The discovery of transgene silencing in plants and double-stranded RNA (dsRNA) interference in the worm Caenorhabditis elegans has led to the latest revolution in molecular biology, RNA interference (RNAi). Over 10 years ago it was noted that several transgenic plant lines each containing the same ectopic transgene not only failed to be expressed but also inhibited the expression of the endogenous gene (1). Similarly, a determined Craig Mello and Andy Fire (2), attempting to reduce gene function using antisense RNA in the worm, discovered a minor contaminant in their antisense RNA preparations effectively and repeatedly reduced expression of the endogenous gene. In both cases, dsRNA homologous to the gene of interest was responsible for these observations. In the last 4 years, these discoveries have been extended to include protozoa, fungi, and mammals.
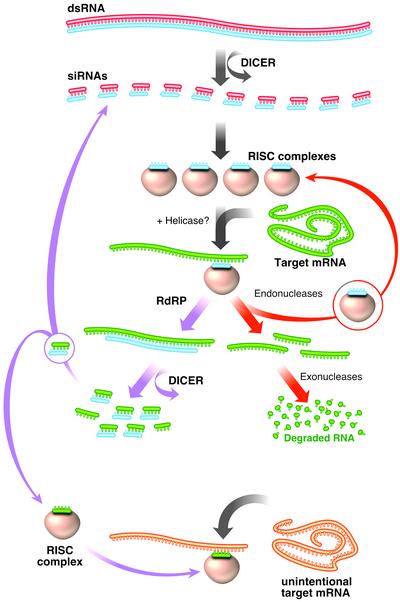
What is RNAi? RNAi is a highly conserved mechanism found in almost all eukaryotes and believed to serve as an antiviral defense mechanism. The molecular details are becoming clear from combined genetic and biochemical approaches (reviewed in refs. 3 and 4). On entry into the cell, the dsRNA is cleaved by an RNase III like enzyme, Dicer, into small interfering (21- to 23-nt) RNAs (siRNAs) (5–8) (Fig. 1). Biochemical evidence indicates the siRNAs are incorporated into a multisubunit protein complex, the RNAi-induced silencing complex (RISC), which directs the siRNA to the appropriate mRNA. New data suggest that the RISC complex may unwind the siRNA to help interactions with the target mRNA (9). Mismatches >1–2 bp within the 21- to 23-nt siRNA effectively disrupt proper degradation of the target mRNA (10, 11).
Fig. 1.
Overview of dsRNA-mediated mRNA degradation. On entry into cells, dsRNA is cleaved by Dicer into 21- to 23-nt siRNAs. siRNAs are complexed with a large multiprotein complex, the RISC. RISC is thought to unwind the siRNA to help target the appropriate mRNA (shown in green). The siRNA/mRNA hybrid is cleaved, releasing the siRNA, and the mRNA is degraded by endo and exonucleases. In worms and plants, the siRNA can also serve as a template for RdRP using the mRNA as a template. Elongation of the siRNA can lead to the production of more siRNAs that could share homology to other genes (shown in orange), causing their degradation, known as transitive RNAi.
In worms, interaction between the siRNA and mRNA can lead to immediate cleavage by Dicer, liberating a new siRNA, and degradation of the mRNA by endo- and exonucleases (Fig. 1). Alternatively, the siRNA can serve as a primer for an RNA-dependent RNA polymerase (RdRP), creating many more siRNAs. The action of an RdRP could explain the catalytic mechanism of RNAi, because only a few dsRNA molecules are required to degrade a much larger population of mRNAs, and RNAi is inheritable (2). Two important pieces of data support the idea that an RdRP plays an important role in RNAi. One, worms with mutations in novel RdRPs are not able to perform RNAi (12, 13). Two, elegant studies using fusion genes of unc-22 and GFP showed that dsRNA directed against GFP could effectively lead to the degradation of the endogenous unc-22 (13) (Fig. 2). These experiments also showed the polarity of the RdRP must be 5′ to 3′ on the antisense strand because unc-22 fused to the 5′ end of GFP caused degradation of the endogenous unc-22, but fusion to the 3′ end did not. These experiments displayed a new level of specificity not previously appreciated outside the plant kingdom (14). Mainly, siRNA targeting is very specific; however, elongation of the siRNA using the mRNA as a template could lead to nonspecific interference by sequences homologous to other genes, known as transitive RNAi.
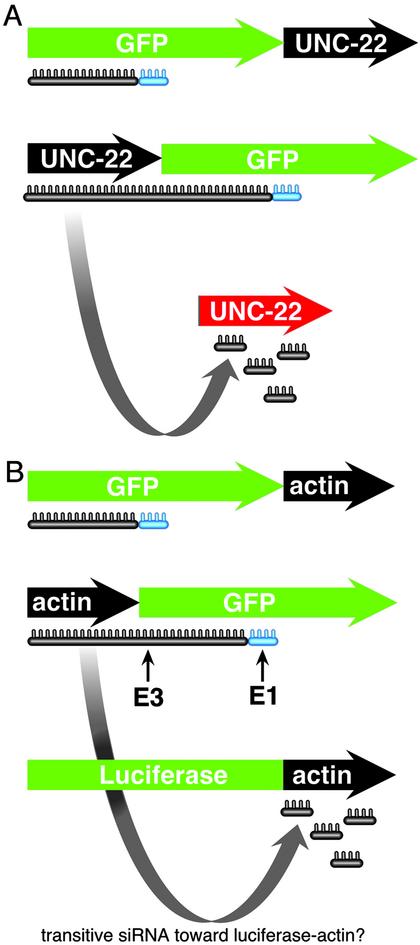
Fig. 2.
Formal test of transitive RNAi in worms and human 293 cells. (A) In worms, Sijen et al. (13) used GFP–unc-22 fusion genes to test whether mRNA from the endogenous unc-22 gene (shown in red) could be degraded. When dsRNA directed toward GFP was used, the endogenous unc-22 could be degraded only when unc-22 was fused 5′ to the GFP DNA sequences. Fusion at the 3′ end did not cause degradation of the endogenous unc-22 mRNA. (B) Chi et al. (16) used a similar approach to create human kidney cells (293 cells) expressing gene fusions of actin–GFP and actin–luciferase. siRNA directed against GFP caused degradation of the actin–GFP fusion but did not lead to elongation of the siRNA into the actin sequences and subsequent degradation of the actin–luciferase gene fusion. Two siRNAs were used, E1 and E3. E1 is within 20 bp of the fusion junction.
At present, this phenomenon is particular to plants and nematodes and does not appear to be a concern for mammalian systems, because there is no easily identifiable mammalian RdRP homolog. Additionally, in vitro reconstitution of mammalian RNAi activity does not require an RNA polymerase (8, 9) and RNAi in mouse oocytes is not blocked by drugs that interfere with RNA polymerases (15). Lastly, two reports in this issue of PNAS, using DNA microarray analysis, indicate that there is no significant decline of endogenous mRNAs in mammalian cells subjected to siRNA (16, 17).
In one set of experiments, Chi et al. (16) used a DNA microarray containing 43,000 DNA elements (36,000 genes) to address the specificity of siRNA. They asked whether global changes in mRNA levels could occur because of the active process of RNAi directed against an exogenous gene, GFP, in human embryonic kidney cells (293 cells). Remarkably, in each of three experimental settings, there was no statistically consistent change. Therefore, two conclusions can be drawn from this experiment. One, if transitive RNAi does exist in mammals, it is not robust enough in this experimental setting to cause nonspecific degradation of any mRNA detectable on their DNA microarray. However, it should be noted that although there was no clear decrease of mRNAs found after 48 and 72 h of siRNA exposure, it is not clear how long it might take a cell to mount such a response. Two, there appears to be no active up-regulation of cellular genes in response to RNAi. However, it is possible that the RNAi machinery is acutely up-regulated in response to siRNA, but not maintained after 48 h.
Chi et al. (16) also recreate the elegant worm experiments of Sijen et al. (13) to formally test transitive RNAi in 293 cells. In this experiment, Chi et al. created a fusion gene of actin DNA sequences with GFP DNA sequences (Fig. 2). Within the same cell, they engineered another fusion gene between actin and luciferase. If the siRNA were able to spread to nearby sequences, it would create new siRNAs homologous to actin, causing the subsequent degradation of the actin–luciferase fusion gene. This did not happen. What did happen is that the actin–GFP fusion gene was degraded, but the actin-luciferase gene remained intact. Even more compelling, siRNA directed to within 20 nt of the actin–GFP fusion junction did not lead to degradation of the actin–luciferase fusion. This result clearly indicates that 293 cells lack transitive RNAi.
In a tour de force, Semizarov et al. (17) reasoned that if siRNA caused nonspecific effects, then comparison of three different siRNA experiments targeting three different genes should share some commonality within their transcriptional profiles. They did not. The expression profiles of lung carcinoma cells treated with siRNA directed toward Rb, AKT1, or Plk1 were unrelated. Therefore, similar to the Chi et al. study, nonspecific degradation of cellular mRNAs by transitive RNAi was undetectable, and there appears to be no transcriptional regulation of cellular gene in response to siRNAs. Equally important, Semizarov et al. discovered that high doses of siRNAs did indeed induce many nonspecific genes, but lower doses did not. Therefore, more is not always better.
Why do worms have transitive RNAi and mammals do not? In mammals, long dsRNAs induce the IFN response, thereby shutting down translation, inducing RNaseL, and apoptosis. Lower eukaryotes lack this response. One possibility may be a competition between those mechanisms required for transitive RNAi, mainly an RdRP, and the IFN response. It is quite possible that extension of a siRNA, using the mRNA as a template, could create a long dsRNA recognized by the IFN response. Worms, which lack an IFN response, are capable of transitive RNAi and can take up kilobase-sized dsRNA, but mammals cannot. The evolution of the IFN response possibly came at the expense of the RdRP.
Are we ready for the first siRNA therapy? No. Although we are closer to the clinical setting than previously anticipated, several key questions need attention. It is becoming clear that siRNAs can target the appropriate transcript with great fidelity, and this holds promise to inactive mRNAs of disease alleles without affecting the normal allele. If indeed mammalian cells lack an RdRP, then the issue of transitive RNAi is no longer a problem. Although the articles described here indicate that kidney and lung cells are not capable of transitive RNAi, we do not know whether other cell types (postmitotic for instance) will have the same transcriptional response to RNAi or possess an RdRP that could be missing in these cell types. What is clear, however, is that we must hold all siRNA-based therapies to the same set of criteria used in these studies before embarking on clinical trials.
Of greater concern is that we do not know whether cells undergoing RNAi are as healthy as their unaffected counterparts. A few pieces of data suggest that they should not. One, siRNAs and members of the RNAi machinery play a role in chromosome architecture in several organisms (reviewed in refs. 18 and 19). Two, the RNAi machinery can be saturated (20). Therefore, it is conceivable that a cell devoting much of its RNAi machinery to the process of mRNA degradation could become defective in chromosome function or be more susceptible to viral infection. Finally, worms and green algae with defects in the RNAi machinery have a higher incidence of transposon hopping, which in turn could lead to increased mutagenesis (21–23). It is unknown whether this function is conserved in humans, but, if so, siRNA could lead to genome instability and cancer later in life.
A competitive growth experiment will be necessary to clarify this issue. In this experiment, a cell culture expressing a reporter gene is divided in half. One half is treated with siRNA directed toward the reporter and the other half sham treated. The two halves are cocultured for several divisions (n = 10–20). If there is indeed a negative impact on cell physiology by RNAi, then those cells treated with siRNA against the reporter will be under represented after 10–20 divisions. If there is no impact, then an equal number of siRNA treated and sham treated cells will be present.
Also of concern is the question of confinement. We do not know whether siRNA in one cell type can spread to neighboring cells not intended to receive the siRNA. siRNA is not actively taken up in cultured cells; however, it is not clear what happens in an intake animal. Precedence in worms and plants indicates that this type of mechanism does exist and may serve as a systematic method to “immunize” the entire organisms against potential viral infection. Therefore, siRNA directed against your favorite oncogene could reduce tumor growth, but could also affect normal cell division in the body.
Although these questions need to be addressed, the therapeutic use for siRNA looks promising. With the rich resource of experiments used to determine the efficacy of antisense RNA, viral vector delivery systems (24–27) and the stability of dsRNA, it will not be too long in the future that we see the first therapy using this revolutionary technology.
Acknowledgments
I thank Matthew Weitzman, Jan Karlseder, and Tony Hunter for insightful discussions and Jamie Simon for design and construction of the figures.
References
- 1.Napoli, C., Lemieux, C. & Jorgensen, R. (1990) Plant Cell 2, 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 3.Tijsterman, M., Ketting, R. F. & Plasterk, R. H. (2002) Annu. Rev. Genet. 36, 489–519. [DOI] [PubMed] [Google Scholar]
- 4.Tabara, H., Yigit, E., Siomi, H. & Mello, C. C. (2002) Cell 109, 861–871. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 6.Ketting, R. F., Fischer, S. E., Bernstein, E., Sijen, T., Hannon, G. J. & Plasterk, R. H. (2001) Genes Dev. 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knight, S. W. & Bass, B. L. (2001) Science 293, 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zamore, P. D., Tuschl, T., Sharp, P. A. & Bartel, D. P. (2000) Cell 101, 25–33. [DOI] [PubMed] [Google Scholar]
- 9.Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R. & Tuschl, T. (2002) Cell 110, 563–574. [DOI] [PubMed] [Google Scholar]
- 10.Elbashir, S. M., Martinez, J., Patkaniowska, A., Lendeckel, W. & Tuschl, T. (2001) EMBO J. 20, 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 12.Smardon, A., Spoerke, J. M., Stacey, S. C., Klein, M. E., Mackin, N. & Maine, E. M. (2000) Curr. Biol. 10, 169–178. [DOI] [PubMed] [Google Scholar]
- 13.Sijen, T., Fleenor, J., Simmer, F., Thijssen, K. L., Parrish, S., Timmons, L., Plasterk, R. H. & Fire, A. (2001) Cell 107, 465–476. [DOI] [PubMed] [Google Scholar]
- 14.Voinnet, O., Vain, P., Angell, S. & Baulcombe, D. C. (1998) Cell 95, 177–187. [DOI] [PubMed] [Google Scholar]
- 15.Stein, P., Svoboda, P., Anger, M. & Schultz, R. M. (2003) RNA 9, 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chi, J.-T., Chang, H. Y., Wang, N. N., Chang, D. S., Dunphy, N. & Brown, P. O. (2003) Proc. Natl. Acad. Sci. USA 100, 6343–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Semizarov, D., Frost, L., Sarthy, A., Kroeger, P., Halbert, D. N. & Fesik, S. W. (2003) Proc. Natl. Acad. Sci. USA 100, 6347–6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailis, J. M. & Forsburg, S. L. (2002) Genome Biol. 3, 1035.1–1035.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dernburg, A. F. & Karpen, G. H. (2002) Cell 111, 159–162. [DOI] [PubMed] [Google Scholar]
- 20.Kamath, R. S., Martinez-Campos, M., Zipperlen, P., Fraser, A. G. & Ahringer, J. (2000) Genome Biol. 2, 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ketting, R. F., Haverkamp, T. H., van Luenen, H. G. & Plasterk, R. H. (1999) Cell 99, 133–141. [DOI] [PubMed] [Google Scholar]
- 22.Ketting, R. F. & Plasterk, R. H. (2000) Nature 404, 296–298. [DOI] [PubMed] [Google Scholar]
- 23.Wu-Scharf, D., Jeong, B., Zhang, C. & Cerutti, H. (2000) Science 290, 1159–1162. [DOI] [PubMed] [Google Scholar]
- 24.Kootstra, N. A. & Verma, I. M. (2003) Annu. Rev. Pharmacol. Toxicol. 43, 413–439. [DOI] [PubMed] [Google Scholar]
- 25.Tiscornia, G., Singer, O., Ikawa, M. & Verma, I. M. (2003) Proc. Natl. Acad. Sci. USA 100, 1844–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart, S. A., Dykxhoorn, D. M., Palliser, D., Mizuno, H., Yu, E. Y., An, D. S., Sabatini, D. M., Chen, I. S., Hahn, W. C., Sharp, P. A., et al. (2003) RNA 9, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia, H., Mao, Q., Paulson, H. L. & Davidson, B. L. (2002) Nat. Biotechnol. 20, 1006–1010. [DOI] [PubMed] [Google Scholar]