Abstract
Objective: The objective was to develop and test search strategies to identify diagnostic articles recorded on EMBASE.
Methods: Four general medical journals were hand searched for diagnostic accuracy studies published in 1999. Identified studies served as a gold standard. Candidate terms for search strategies were identified using a word-frequency analysis of their abstracts. According to the frequency of identified terms, searches were run for each term independently. Sensitivity, precision, and number needed to read (NNR) (1/precision) of every candidate term were calculated. Terms with the highest “sensitivity*precision” product were used as free-text terms and combined into a final strategy using the Boolean operator “OR.”
Results: The most frequently occurring eight terms (sensitiv* or detect* or accura* or specific* or reliab* or positive or negative or diagnos*) produced a sensitivity of 100% (95% confidence interval [CI] 94.1 to 100%) and an NNR of 27 (95% CI 21.0 to 34.8). The combination of the two truncated terms sensitiv* or detect* gave a sensitivity of 73.8% (95% CI 60.9 to 84.2%) and an NNR of 5.7 (95% CI 4.4 to 7.6).
Conclusions: The identified search terms offer the choice of either reasonably sensitive or precise search strategies for the detection of diagnostic accuracy studies in EMBASE. The terms are useful both for busy health care professionals who value precision and for reviewers who value sensitivity.
INTRODUCTION
When producing systematic reviews, researchers should try to identify as much empirical evidence as possible to inform the review question. Usually, the major biomedical databases such as MEDLINE and EMBASE are the starting points when trying to identify this evidence. However, information retrieval in such databases can become very time consuming because searches usually identify many irrelevant articles (low retrieval precision). In recent years, researchers have adopted various approaches to the development of search strategies to identify different types of studies (therapy, prognosis, diagnosis, and etiology) and different study designs [1–5]. Search strategies to identify diagnostic studies have also been developed [6–9].
For example, in 1994, Haynes and coworkers published a MEDLINE search filter for diagnosis [10], which is now publicly available in PubMed (Clinical Queries) [11]. However, differences in indexing hampered the straightforward use of this filter in EMBASE [12]. For example, the suggested MEDLINE Medical Subject Headings (MeSH) term “Sensitivity and Specificity” was only entered into the EMBASE EMTREE thesaurus in 2001. Alternatively, EMTREE provides the controlled vocabulary term “diagnostic accuracy.” The authors developed and tested search strategies to identify diagnostic articles recorded on EMBASE.
METHODS
One reviewer (Kronenberg) hand searched all issues published in 1999 of the New England Journal of Medicine, The Lancet, JAMA, and British Medical Journal (BMJ). The journals used in this study are indexed cover to cover in EMBASE. An article was deemed to be about diagnostic accuracy if at least one test was compared with a reference standard. A test was defined as any procedure used to change the estimate of the likelihood of disease presence. This definition included history taking, physical exam, and more advanced tests. All references of diagnostic studies identified (gold standard) were stored in a Reference Manager file.* Articles that were not diagnostic studies were excluded.
The result of the hand search was assumed to be perfect and reflected the true number of diagnostic accuracy studies in our total set or universe. The challenge for any automated search is to find all the references to accuracy studies (100% sensitivity) and at the same time not to find references to any other studies (100% specificity).
To assess the reproducibility of the hand search, a second reviewer (Bachmann) independently duplicated the hand search in a randomly selected 10% of all issues. The 10% sample was determined by numbering all references in the four journals sequentially, and 10% of references were then randomly selected using the Statistix software.†
The gold standard references were identified in EMBASE (Datastar version) using the accession number, a unique identifier for a specific record. A strategy combining all accession numbers using the Boolean connector “OR” was saved. Thus, a search in EMBASE would uniquely identify the gold standard references. The number of references in EMBASE was reduced to the subset of all references (6,143) that were published in the four chosen journals in 1999 to proxy a “universe” of searchable articles.
Because our primary aim was to define a search strategy using the offered thesaurus terms, we ran a first search with the EMTREE thesaurus term “Diagnostic Accuracy.” This term was considered an equivalent of the MEDLINE term “Sensitivity and Specificity” by the authors. This search identified 94% of the gold standard studies but was also associated with a low precision of 4.2%. Because further terms added to this EMTREE term would increase the sensitivity but would also further decrease precision, this preliminary finding suggested that we explore the effects focusing only on text words.
Realizing that the identification of relevant text words might be subjective and be associated with substantial risk of bias, we decided to apply the method of Boynton and coworkers [13] who selected potentially useful text words through the process of word frequency analysis.
We performed the frequency analysis of the occurrence of each word in each reference using the Idealist bibliographic software package.‡ The ListIndex function in the software was used to determine the frequency of occurrence of all the words in the titles, abstracts, and subject index. An example for words starting with the letter “d” is provided in Table 1.
Table 1 Example of term frequencies for the letter “d” as provided by the List Index function
The list was transferred to a Microsoft Excel file. To specifically select terms semantically associated with diagnostic accuracy, two reviewers (Estermann and Bachmann) excluded numbers, single letters, author names and institutions, register numbers, and journal names. Terms were also excluded if they were general medical language, for example, organ names or diseases, population of interest, or the word “study.” We considered these words not helpful in focusing a search on diagnostic accuracy studies. If the two reviewers disagreed on excluding a term, it was included. All included expressions were sorted alphabetically.
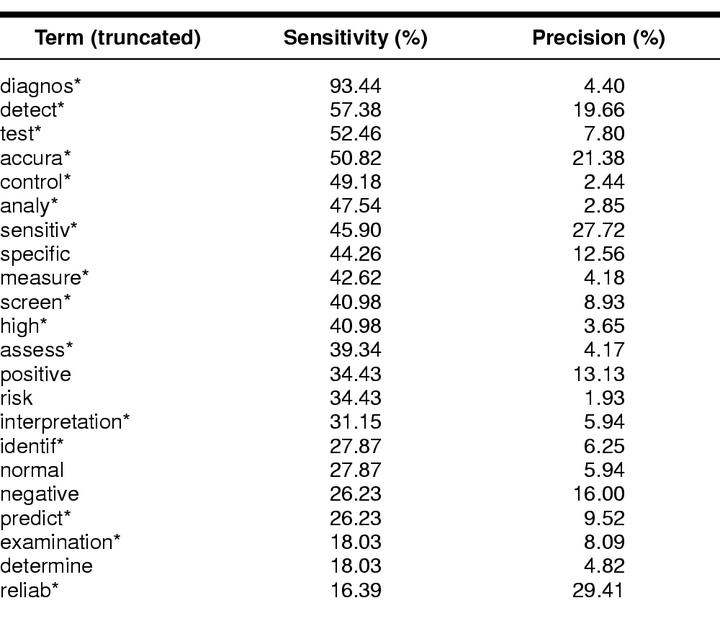
If terms differed only in the ending (e.g., diagnosis, diagnose, diagnostic, diagnostics), we decided to use the truncated term (e.g., “diagnos*”). According to the frequency of the identified most-frequent (truncated) terms, twenty-three searches were run for each term independently (Table 2). Sensitivity (retrieved articles as a proportion of all gold standard diagnostic articles), precision (gold standard diagnostic articles as a proportion of all retrieved articles), and number needed to read (NNR = 1/precision) of each text word were then calculated. Sensitivity is the number of electronically retrieved citations as a proportion of the number of truly relevant full papers (or diagnostic accuracy studies in this paper). The term is often used in medical research on diagnostic tests where it reflects the proportion of persons with a non-normal test result among all patients with some target disease as established by a gold standard reference test. Precision is the number of relevant full papers as a proportion of the number of electronically retrieved citations. In a clinical context, this quantity is often called “positive predictive value,” the proportion of persons with the target disease among persons with a non-normal test result. In addition, we coined the term NNR as an analogy to the number needed to treat (NNT) to describe the number of irrelevant references that have to be screened to find one of relevance. The NNR refers to the number of titles or abstracts necessary to read and ponder to find a reference to another relevant study in the set of retrieved references.
Table 2 List of twenty-three (truncated) terms with corresponding sensitivities and precisions if searched as a single term
Next, the product of sensitivity and precision was computed for each of the text words. We decided to calculate this figure because we wanted to identify those terms most balanced for sensitivity and precision. We thought that only terms contributing both to sensitivity and to precision were useful in building an efficient strategy.
The ten terms with the highest sensitivity-precision product were combined using “OR” to produce a series of search strategies. The sensitivities and precisions of these cumulative search strategies were then calculated.
RESULTS
The hand searches identified sixty-one articles (gold standard) as citations of a diagnostic accuracy study from a pool of 6,143 references in the four selected journals. We assumed these sixty-one citations to be the true number of diagnostic papers in the set of 6,143 references. The twenty-three truncated terms with the highest frequency according to the ListIndex function (Idealist) are listed in Table 3. The term “low” was removed because it is part of many author names.
Table 3 Development of eight search strategies with stepwise adding of terms
The calculation of the sensitivity-precision products led to a new order of terms. The consecutive connection of these terms with the Boolean operator “OR” produced the final set of search strategies. Their performance is shown in Table 3.
After the addition of the term “diagnos*,” the sensitivity in our test set reached 100%. Every additional term then only produced a decrease of retrieval precision. The combination of the first six terms resulted in a sensitivity of 91.8% (95% confidence interval [CI] 81.9 to 97.3%) and an NNR of 10.9 (95% CI 8.5 to 14.3). That is, almost eleven articles have to be read to identify one on diagnostic accuracy. This strategy seemed to be a good compromise with a high sensitivity and a reasonable precision.
By adding the two terms “negative” and the truncated expression diagnos* to search strategy 6, sensitivity reaches 100% (95% CI 94.1 to 100%) and an NNR of twenty-seven (95% CI 21.0 to 34.8). This strategy could be appropriate for systematic reviews.
The combination of the two truncated terms sensitiv* or detect* resulted in a sensitivity of 73.8% (95% CI 60.9 to 84.2%) and a NNR of 5.7 (95% CI 4.4 to 7.6). This latter strategy might be useful for busy clinicians, who may be interested in achieving reasonable sensitivity while avoiding sifting through hundreds of papers.
Figure 1 provides the detailed search strategies for three commonly used EMBASE interfaces for a reasonably precise and a comprehensive search strategy.
Figure 1.
Description of search strategy syntax for three commonly used interfaces
DISCUSSION
In contrast to Haynes and coworkers [14], we included diagnostic articles published in the comment, correspondence, and editorial sections to increase the likelihood of estimating precision correctly. Additionally, we focused on the most relevant indices, that is, sensitivity and precision, ignoring the less useful parameters of specificity and accuracy. In contrast to Haynes and coworkers [15], we did not find any advantage of combining EMTREE terms with text words.
For example, a search with the EMTREE term “DIAGNOSIS # (explode)” achieved 93.7% sensitivity and 4.0% precision (NNR = 25). The addition of text words with the Boolean operator “OR” would increase the sensitivity but at the cost of worse precision. In our search strategy, however, searches with sensitivities of about 90% were associated with precision of about 10%.
Our aim was to build useful search strategies for systematic reviews requiring very high sensitivity. The precision of search strategies, however, is important for busy health professionals but cannot be fully neglected in systematic reviews either. Search strategies should be evaluated in a context of time investments (cost) and consequences (of missing useful papers) [16]. In analogy to the assessment of the impact of language restrictions on summary measures in systematic reviews [17], the impact of using search strategies with lower sensitivity and higher precision on the summary measures in diagnostic reviews could be evaluated. Finally, this method could be applied to build optimal search strategies to detect diagnostic accuracy research for MEDLINE.
In our study, we hand searched four important general medical journals for the year 1999 to find diagnostic studies. The restriction to four important general medical journals might limit the generalizability of the search strategy. The restriction to 1999 was reflected in the width of the confidence intervals for sensitivity. We did not measure the test/retest reliability of our final strategies. Independent reassessment of the search performances in another set of gold standard articles would be useful.
Some terms such as “low” had to be removed from further analysis. Had we been able to analyze frequencies not by words, but by phrase, we would have been able to identify the proportion of those terms that were used in the diagnostic context (e.g., low accuracy). Including those terms could have been potentially relevant for the filter.
CONCLUSION
The identified search terms allow the choice of either reasonably sensitive or reasonably precise search strategies for the detection of diagnostic accuracy studies in EMBASE. This strategy is useful both for busy health care professionals who value precision and for reviewers who value sensitivity. In practice, clinicians combine the strategies proposed here with terms indicating the specific area of interest, very often a particular disease.
ACKNOWLEDGMENT
We thank Julie Glanville, from the Centre for Reviews and Dissemination in York, United Kingdom, for providing the word frequency analysis and for commenting on an earlier draft.
Table 1 Continued
Footnotes
* Information about Reference Manager 9.5, used in this study, may be viewed at http://www.refman.com.
† Information about Statistix 7, used in this study, may be viewed at http://www.statistix.com.
‡ Information about Blackwell Idealist, used in this study, may be viewed at http://www.blackwell-science.com/Products/IDEALIST/DEFAULT.HTM.
Contributor Information
Lucas M. Bachmann, Email: lucas.Bachmann@evimed.ch.
Pius Estermann, Email: ester@uszbib.unizh.ch.
Corinna Kronenberg, Email: ac-kronenberg@bluewin.ch.
Gerben ter Riet, Email: g.terriet@amc.uva.nl.
REFERENCES
- Haynes RB, Wilczynski N, McKibbon KA, Walker CJ, and Sinclair JC. Developing optimal search strategies for detecting clinically sound studies in MEDLINE. J Am Med Inform Assoc. 1994 Nov–Dec; 1(6):447–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanville J. Identification of research (phase 3): conducting the review (stage II). [Web document]. In: Undertaking systematic reviews of research on effectiveness. CRD report no. 4. 2001;3–11. [rev. Mar 2001; cited 22 Jun 2002]. <http://www.york.ac.uk/inst/crd/report4.htm>. [Google Scholar]
- Jadad AR, McQuay HJ. A high-yield strategy to identify randomized controlled trials for systematic reviews. Online J Curr Clin Trials 1993 Feb 27;<011>Doc No 33:3973 words. [PubMed] [Google Scholar]
- Boynton J, Glanville J, McDaid D, and Lefebvre C. Identifying systematic reviews in MEDLINE: developing an objective approach to search strategy design. Information Science. 1998 Summer; 24(3):137–54. [Google Scholar]
- Allison JJ, Kiefe CI, Weissman NW, Carter J, and Centor RM. The art and science of searching MEDLINE to answer clinical questions. finding the right number of articles. Int J Technol Assess Health Care. 1999 Spring; 15(2):281–96. [PubMed] [Google Scholar]
- Deville WL, Bezemer PD, and Bouter LM. Publications on diagnostic test evaluation in family medicine journals: an optimal search strategy. J Clin Epidemiol. 2000 Jan; 53(1):65–9. [DOI] [PubMed] [Google Scholar]
- McKibbon KA, Walker Dilks CJ. Beyond ACP Journal Club: how to harness MEDLINE for diagnostic problems. ACP J Club. 1994 Sep–Oct; (121Suppl 2): A. 10–2. [PubMed] [Google Scholar]
- Irwig L, Tosteson AN, Gatsonis C, Lau J, Colditz G, Chalmers TC, and Mosteller F. Guidelines for meta-analyses evaluating diagnostic tests. Ann Intern Med. 1994 Apr 15; 120(8):667–76. [DOI] [PubMed] [Google Scholar]
- Cochrane Methods Working Group on Systematic Review of Screening and Diagnostic Tests. Screening and diagnostic tests: recommended methods. [Web document]. [rev. 8 Feb 1998; cited 22 Jun 2002]. <http://www.cochrane.org/cochrane/sadtdoc1.htm>. [Google Scholar]
- Haynes . op. cit. [Google Scholar]
- National Library of Medicine. PubMed. [Web document]. Bethesda, MD: The Library, 2001. [rev. 10 Oct 2001; cited 22 Jun 2002]. <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi>. [Google Scholar]
- Embase. [Web document] [rev. 1 Apr 2000; cited 22 Jun 2002] <http://library.dialog.com/bluesheets/html/bl0072.html/>. [Google Scholar]
- Boynton . op. cit. [Google Scholar]
- Haynes . op. cit. [Google Scholar]
- Haynes . op. cit. [Google Scholar]
- Jadad . op. cit. [Google Scholar]
- Egger M, Zellweger-Zahner T, Schneider M, Junker C, Lengeler C, and Antes G. Language bias in randomised controlled trials published in English and German. Lancet. 1997 Aug 2; 350(9074):326–9. [DOI] [PubMed] [Google Scholar]