 1998. [Google Scholar]
- 22.Schleper C, DeLong EF, Preston CM, Feldman RA, Wu KY, Swanson RV. J Bacteriol. 1998;180:5003–5009. doi: 10.1128/jb.180.19.5003-5009.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schleper C, Swanson RV, Mathur EJ, DeLong EF. J Bacteriol. 1997;179:7803–7811. doi: 10.1128/jb.179.24.7803-7811.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLong EF, King LL, Massana R, Cittone H, Murray A, Schleper C, Wakeham SG. Appl Environ Microbiol. 1998;64:1133–1138. doi: 10.1128/aem.64.3.1133-1138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schouten S, Hopmans EC, Pancost RD, Damste JS. Proc Natl Acad Sci USA. 2000;97:14421–14426. doi: 10.1073/pnas.97.26.14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konstantinidis KT, Tiedje JM. J Bacteriol. 2005;187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Konstantinidis KT, Tiedje JM. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang R, Zhang CT. Archaea. 2005;1:335–346. doi: 10.1155/2005/509646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelman Z. Trends Biochem Sci. 2000;25:521–523. doi: 10.1016/s0968-0004(00)01687-x. [DOI] [PubMed] [Google Scholar]
- 30.Smith TF, Gaitatzes C, Saxena K, Neer EJ. Trends Biochem Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 31.Hutchins AM, Holden JF, Adams MW. J Bacteriol. 2001;183:709–715. doi: 10.1128/JB.183.2.709-715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattar S, Scharf B, Kent SB, Rodewald K, Oesterhelt D, Engelhard M. J Biol Chem. 1994;269:14939–14945. [PubMed] [Google Scholar]
- 33.Scharf B, Engelhard M. Biochemistry. 1993;32:12894–12900. doi: 10.1021/bi00210a043. [DOI] [PubMed] [Google Scholar]
- 34.Cubonova L, Sandman K, Hallam SJ, Delong EF, Reeve JN. J Bacteriol. 2005;187:5482–5485. doi: 10.1128/JB.187.15.5482-5485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sugahara J, Yachie N, Sekine Y, Soma A, Matsui M, Tomita M, Kanai A. In Silico Biol. 2006;6:0039. [PubMed] [Google Scholar]
- 36.Marck C, Grosjean H. RNA. 2003;9:1516–1531. doi: 10.1261/rna.5132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. Proc Natl Acad Sci USA. 2005;102:8933–8938. doi: 10.1073/pnas.0502350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshinari S, Fujita S, Masui R, Kuramitsu S, Yokobori S, Kita K, Watanabe Y. Biochem Biophys Res Commun. 2005;334:1254–1259. doi: 10.1016/j.bbrc.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 39.Calvin K, Hall MD, Xu F, Xue S, Li H. J Mol Biol. 2005;353:952–960. doi: 10.1016/j.jmb.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 40.Yoshinari S, Itoh T, Hallam SJ, Delong EF, Yokobori SI, Yamagishi A, Oshima T, Kita K, Watanabe YI. Biochem Biophys Res Commun. 2006;346:1024–1032. doi: 10.1016/j.bbrc.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 41.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 42.Cohan FM. Syst Biol. 2001;50:513–524. doi: 10.1080/10635150118398. [DOI] [PubMed] [Google Scholar]
- 43.Cohan FM. Annu Rev Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- 44.DeLong EF, Wu KY, Prezelin BB, Jovine RV. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 45.Pace NR. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 46.Barns SM, Delwiche CF, Palmer JD, Pace NR. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeLong EF. Curr Opin Genet Dev. 1998;8:649–654. doi: 10.1016/s0959-437x(98)80032-4. [DOI] [PubMed] [Google Scholar]
- 48.Holmes B, Blanch H. Mar Biol. 2006 Jul 4; doi: 10.1007/s00227-006-0361-x. [DOI] [Google Scholar]
- 49.Margot H, Acebal C, Toril E, Amils R, Fernandez Puentes J. Mar Biol. 2002;140:739–745. [Google Scholar]
- 50.Diaz MC, Ward BB. Mar Ecol Prog Ser. 1997;156:97–107. [Google Scholar]
- 51.Stein JL, Marsh TL, Wu KY, Shizuya H, DeLong EF. J Bacteriol. 1996;178:591–599. doi: 10.1128/jb.178.3.591-599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 53.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rasko DA, Myers GS, Ravel J. BMC Bioinformatics. 2005;6:2. doi: 10.1186/1471-2105-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guindon S, Gascuel O. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.