Abstract
The assessment of biomaterial compatibility relies heavily on the analysis of macroscopic cellular responses to material interaction. However, new technologies have become available that permit a more profound understanding of the molecular basis of cell–biomaterial interaction. Here, both conventional phenotypic and contemporary transcriptomic (DNA microarray-based) analysis techniques were combined to examine the interaction of cells with a homologous series of copolymer films that subtly vary in terms of surface hydrophobicity. More specifically, we used differing combinations of N-isopropylacrylamide, which is presently used as an adaptive cell culture substrate, and the more hydrophobic, yet structurally similar, monomer N-tert-butylacrylamide. We show here that even discrete modifications with respect to the physiochemistry of soft amorphous materials can lead to significant impacts on the phenotype of interacting cells. Furthermore, we have elucidated putative links between phenotypic responses to cell–biomaterial interaction and global gene expression profile alterations. This case study indicates that high-throughput analysis of gene expression not only can greatly refine our knowledge of cell–biomaterial interaction, but also can yield novel biomarkers for potential use in biocompatibility assessment.
The macroscopic response of cells in contact with soft amorphous materials in vitro has long been of considerable interest to the scientific and medical community. For example, examination of such responses still remains the basis of early-stage assessment within the medical device industry of the viability of implants. Based on experience of these phenomena, criteria for the likely compatibility of materials in contact with animal tissue have been evolved, despite the apparent complexity of in vivo responses (1).
On the other hand, the pace of development of soft amorphous materials has been remarkable. It is now possible to contemplate the development of functional materials with highly controlled structural and mechanical properties, even based on quite fundamental principles (2). However, these developments have barely begun to be harnessed in terms of practical applications in biomedicine. Indeed, the relative number of new biocompatible materials entering clinical use is negligible. There seem to be at least two important technical barriers to making progress in this context.
First, the methods by which materials are routinely screened for suitability in biological settings are still greatly limited. The time-consuming nature and expense of the current spectrum of assays, together with the failure of a certain proportion of promising biomarkers to successfully predict long-term biocompatibility, represent a major barrier to significant development of the field (3). From a broader perspective, one notes that there are few fast-screening or “combinatorial” approaches to material suitability, such as those that have been pioneered in the pharmaceutical industry for lead compounds.
Second, and perhaps more deeply limiting, there is still a limited understanding of the key material surface control parameters that lead to prescribed cell–material responses. Surface characteristics such as hydrophobicity, surface energy, surface texture or patterning at various length scales, surface charge, and chemical composition are all known to play key roles in governing cell adhesion and growth (4). However, there is as yet no clear physiochemical intellectual framework within which to facilitate the development of materials that are likely to lead to useful interactions with cells.
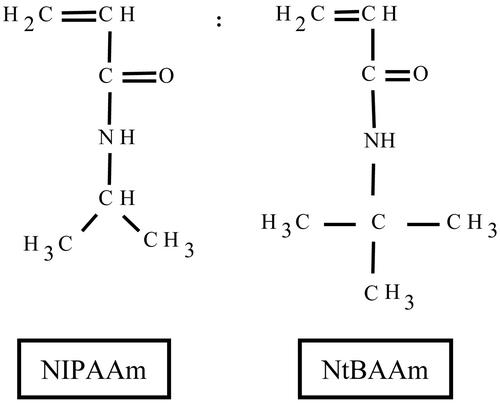
Herein, we demonstrate the utility of applying DNA microarray-based technology to provide an integrated view of cell–biomaterial interaction. Specifically, we examined the phenotypic response of a range of cell types to interaction with a series of copolymer films derived from N-isopropylacrylamide (NIPAAm). We used a series of increasingly hydrophobic copolymer films, comprised of NIPAAm and N-tert-butylacrylamide (NtBAAm) with the following ratios of NIPAAm to NtBAAm (wt/wt): 85:15, 65:35, and 50:50 (Fig. 1). We chose the well-established parameter of hydrophobicity to illustrate that significant insight into the influence of surface characteristics on cellular response may be attained via high-throughput analysis of gene expression. Through increasing addition of the more hydrophobic monomer NtBAAm, we were able to alter the bulk hydrophobic nature of these copolymer films. This alteration in bulk hydrophobicity was confirmed by microcalorimetric measurements and is reflected in the lowering of the lower critical solution temperature, which ranges from 25.1°C for the most hydrophilic (85:15) copolymer, down to 9.8°C for the most hydrophobic (50:50) copolymer (5).
Fig. 1.
Structure of NIPAAm and NtBAAm monomers. Variations in the ratio of NtBAAm to NIPAAm make it possible to modulate the adhesivity of the copolymer surface to living cells by altering surface hydrophobicity, as previously confirmed by microcalorimetry studies (5).
NIPAAm-based copolymer films have been previously used as adaptive cell culture substrates (5, 6) and, latterly by us and other groups, as prototypical drug delivery systems (7, 8). These “responsive” copolymers, although not known for their biocompatibility, are relatively benign with respect to interaction with cells, and make for an interesting case study from a practical point of view. Our results show that gene expression-profiling technology may greatly assist in defining the molecular basis of observed macroscopic cellular responses after biomaterial interaction.
Materials and Methods
Copolymer Synthesis. NIPAAm (99%; Acros Organics, Fairlawn, NJ) and NtBAAm (purum; Fluka) were recrystallized from hexane and dried at room temperature in vacuum.
The NIPAAm:NtBAAm copolymer series was obtained from T. Golubeva and A. Gorelov (Department of Chemistry, University College Dublin), and were synthesized as outlined (5).
Copolymer Film Preparation. All films were prepared from 5% (wt/wt) solutions of each copolymer in 100% ethanol and allowed to evaporate at room temperature to dryness in a sterile flow hood to create films 5 μm thick. Cell attachment and growth assays were performed by using 96-well flat-bottomed plates (Nunc), whereas cell spreading and morphometric studies were carried out by using 24-well flat-bottomed plates (Nunc) and LAB TEKII chamber slides (Nunc), respectively. Untreated tissue culture polystyrene (TCP) served as a control substratum. The chamber slides used for microscopy studies are modified to facilitate cell culture and thus mimic the effect of TCP.
Contact Angle Measurement. Static contact angle measurements for a water droplet on the polymer films were performed by using a contact angle goniometer (A1100 Ramé Hart, Mountain Lakes, NJ). These experiments were carried out at 37°C (above the lower critical solution temperature of the copolymer films) by using a temperature control cell. Between five and eight contact angle measurements were performed for each surface. For contact angle experiments, copolymer films were prepared on a microscope object glass by using the method outlined in the previous section. All static experiments were performed by using Millipore-filtered water.
Cell Culture. HeLa (human epithelial adenocarcinoma cells) and MRC-5 (human fetal lung SV-40-transformed fibroblasts) cell lines, as well as 1BR3 (primary human adult skin fibroblasts) cells, were obtained from the European Collection of Animal Cell Cultures (www.ecacc.org.uk). In addition, adhesion and growth studies were performed on primary human aortic smooth muscle cells (HASMC) obtained from Clonetics (BioWhittaker). Relevant cell culture media and associated supplements were as detailed by the respective distributors. All cell types were maintained at 37°C under 95% air/5% CO2.
Cell Attachment and Growth Assays. By using 96-well plates, three entire columns (eight wells each) were coated with NIPAAm: NtBAAm copolymers with the following ratios (50:50, 65:35, and 85:15). A single suspension of cells was prepared by trypsin/EDTA treatment. Wells were seeded with 200 μl of cell suspension (1.25 × 103 cells per ml). Plates were incubated under standard cell culture conditions for 24 and 96 h postseeding, with fresh medium being added after 48 h. At relevant time points, wells were rinsed with prewarmed PBS to remove unattached cells. Plates were subsequently placed at –70°C for future analysis. Cell adhesion and growth were quantified by using a CyQUANT nucleic acid-sensitive fluorescence assay kit (Molecular Probes). Calibration curves showed fluorescence measurements to be proportional to cell number. Cell attachment was quantified by using a Wallac (Gaithersburg, MD) Victor2 1420 Multilabel HTS Counter (absorbance maximum 485 nm, emission maximum 535 nm).
Cell Spreading Assay. Each precoated well of a 24-well plate was seeded with 1 ml of cell suspension (2 × 104 cells per ml) and incubated under standard cell culture conditions for 3 or 24 h. Benefiting from the transparent nature of the copolymer films, cells were viewed with an inverted phase contrast Nikon phase contrast microscope, and digital images were taken by using a KY-F55B 3-charge-coupled device color video camera (JVC) at 1,024 × 768 pixel resolution. Images were subsequently analyzed by using SCION IMAGE software (Scion, Fredrick, MD), which is based on NIH IMAGE.
Cell Morphology Assay. Precoated chamber slides were seeded with 1 ml of cell suspension (2 × 104 per ml) and incubated under standard cell culture conditions for 24 h. Cells were fixed in a prewarmed 3.7% (vol/vol) solution of formaldehyde in PBS. After fixation, cells were stained for 20 min by using 0.1% (wt/vol) crystal violet in PBS. Cells were mounted and viewed at magnification ×20 with a Zeiss Axioplan imaging microscope and images were obtained by using Zeiss AxioCam at a resolution of 1,300 × 1,030 pixels.
For fluorescence-based localization of actin, cells were initially rinsed with PBS, followed by fixation using 3.7% formaldehyde in PBS and, permeabilized by using 0.5% Triton X-100 in PBS for 10 min. Next, cells were incubated in rhodaminephalloidin (Molecular Probes), which labels F-actin. Fluorescence images were collected as above by using the Zeiss microscope system.
Gene Expression Analysis. Affymetrix Human Cancer G110 GeneChip arrays (2,059 probe sets corresponding to 1,201 unique UniGene clusters) were used in this study. Specifically, HeLa cells (1 × 106) were seeded onto six-well plates plus/minus precoating with the various copolymer films and left to adhere for 24 h. After this, total RNA was isolated from adherent cells by using RNeasy Mini Kit spin columns (Qiagen, Chatsworth, CA). Double-stranded (ds) cDNA was prepared in each case from 10 μg of total RNA by using a T7 promoter/oligo dT primer. Biotinylated in vitro transcribed cRNA probes were prepared from each ds cDNA preparation, which formed the basis of the hybridization mixtures added to the GeneChip arrays. Hybridized arrays were stained and washed before being scanned according to procedures developed by the manufacturer (Affymetrix, High Wycombe, U.K.).
Scanned output files were visually inspected for hybridization artifacts. To identify differentially expressed transcripts, pairwise comparison analyses were carried out by using Affymetrix MICROARRAY SUITE V. 4.0 and Microsoft ACCESS.
Results
We have previously demonstrated that alteration of the bulk hydrophobic nature of NIPAAm:NtBAAm copolymer films affects cellular interaction (5). It is not trivially obvious that an increase in bulk hydrophobic content should be reflected on the surface of cast films. Here, contact angle studies have been carried out to determine the impact of differing NtBAAm to NIPAAm ratios on the surface hydrophobicity of NIPAAm: NtBAAm copolymer films. It was found that the contact angle of a spotted water droplet on NIPAAm:NtBAAm copolymer films increased with the addition of NtBAAm. In more detail, long-time exposure (180 s) static contact angle measurements on 50:50 and 85:15 NIPAAm:NtBAAm copolymer films were found to be 56.53° (n = 4) and 52.98° (n = 6), respectively. This result indicates that the increased bulk hydrophobicity previously observed in this copolymer series resulting from the addition of NtBAAm is reflected in a simultaneous increase in the surface hydrophobicity of these cast films.
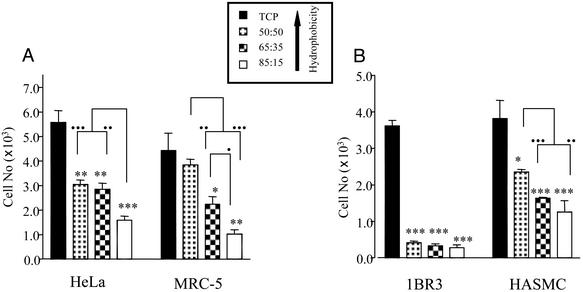
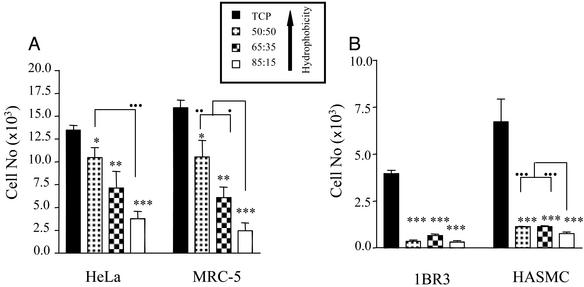
To study cell–polymer interaction, transparent NIPAAm: NtBAAm films were cast onto TCP (5, 6). Cells were seeded onto TCP surfaces (with or without precoating with copolymer film) and left for specific time periods before assessment of cell adhesion and growth. All cell types examined (HeLa, MRC-5, 1BR3, and HASMC) displayed a marked reduction in ability to adhere (24 h postseeding) to the various copolymer films in comparison to TCP (Fig. 2). Moreover, the extent of cell growth (up to 96 h) on these surfaces was also compromised (Fig. 3). This result indicates that TCP is more favorable to cell interaction than any of the copolymer films examined.
Fig. 2.
Cell adhesion on TCP or various NIPAAm:NtBAAm copolymer films. Adhesion was quantified by determining cell numbers on the different surfaces 24 h postseeding. (A) Adhesion of two transformed cell lines, HeLa (epithelial) and MCR-5 (fibroblast). (B) Adhesion of two primary cell types, 1BR3 (fibroblast) and HASMC (aortic smooth muscle). Error bars indicate SEM (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.005, respectively, as evaluated by unpaired Student's t test with respect to difference between adhesion determined on the various copolymer films in comparison with TCP. •, ••, and ••• indicate significant differences (P < 0.05, P < 0.01, or P < 0.005, respectively; unpaired Student's t test) between individual copolymer films.
Fig. 3.
Cell growth on TCP or various NIPAAm:NtBAAm copolymer films. The extent of growth was quantified by determining cell numbers on the different surfaces 96 h postseeding. (A) Adhesion of two transformed cell lines, HeLa (epithelial) and MCR-5 (fibroblast). (B) Adhesion of two primary cell types, 1BR3 (fibroblast) and HASMC (aortic smooth muscle). Error bars indicate SEM (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.005, respectively, as evaluated by unpaired Student's t test with respect to difference between the extent of growth determined on the various copolymer films in comparison with TCP. •, ••, and ••• indicate significant differences (P < 0.05, P < 0.01, or P < 0.005, respectively; unpaired Student's t test) between individual copolymer films.
However, incorporation of the hydrophobic monomer, NtBAAm, into the copolymer film improved the adhesion and growth of all cell types examined (Figs. 2 and 3), illustrating that surface hydrophobicity is an important determinant of cellular response. Minimal cell attachment and growth was observed on 85:15 copolymer films, with maximal attachment to 50:50 copolymer film (when TCP is excluded). This relationship between surface hydrophobicity and cellular response was independent of cell type (epithelial or fibroblast) or status (primary or transformed).
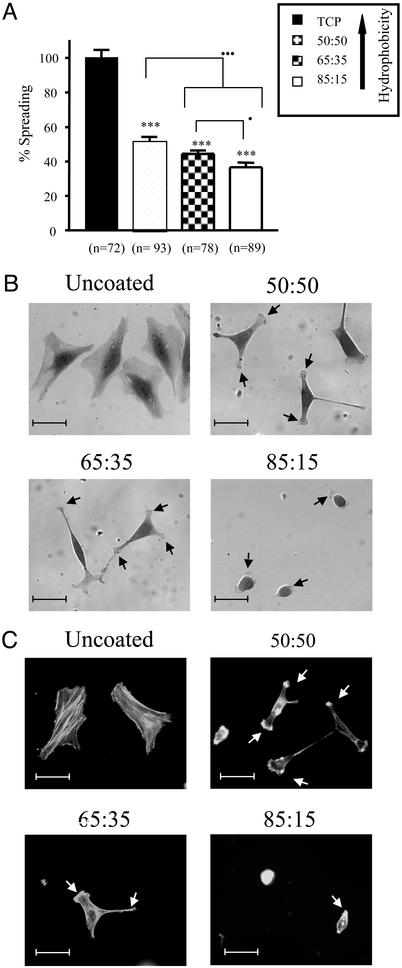
To more precisely examine the phenomenon of reduced cell adhesion and growth seen on the copolymer films, adhered HeLa cells were examined by light microscopy at 3 and 24 h postseeding. Analysis of cell spreading revealed a significant reduction in the average spread area of cells adhering to copolymer film in comparison with TCP at both time points (Fig. 4A). Indeed, cells seeded onto the copolymer films did not display any evidence of cell spreading until 3 h postseeding (data not shown), with no significant differences observed between the various copolymers at this time point. Twenty-four hours postseeding, increased content of NtBAAm in the copolymer film was associated with greater cell spreading, thus providing a link between surface hydrophobicity and cell spreading.
Fig. 4.
Inhibition of cell spreading on NIPAAm:NtBAAm copolymer films. HeLa cells were allowed to adhere and spread on either copolymer film or TCP for 24 h and then fixed with 4% paraformaldehyde. Fixed cells were visualized by light microscopy. (A) The average spread area is expressed relative to the degree of cell spreading observed on TCP. Error bars indicate SEM (with numbers of cells analyzed shown under each bar). *, P < 0.05; **, P < 0.01; ***, P < 0.005, respectively, as evaluated by unpaired Student's t test with respect to difference between cell spreading determined on the various copolymer films in comparison with TCP. • and ••• indicate significant differences (P < 0.05 or P < 0.005, respectively; unpaired Student's t test) between individual copolymer films. (B) Representative images of adherent cells seeded onto either copolymer film or TCP. Cells grown on 85:15 copolymer film for >24 h displayed a stellate morphology similar to that observed with the other copolymer films at 24 h. (C) Modulation of cytoskeletal structure. HeLa cells were allowed to spread on either copolymer film-coated or uncoated Lab-TekII chamber slides for 24 h and then fixed with 4% paraformaldehyde. Fixed cells were stained with rhodamine-phalloidin to detect F-actin. (Scale bar = 50 μm.)
Striking differences were also revealed with respect to gross cellular morphology, depending on the surface type examined. HeLa cells adherent to TCP displayed a polygonal shape, whereas those residing on copolymer films displayed stellate or arborized shapes, generated by the outgrowth of a number of narrow processes (Fig. 4B). Each of these outgrowths terminated in a fan of ruffled membrane (Fig. 4B, indicated by arrows). After a period of several days, cells achieved the same morphological appearance across all copolymer film combinations (data not shown). However, cells seeded on the copolymer films never fully achieved the same appearance or degree of spreading as that observed in the case of cells seeded on TCP.
Because cell spreading is controlled by alterations in cytoskeletal architecture (9), we subsequently visualized the cytoskeletal organization of HeLa cells adherent to TCP or copolymer films by staining for F-actin. Cells adhering to copolymer films displayed an absence of any organized cytoskeletal framework (Fig. 4C). It has been well documented that cell stellation is accompanied by depletion of microfilaments, particularly evidenced by loss of stress fibers (10). Indeed, we observed that the outgrowth of processes in cells spreading on copolymer films was accompanied by strong staining for F-actin at the leading edge of the growing processes (Fig. 4C, arrowheads), a phenomenon previously associated with cell stellation (10).
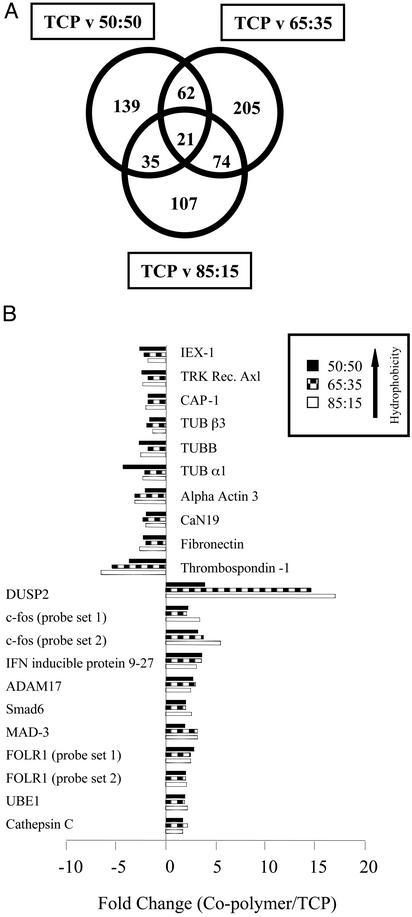
It has been postulated that alterations in cell shape can modulate gene expression (11). Through the use of an oligonucleotide array-based gene expression profiling approach, we investigated the relationship between global gene expression profiles and phenotypic cellular responses to copolymer film interaction. The expression of 1,785 human transcripts was assessed in HeLa cells grown for 24 h on either TCP or the various copolymer films (Fig. 5). Over half of all transcripts represented by the DNA microarray were expressed in the adhered cells (ranging between 748 and 975 expressed transcripts). Comparative analysis of the obtained gene expression profiles revealed that 21 transcripts were consistently differentially expressed between the copolymer film- and TCP-exposed cells.
Fig. 5.
Differential gene expression in HeLa cells in response to interaction with copolymer films. HeLa cells were seeded onto either TCP or the various copolymer films and left for 24 h before DNA microarray-based gene expression profile analysis. (A) Venn diagram illustrating results from comparative analysis of different gene expression profiles. Each circle represents a copolymer film/TCP comparison. Figures represent the number of human transcripts differentially expressed between particular comparisons. (B) A graphical illustration of fold change values associated with the central subset of 21 differentially expressed transcripts. Transcript names were derived from GeneCards (http://bioinformatics.weizmann.ac.il/cards). As an illustration of experimental reproducibility, the altered expression levels of c-fos and FOLR1 were verified by two independent probe sets (probe sets 1 and 2). Differential gene expression of a subset of these transcripts was further confirmed by quantitative RT-PCR amplification (data not shown).
The associated activities of members within this subset cover a wide variety of cellular functions, including extracellular matrix production (thrombospondin-1, fibronectin), cytoskeletal organization (CAP-1), mitogenic signal transduction (Smad 6), and cytokine release (ADAM17). In addition, a number of genes were identified whose expression differences closely correlated with modulation in surface hydrophobicity, including thrombospondin-1, c-fos, and DUSP2. Of particular interest is the elucidation of relationships between transcriptome alterations and the observed phenotypic responses of cells to copolymer interaction.
Biomaterial-dependent modification of extracellular matrix production and organization is used in the assessment of biocompatibility prediction (12). Fibronectin and thrombospondin-1 are extracellular matrix proteins known to promote cell adhesion (13, 14). Cells exposed to copolymer films displayed down-regulation of fibronectin (–2.3-, –2.0-, and –2.6-fold; 50:50, 65:35, 85:15 NIPAAm:NtBAAm, respectively) and thrombospondin-1 (–3.7-, –5.4-, and –6.5-fold; order of copolymer films as previously shown) mRNA expression. These results are consistent with the reduction in cell adhesion observed in copolymer film-exposed cells (Fig. 2). Reduced thrombospondin-1 activity is further supported by the observation of increased DUSP2 (+3.9-, +14.6-, and +17.0-fold; order as previously shown) mRNA expression (known to be inhibited by thrombospondin-1 at the transcriptional level; ref. 15).
Altered extracellular matrix structure can have pleiotropic effects on cellular morphology, spreading, and cytoskeletal arrangement (16). Phenotypic observations revealed aberrant stellate cell morphology and loss of microfilament fibers in response to copolymer interaction (Fig. 4 B and C). Parallel gene expression analysis showed down-regulation of a subset of genes (TUBB4, TUBB, TUBA1, and smooth muscle α–actin) that play important roles in cytoskeletal organization, cell spreading, and morphology (Fig. 5; ref. 17). In addition, we observed reduced expression of CAP-1 mRNA (–1.8-, –1.8-, and –2.0-fold; order as previously shown) in cells exposed to copolymer film. CAP-1 is associated with increased levels of cytosolic cAMP, microfilament disassembly, and altered cell morphology (18). Interestingly, cAMP is currently being investigated with respect to cell–biomaterial interaction (18). Reduced expression of CAP-1 mRNA may thus prove to be indicative of biomaterialinduced alterations in the structural arrangement of the cytoskeleton and related morphological changes.
Together with analysis of phenotypic alterations, biochemical methods have been used to determine the compatibility of novel biomaterials. Bioincompatibilty is often associated with increased release of proinflammatory cytokines (19). For example, in vitro studies have demonstrated that surface chemistry may influence the production and secretion of two known proinflammatory cytokines IL-1β and tumor necrosis factor (TNF)-α (18). ADAM17 is a member of a new class of sheddases (20) that is responsible for the proteolytic release of TNF-α and transforming growth factor-α, along with several other cell surface proteins, IL-1 receptor type II, including p75 TNF-receptor, p55 TNF-receptor, and amyloid precursor protein (21). ADAM17 mRNA expression was consistently up-regulated in cells grown on copolymer films (+2.7-, +3.0-, and +2.5-fold; order as previously shown). Increased expression of ADAM17 may either result in, or be reflective of, an inflammatory response to copolymer film interaction. In summary, ADAM17 might be a promising marker for biocompatibility assessment.
Discussion
NIPAAm has previously been used as an adaptive cell culture substrate (6), despite its poor initial cell adhesion qualities. As reported earlier and also here, we can improve NIPAAm as a cell culture substrate through the addition of a more hydrophobic, yet structurally similar, monomer NtBAAm (5). The synthesis of such a series demonstrates how one can alter a single surface characteristic (without significantly altering the bulk properties of the polymer) and subsequently affect cellular phenotypic responses. Our current findings further demonstrate that alteration in cell adhesion and growth due to increasing surface hydrophobicity is independent of cell type and status.
Here, we have illustrated a link between substratum adhesivity and hydrophobicity and consequent relationship to the rate of cell spreading, with the most hydrophilic surfaces demonstrating the least extent of cell spreading 24 h postseeding. After several days of growth, HeLa cells on all surfaces acquired the same morphology, which was distinct from that observed on TCP. This result suggests that, whereas surface hydrophobicity has an initial effect on cell spreading and morphology, other surface characteristics or combinations thereof, that are common to all NIPAAm:NtBAAm copolymer films, seem to ultimately determine cellular morphology in the long-term. Cytochemical analysis indicated that the reduced cell spreading and stellate morphological appearance of HeLa cells on copolymer films may be due to a lack of cytoskeletal organization. In support of this concept, such stellate shapes have been previously demonstrated to be due to a depletion of actin microfilaments and have been associated with outgrowths terminating in a fan of ruffled membrane (10).
The identification of associations between gene expression alterations and biological processes is an emerging theme in biomedical research. In the past few years, there have been enormous advances in the ability to perform high-throughput analysis of gene expression. DNA microarrays, in particular, have become increasingly useful tools in providing global insights into the molecular basis of cellular responses to various physiological stimuli and toxic insults. Previously, investigators have demonstrated the influence of surface physiochemical properties on gene expression via the use of quantitative RTPCR amplification (22). However, the use of quantitative RTPCR is limited with respect to the number of transcripts that can be reasonably assessed for any one material. By employing a DNA microarray-based gene expression profiling approach, we were able to investigate the expression of hundreds to thousands of genes simultaneously in cells interacting with copolymer film.
Of the 21 transcripts that were identified as being differentially expressed between copolymer and TCP comparisons, a number of these genes either have been or are currently being investigated as markers of material biocompatibility, including fibronectin, thrombospondin-1, and c-fos. In addition, candidate biomarkers for compatibility prediction were revealed, such as ADAM17.
This study illustrates the enormous potential offered by the application of DNA microarray-based gene expression profiling, combined with conventional cellular phenotypic analysis, to provide a comprehensive and integrated overview of cell–biomaterial interaction. In addition, we have shown how discrete modifications with respect to the physiochemistry of soft amorphous materials can lead to significant impacts on the phenotype of interacting cells. This case study sets the scene for the development of high-throughput biological assessment of novel biomaterials.
Acknowledgments
The authors would like to acknowledge financial support from Enterprise Ireland and the Health Research Board of Ireland.
Abbreviations: NIPAAm, N-isopropylacrylamide; NtBAAm, N-tert-butylacrylamide; TCP, tissue culture polystyrene; HASMC, human aortic smooth muscle cell.
References
- 1.Rihova, B. (1996) Adv. Drug Delivery Rev. 21, 157–176. [Google Scholar]
- 2.Dawson, K. A. (2002) Curr. Opin. Colloid Interface Sci. 7, 218–227. [Google Scholar]
- 3.Tang, L. & Eaton, J. W. (1995) Am. J. Clin. Pathol. 103, 466–471. [DOI] [PubMed] [Google Scholar]
- 4.Ito, Y. (1999) Biomaterials 20, 2333–2342. [DOI] [PubMed] [Google Scholar]
- 5.Rochev, Y., Golubeva, T., Gorelov, A., Allen, L. T., Gallagher, W. M., Selezneva, I., Gavrilyuk, B. & Dawson, K. (2001) Prog. Colloid Polym. Sci. 118, 153–156. [Google Scholar]
- 6.Rollason, G., Davies, J. E. & Sefton, M. V. (1993) Biomaterials 14, 153–155. [DOI] [PubMed] [Google Scholar]
- 7.Kikuchi, A., Okuhara, M., Karikusa, F., Sakurai, Y. & Okano, T. (1998) J. Biomaterial Sci. Polym. Ed. 9, 1331–1348. [DOI] [PubMed] [Google Scholar]
- 8.Doorty, K. B., Golubeva, T. A., Gorelov, A. V., Rochev, Y. A., Allen, L. T., Dawson, K. A., Gallagher, W. M. & Keenan, A. K. (2003) Cardiovasc. Pathol. 12, 105–110. [DOI] [PubMed] [Google Scholar]
- 9.Cramer, L. P. & Mitchison, T. J. (1995) J. Cell Biol. 131, 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards, J. G., Campbell, G., Carr, M. & Edwards, C. C. (1993) J. Cell Sci. 104, 399–407. [DOI] [PubMed] [Google Scholar]
- 11.Maniotis, A. J., Chen, C. S. & Ingber, D. E. (1997) Proc. Natl. Acad. Sci. USA 94, 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grill, V., Sandrucci, M. A., Di Lenarda, R., Cadenaro, M., Narducci, P., Bareggi, R. & Martelli, A. M. (2000) J. Biomed. Mater. Res. 52, 479–487. [DOI] [PubMed] [Google Scholar]
- 13.Nuttelman, C. R., Mortisen, D. J., Henry, S. M. & Anseth, K. S. (2001) J. Biomed. Mater. Res. 57, 217–223. [DOI] [PubMed] [Google Scholar]
- 14.Lawler, J. (2000) Curr. Opin. Cell Biol. 12, 634–640. [DOI] [PubMed] [Google Scholar]
- 15.Li, Z., He, L., Wilson, K. & Roberts, D. (2001) J. Immunol. 166, 2427–2436. [DOI] [PubMed] [Google Scholar]
- 16.Tziampazis, E., Kohn, J. & Moghe, P. V. (2000) Biomaterials 21, 511–520. [DOI] [PubMed] [Google Scholar]
- 17.Walczak, C. E. (2000) Curr. Opin. Cell Biol. 12, 52–56. [DOI] [PubMed] [Google Scholar]
- 18.Moriyama, K. & Yahara, I. (2002) J. Cell Sci. 115, 1591–1601. [DOI] [PubMed] [Google Scholar]
- 19.Kalltorp, M., Oblogina, S., Jacobsson, S., Karlsson, A., Tengvall, P. & Thomsen, P. (1999) Biomaterials 20, 2123–2137. [DOI] [PubMed] [Google Scholar]
- 20.Kheradmand, F. & Werb, Z. (2002) Bioessays 24, 8–12. [DOI] [PubMed] [Google Scholar]
- 21.Black, R. A. (2002) Int. J. Biochem. Cell Biol. 34, 1–5. [DOI] [PubMed] [Google Scholar]
- 22.Kato, S., Akagi, T., Sugimura, K., Kishida, A. & Akashi, M. (2000) Biomaterials 21, 521–527. [DOI] [PubMed] [Google Scholar]