Abstract
Ras is a guanine nucleotide-binding protein that cycles between inactive GDP-bound and active GTP-bound states to regulate a diverse array of cellular processes, including cell growth, apoptosis, and differentiation. The guanine nucleotide-bound state of Ras is tightly maintained by regulatory factors to promote regulated growth control. A class of regulatory molecules that lead to Ras activation are guanine nucleotide exchange factors (GEFs). Ras GEFs bind to Ras and facilitate GDP release, followed by GTP incorporation and Ras activation. Nitric oxide (NO) has also been shown to promote guanine nucleotide exchange (GNE) on Ras and increase cellular Ras-GTP levels, but the process by which NO-mediated GNE occurs is not clear. We initiated NMR structural and biochemical studies to elucidate how nitrosylation of Ras might lead to enhanced GNE. Surprisingly, our studies show that stable S-nitrosylation of Ras at Cys-118, does not affect the structure of Ras, its association with the Ras-binding domain of Raf (a downstream effector of Ras), or GNE rates relative to non-nitrosylated Ras. We have found, however, that the actual chemical process of nitrosylation, rather than the end-product of Ras S-nitrosylation, accounts for the enhanced GNE that we have observed and that has been previously observed by others.
Keywords: guanosine triphosphatase, redox, cancer, reactive oxygen species, reactive nitrogen species
Nearly 20 years ago, nitric oxide (NO) was identified as a vaso-relaxation factor and was found to mediate its effects on smooth muscle by coordinating the heme moiety of guanylate cyclase, leading to enhanced production of cGMP (1). It was later shown that increased levels of cGMP modulate the function of other physiological targets, such as protein kinases, phosphodiesterases, and ion channels, causing vaso-relaxation (as reviewed in refs. 1–3). NO has also been shown to modulate cGMP-independent pathways by modifying a multitude of cellular biomolecules containing reactive metal centers, as well as cysteine residues (4–6). Although NO is able to modulate the activity of tyrosine kinases, serine/threonine kinases, guanosine triphosphatases (GTPases), and phosphatases, the molecular basis of NO modification and the subsequent effect on enzyme activity are not completely understood. NO is generally believed to exert its effects on biomolecules by direct modification of a residue important for protein structure or enzymatic catalysis.
The Ras GTPase is a protein product of the most commonly mutated oncogene in all of human cancer (ras), and has been identified as a target for S-nitrosylation by NO (7). Ras is a molecular switch that cycles between an inactive GDP-bound state and an active GTP-bound state, to regulate a number of cellular processes, including cell growth, differentiation and apoptosis (for reviews, see refs. 8–10). In its oncogenic form, Ras is populated in a GTP-bound state and is constitutively active. The chronic activation of Ras leads to persistent activation of downstream signaling cascades to promote cellular transformation. In normal tissues, however, regulatory proteins tightly maintain the activity of Ras (as reviewed in refs. 11–13). These regulatory proteins, termed guanine-nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs), bind to Ras and either facilitate GDP release followed by GTP incorporation and, therefore, Ras activation (GEFs), or enhance GTP hydrolysis to GDP and, hence, Ras inactivation (GAPs). Several lines of evidence show Ras to be modified and activated by NO in vivo (7, 14, 15). First, Lander et al. (7) showed that human T cells treated with NO gas contain an increased percentage of Ras in the GTP-bound form. Additionally, in vitro experiments showed that purified H-Ras exchanged guanine nucleotide more rapidly in the presence of NO gas (7). It was later shown that Cys-118 was the Ras residue modified by NO and that a C118S Ras variant, while otherwise normal (16), cannot be S-nitrosylated and does not respond to NO in vitro or in vivo (14). Other in vivo studies have shown that activation of neuronal-NO synthase (nNOS) in rodent primary cortical neuronal cultures, by addition of N-methyl-D-aspartate (NMDA), populates Ras in its active GTP-bound state (15). Moreover, the addition of NMDA to nNOS-deficient (nNOS–/–) cultures does not lead to activation of Ras (15).
The observed enhancement of guanine nucleotide exchange (GNE) on treatment with NO was attributed to a structural change in Ras, resulting in a conformation that binds the guanine nucleotide with lower affinity (7). This mechanism for NO action was deduced by analogy to other systems where NO modification results in either the perturbation of a catalytic residue or the structural alteration of the modified protein. Inspection of the Ras structure, however, provides little support for either of these modes of NO action. Cys-118 is the most solvent-exposed cysteine in Ras, decreasing the likelihood that side-chain modifications will induce significant structural perturbations. Moreover, even though Cys-118 is located in the NKXD guanine nucleotide-binding motif, it occupies the hypervariable position and does not form specific contacts with the guanine nucleotide or any of the other residues in the guanine nucleotide-binding pocket (17, 18). Additionally, mutation of Cys-118 to serine results in a variant of Ras that does not differ significantly from wild-type Ras in structure or GNE (16). The lack of involvement of the Cys-118 side chain in guanine nucleotide coordination and guanine nucleotide-binding site architecture makes it difficult to understand how the addition of a nitroso group could alter the guanine nucleotide-binding properties or the structure of Ras. To better define the effects of S-nitrosylation on Ras structure and activity, we generated stably and stoichiometrically S-nitrosylated Ras and used NMR structural and biochemical studies to elucidate how NO activates Ras.
Materials and Methods
Chemicals and Reagents. All chemicals, unless otherwise noted, were purchased from Sigma. [3H]GDP was purchased from NEN.
Ras(1–166) Purification. Wild-type Ras(1–166) and Ras(1–166)C118S were purified as described (19).
Determination of Nitroso Content. Nitroso contents were determined by the Saville assay essentially as described (20).
S-Nitrosylation of Ras. Equal volumes of 100 mM L-cysteine or glutathione in 0.25 M HCl and 100 mM NaNO2 in dH2O were mixed and incubated for 10 min at room temperature in the dark. After incubation, reaction mixtures were diluted 4:1 with buffer (20 mM Tris, pH 7.4/l mM diethylenetriamine pentaacetic acid), and the pH was adjusted to pH 7.4 with NaOH. After determination of nitroso content by using the Saville assay (20), the nitroso donor S-nitroso-cysteine (Cys-NO) was added in 100-fold excess to Ras in reaction buffer and incubated for 30 min at room temperature in the dark. After removal of excess donor with a PD-10 desalting column (Pharmacia), the recovered Ras was assayed for nitroso content before being used in assays.
Mass Spectrometric Analysis. Measurements were made on a Micromass Quattro LC (Altrincham, U.K.) triple quadrupole mass spectrometer equipped with a pneumatically assisted electrostatic ion source operating at atmospheric pressure and in a positive ion mode. Samples were diluted with an equivolume of acetonitrile containing 1% formic acid. Analyses were made by loop injection into a stream of 50% aqueous acetonitrile flowing at 10 μl/min. Spectra were acquired in the multichannel analyzer mode from m/z 600–1,400 (scan time 5 s). The mass scale was calibrated by using the multiply charged envelope of the horse heart myoglobin (Mr 16951.48). The raw mass spectra were transformed to a molecular mass scale by using a maximum entropy-based method that uses the MEMSYS5 program (MaxEnt Solutions, Cambridge, U.K.) and is part of the Micromass MASSLYNX software suite. Transformation was performed from 800–1,400 m/z by using a resolution of 1 atomic mass unit.
Ras-Binding Domain of Raf-1 (Raf-RBD) Binding Assay. Fluorescence anisostropy measurements were conducted on a 2.5-ml sample of 0.5 μM Ras-Mant-GDP, Ras-Mant-GMPPNP, S-nitrosylated Ras-Mant-GDP, or S-nitrosylated Ras-Mant-GMPPNP in 20 mM Tris, 50 mM NaCl, 5 mM MgCl2 by using a Perkin–Elmer LS50B spectrophotometer at 25°C. GST-tagged Raf-RBD fragment (Raf-1 residues 50–131) was titrated into the Ras, and the resulting increase in anisotropy was recorded as a function of Raf-RBD concentration. Equilibrium dissociation constants, KDs, were determined by fitting the data to a hyperbola (Y = Bmax·X/(KD + X). In this equation, Y is the anisotropy value normalized from 0 to 1, Bmax is the anisotropy value at saturation (1 for normalized anisotropy values), X is the Raf-RBD concentration, and KD is the equilibrium dissociation constant.
GNE Assay. A 500-μl sample of 5 μM Ras(1–166), S-nitrosylated Ras(1–166), or Ras(1–166)C118S in 20 mM Tris (pH 7.4), 50 mM NaCl, and 5 mM MgCl2, with either 500 μM Cys-NO, 500 μM S-nitroso-glutathione (GS-NO), 5 mM EDTA, 1 μM mSOS (murine Son of Sevenless), or buffer control, was prepared. Assays were initiated by the addition of [3H]GDP, and aliquots were removed at specific time points and spotted onto nitrocellulose filters. The filters were then washed with a 2-ml reaction buffer three times. Radioactivity was assayed in a Beckman-Coulter scintillation counter, by using EcoScint fluid (Fisher).
NMR Structural Studies. NMR experiments were recorded on a Varian Inova 600 (600 MHz) spectrometer at 25°C. 2D 1H–15N heteronuclear single quantum coherence spectroscopy (HSQC) experiments were performed with pulsed field gradient and water flip-back methods described (21). HSQC experiments were acquired with 1,024 × 128 complex data points and a spectral width of 8,000 Hz for the 1H dimension and 1,709 Hz for the 15N dimension. 15N-edited 3D nuclear Overhauser enhancement (NOE) experiments were acquired with 1,024 × 48 × 32 complex data points with a mixing time of 120 ms. NMR data were processed and analyzed by using the program FELIX V. 97.2 (Accelrys, San Diego).
Results
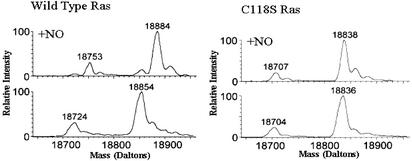
To investigate how treatment of Ras with NO leads to activation of Ras, we conducted a series of structural and biochemical studies on S-nitrosylated Ras. Structural studies were initiated by preparing stably and stoichiometrically S-nitrosylated H-Ras. Mass spectrometric analysis was used to characterize Ras-species generated by Ras S-nitrosylation. Mass spectrometric results described in Fig. 1 show that addition of Cys-NO to Ras(1–166) produces a nearly complete, stoichiometric modification of Ras by NO whereas the Ras(1–166)C118S variant is insensitive to Cys-NO treatment. These results indicate that Ras is specifically modified by NO, because the increase in mass of the Ras protein on treatment with Cys-NO corresponds to the mass of an NO moiety. Moreover, our results indicate that NO modification of Ras is specific to residue Cys-118, because the mass of the Ras(1–166)C118S variant is not altered by Cys-NO treatment. This finding was later confirmed by NMR. These mass spectrometric results also eliminate the possibility that unanticipated side-reactions or by-products of Cys-NO treatment induce Ras modification.
Fig. 1.
Mass spectral data of Ras(1–166) and Ras(1–166)C118S before and after treatment with Cys-NO. (Lower Left) Mass chromatogram representative of Ras(1–166). The larger peak corresponds to Ras(1–166), whereas the smaller peak corresponds to a loss of the N-terminal methionine and hence Ras(2–166). (Upper Left) Mass chromatogram of Ras(1–166) after treatment with Cys-NO. Note the gain of 30 mass units for both the Ras(1–166) and the Ras(2–166). A small shoulder to the left of the S-nitrosylated Ras(1–166) peak corresponds to a small population of unmodified Ras(1–166). The mass chromatograms in Right represent the Ras(1–166)C118S variant of Ras. The approximate 16 mass unit shift in molecular weight in the mass of Ras from Lower Right relative to the mass of Ras as determined in Lower Left signifies the C118S mutation. The Ras(1–166)C118S variant does not gain mass after exposure to Cys-NO as can be seen in Upper Right.
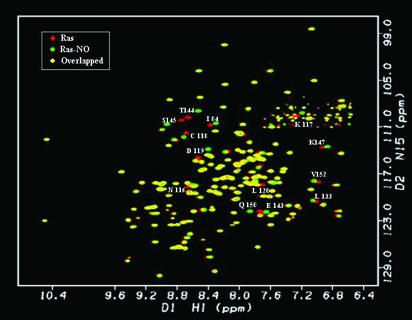
After preparing S-nitrosylated Ras, we conducted NMR studies to evaluate whether the structure of S-nitrosylated Ras differs from wild-type Ras. Both 2D 1H–15N HSQC and 3D 15N-edited NOE data were acquired to elucidate whether chemical shift and/or structural differences can be detected between S-nitrosylated Ras and unmodified Ras. A 1H–15N HSQC is a 2D NMR experiment that selects for protons attached to 15N nuclei (22). The ability to selectively observe amide resonances provides a site-specific probe for every amino acid in the protein except for proline, a spectral fingerprint of Ras. Comparison of the chemical shift changes between the HSQC spectrum of unmodified Ras and the spectrum of NO-labeled Ras show minor chemical shift differences (Fig. 2) that are restricted to residues proximal to Cys-118 in three-dimensional space (Fig. 3). The small, localized chemical shift differences between NO-modified and unmodified Ras indicate that Ras does not undergo a large-scale structural perturbation on modification of Cys-118 by NO. Although the limited chemical shift perturbations in the HSQC spectra are unlikely to reflect significant structural changes, the qualitative nature of the 1H–15N HSQC perturbation map does not eliminate the possibility of local conformational rearrangement occurring around Cys-118. Therefore, we collected 3D 15N-edited NOESY data on NO-modified Ras. This experiment detects through-space proton dipoles coupled to amide protons and provides a measure of inter-proton distances (23). We compared NOE spectra between unmodified and S-nitrosylated Ras, and found only small and local NOE differences (Fig. 4). As yet, it is not clear whether these observed differences reflect any NO-induced structural change in Ras, because most of the changes in the NOE resonances near the site of nitrosylation (i.e., Cys-118 and Ser-145) are likely due to shifts in the NOE resonances caused by a modified chemical environment (R-SH to R-S-NO) rather than an altered Ras structure. Overall, there are relatively few differences in the NOE resonances in nitrosylated Ras relative to non-nitrosylated Ras. The magnesium binding site in Ras, as illustrated by the slices for Gly-13 and Lys-16, and the effector binding region, as depicted by Asp-33, do not show NO-induced changes in the NOE resonances. These results are consistent with our HSQC results, indicating that no notable structural perturbations occur in Ras on S-nitrosylation of Cys-118.
Fig. 2.
1H–15N-HSQC of Ras(1–166) before and after treatment with Cys-NO. NMR overlay of Ras alone (red) and S-nitrosylated Ras (green). The HN resonances that overlay (yellow) indicate no differences in structure or chemical environment. Most HN resonance shifts are small and correspond to residues localized to the regions around residue Cys-118.
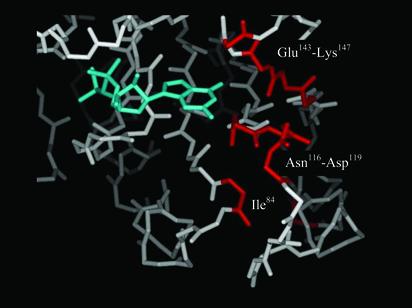
Fig. 3.
The regions of Ras affected by S-nitrosylation. Model of Ras(1–166)-GDP (PDB 1CRQ) displaying the nucleotide-binding pocket. The guanine nucleotide is colored cyan. The amino acid residues that correspond to HN resonances that showed differences in chemical shift between unmodified Ras and S-nitrosylated Ras are highlighted in red.
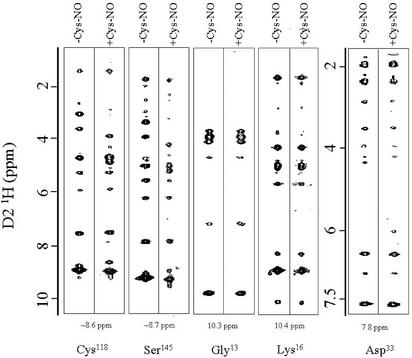
Fig. 4.
NOE slices of Ras-GDP and S-nitrosylated-Ras-GDP. 15N-edited 1H–1H-NOE data were collected on non-nitrosylated and S-nitrosylated Ras-GDP. The NOE data suggest only minor structural perturbation on nitrosylation. As representative data, the 1H–1H-NOE slices for residues Cys-118 (15N plane = 111.4, unmodified Ras; 112.0, S-nitrosylated Ras), Ser-145 (15N plane = 110.8 unmodified Ras; 110.4 S-nitrosylated Ras), Gly-13 (15N plane = 111.8, unmodified Ras; 111.8, S-nitrosylated Ras), Lys-16 (15N plane = 123.6, unmodified Ras; 123.6, S-nitrosylated Ras), and Asp-33 (15N plane = 125.8, unmodified Ras; 125.8, S-nitrosylated Ras) are shown. Note that, even though the D1 1H and D3 15N chemical shift values are altered by nitrosylation (for 118Cys and 145Ser), the 1H–1H NOE patterns (i.e., number and type of NOEs) are similar.
Although our data indicate that S-nitrosylation of Ras does not significantly alter the structure of the protein, we conducted studies to determine whether stable nitrosylation of Ras alters binding interactions with the Ras binding domain of Raf (Raf-RBD). These studies were initiated because of reports indicating that nitrosylation of Ras leads to increased coimmunoprecipitation of two of its effectors, the Raf kinase and phosphatidylinositol 3-kinase (24, 25). However, it was not determined whether the recruitment of effectors to Ras was mediated through an increase in Ras-GTP levels or through an increase in effector affinity that is independent of the guanine nucleotide bound state.
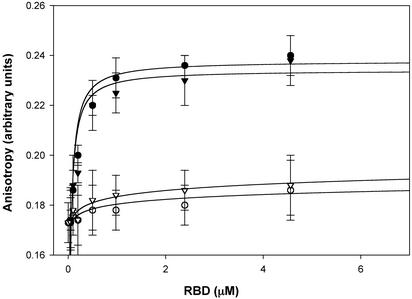
To differentiate between these two possibilities, we compared the amount of unmodified Ras and S-nitrosylated Ras that was associated with the Raf-RBD using a fluorescence based assay. In this assay, Ras is labeled with a fluorescent analog of GDP (Mant-GDP) or an analog of GTP (Mant-GMPPNP). GST-Raf-RBD (50–131) is then titrated into the Ras sample, the change in fluorescence anisotropy is measured, and the KDs are determined by fitting the data to a single saturated site. As demonstrated in Fig. 5, there is no difference in the KD of Ras-GMPPNP or S-nitrosylated-Ras-GMPPNP to the Raf-RBD. These results are not surprising, because the residues in Ras whose chemical shifts are affected by S-nitrosylation are far removed from the effector domain (residues 30–40), the region of Ras that interacts with the Raf-RBD (26). Moreover, our NOE data indicate that stable nitrosylation of Ras does not induce structural differences around the effector domain (Fig. 5). These results suggest that the previously observed increase in association of Ras to Raf in the presence of nitrosylating agents is likely a result of increased levels of Ras-GTP rather than altered affinity of S-nitrosylated Ras for Raf.
Fig. 5.
S-Nitrosylation of Ras does not alter its ability to recognize effectors. In this assay, Ras was labeled with a fluorescent analog of either GDP (Mant-GDP) or GTP (Mant-GMPPNP), and anisotropy measurements were made as a function of Raf-RBD concentration. Little binding of the Raf-RBD is observed when Ras is GDP bound (inactive form) to either non-nitrosylated Ras (○) or S-nitrosylated Ras (▿). In contrast, when Ras is complexed with Mant-GMPPNP, the Raf-RBD binds with a KD of ≈20 nM to both non-nitrosylated (•) and S-nitrosylated Ras (▾). These values are consistent with preexisting literature (28).
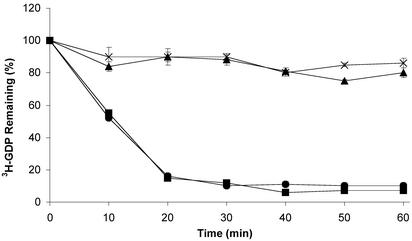
Next, we assayed the effects of S-nitrosylation on the rate of Ras GNE. These assays were conducted under two different conditions. First, GNE assays were performed on stably modified S-nitrosylated Ras. Second, the assays were conducted as described (7), where the nitrosylating agent (Cys-NO or GS-NO) was added to Ras at the beginning of the nucleotide exchange procedure to monitor the affects of the nitrosylation event on GNE. As shown in Fig. 6, neither the intrinsic nor GEF-stimulated GNE rate of Ras-GDP is altered by stable nitrosylation of Ras. In particular, unmodified Ras and S-nitrosylated Ras possess similar, slow intrinsic guanine nucleotide dissociation rates, whereas unmodified Ras and S-nitrosylated Ras treated with the murine guanine-nucleotide exchange factor, mSOS, demonstrate rapid [3H]GDP loss, but in an identical fashion, indicating that the presence of an NO moiety on Cys-118 does not significantly affect either intrinsic or GEF-mediated GNE. To control for the possibility that NO may directly affect SOS activity, we pretreated SOS with NO and performed GNE assays. Pretreatment of SOS with NO did not show any effect on SOS-mediated Ras GNE (data not shown).
Fig. 6.
Stable S-nitrosylation does not affect intrinsic or GEF-mediated guanine nucleotide dissociation. In this assay, Ras or S-nitrosylated Ras was preloaded with [3H]GDP. The assay was initiated by adding either an excess of nonradioactive GDP, or an excess of nonradioactive GDP and a catalytic amount of SOS, and followed as a function of time. As radioactive nucleotide dissociates from Ras, a corresponding loss of radioactivity is observed. The intrinsic rate of guanine nucleotide dissociation for Ras (×) and S-nitrosylated Ras (▴) was comparable. Although addition of mSOS to both untreated and S-nitrosylated Ras enhances the rate of guanine nucleotide dissociation, similar enhancements were observed for both S-nitrosylated Ras (▾) and untreated Ras (•).
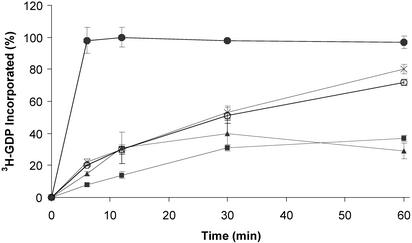
A different NO-mediated effect was observed when conducting the GNE assay under the second condition, where GNE is monitored immediately after the addition of nitrosylating agents. Although the presence of an NO moiety on Cys-118 had little effect on GNE, the introduction of the NO-donating reagents Cys-NO and GS-NO both increased the rate of GNE (Fig. 7), suggesting that the chemical process of S-nitrosylation at Cys-118 may account for the GNE enhancement observed previously. The enhanced rate of GNE in the presence of NO-donating reagents is Cys-118 dependent (Fig. 7), because mutation of Cys-118 to Ser renders Ras insensitive to NO-mediated GNE. C118S Ras exchanges guanine nucleotide normally in the absence of NO, and the addition of Cys-NO does not increase the rate of [3H]GDP incorporation. To exclude the possibility that oxidation or degradation by-products of Cys-NO contribute to the enhanced GNE observed in the presence of Cys-NO, the assay was repeated in the presence of cysteine, nitrite, and nitrate. None of these compounds produced an increase in GNE (data not shown).
Fig. 7.
An NO donor enhances the rate of GNE. This exchange assay was performed by adding [3H]GDP to either wild-type or C118S Ras, alone, or in the presence of EDTA, Cys-NO, or GS-NO, and the incorporation of [3H]GDP was monitored over time. The EDTA serves as a positive control by chelating the magnesium from Ras so that guanine nucleotide affinity is decreased and rapid dissociation and reassociation of the guanine nucleotide can occur. This result is shown by the rapid incorporation of the radiolabeled guanine nucleotide into EDTA-treated Ras (•). Ras in the absence of NO-donating reagents (▾) shows very little guanine nucleotide incorporation during the time course of the experiment, whereas Ras treated with NO-donating reagents Cys-NO (×) or GS-NO (○) shows an increase in the GNE relative to Ras alone. The C118S mutant Ras (▴), which cannot be S-nitrosylated, is insensitive to Cys-NO treatment.
Discussion
It has been established that NO can modify Ras at Cys-118 and act as a positive regulator of Ras signal transduction (7, 14, 15, 27–29). To further elucidate how treatment of Ras with NO donating agents leads to Ras activation in vivo, we conducted structural and biochemical studies on S-nitrosylated Ras. Our mass spectrometric and NMR analyses indicate that addition of Cys-NO to Ras modifies only Cys-118, with S-nitrosylated Ras as the end-product of Cys-NO treatment. No alternative sites of Ras modification by NO or by-products of Cys-NO decomposition were detected under the conditions used in this study. Treatment of Ras with NO-donating agents has been previously shown to enhance Ras GNE (7). Hence, we investigated the possibility that stable nitrosylation of Cys-118 alters Ras structure and subsequently influences Ras GNE. However, our studies revealed that stable S-nitrosylation of Ras at Cys-118 does not cause any significant perturbations in the secondary or tertiary structure of Ras, and does not influence Ras GNE in the presence or absence of the Ras GEF, SOS. Cys-118 is a solvent-exposed residue that makes no contacts with either the guanine nucleotide substrate or other residues in Ras (17, 18); therefore, a large-scale structural perturbation in Ras was neither expected nor observed on S-nitrosylation of Cys-118. The site of S-nitrosylation is removed from the effector binding region (30); however, it was not clear whether more subtle structural alterations in Ras resulting from S-nitrosylation might affect effector binding, because previous studies indicated that Ras nitrosylation leads to increased association of Ras to the Ras effectors Raf-1 and phosphatidylinositol-3 kinase (24, 25). However, the binding affinity of the Raf-RBD to Ras was not influenced by stable nitrosylation of Ras. Although our results indicate that neither Raf-RBD binding nor SOS-mediated GNE of Ras is altered by stable S-nitrosylation of Ras, we cannot exclude the possibility that other GEF(s), GTPase-activating protein(s), or effectors may be specifically altered in their abilities to recognize S-nitrosylated Ras relative to its non-nitrosylated counterpart. However, our observations that stable S-nitrosylation of Ras does not affect the structure or biochemical properties of Ras are consistent with previous studies of a Ras C118S variant of Ras that behaves essentially like wild-type protein except that it is not responsive to NO signaling (16).
Although the presence of an NO moiety on Cys-118 does not affect Ras GNE, our results show that the introduction of NO-donating reagents Cys-NO and GS-NO both increase the rate of Ras GNE, indicating that the chemical process of Ras nitrosylation accounts for the enhanced GNE in the presence of NO reagents, observed previously. In the case of Cys-NO, the affect seems to be specific because neither nitrite, nitrate, nor cysteine promotes GNE exchange on Ras (data not shown).
Consistent with these observations, we have found that NO gas, Cys-NO, and GS-NO can modify Ras through a radical mechanism, where formation of a Ras-radical intermediate reduces the affinity of Ras for its guanine nucleotide substrate, leading to enhanced guanine nucleotide dissociation (J. Heo & S.L.C., unpublished results). Whereas our structural and biochemical studies indicate that the end-product of S-nitrosylation of Ras (Ras-S-NO) does not alter Ras structure, GNE or Raf-RBD binding, the data set forth in this paper indicate that an intermediate of the nitrosylation reaction, rather than the R-S-NO end product, alters Ras structural elements and biochemical properties.
In summary, our results indicate that the addition of NO-donating reagents (Cys-NO and GS-NO) to Ras causes enhanced Ras-GNE, consistent with earlier studies. However, results from this work and our other studies indicate that it is the actual process of S-nitrosylation, rather than the product of S-nitrosylation (Ras-S-NO), that interferes with guanine nucleotide substrate binding, resulting in enhanced guanine nucleotide dissociation (J. Heo & S.L.C., unpublished results). Similar to the effects of GEFs and certain Ras mutations, NO treatment of Ras can lead to enhanced Ras GNE and up-regulation of Ras activity in vivo. However, it is unclear how Ras is regulated by NO in vivo. NO is produced from NOSs (2). NOSs are complex regulatory enzymes that can produce either bursts of NO or constant levels of NO in response to the needs of the cell (31). Ras can in turn be activated in response to cellular conditions where NOS is stimulated and can transduce signals through multiple pathways, leading to changes in cell morphology, gene expression, and apoptosis. Moreover, in vivo activation of Ras by NO may be co-regulated and/or may involve GEF proteins. One can imagine a scenario where GEF proteins may act synergistically with NO to displace the guanine nucleotide from Ras. It will be of interest to further elucidate details of the reaction mechanism involving NO-mediated regulation of Ras activity and how Ras is regulated by NO in vivo.
The effects of NO on GNE may not be unique to Ras. We anticipate that other GTPases containing the NKCD motif, such as Rap, many of the Rabs, and the prokaryotic forms of the elongation factor EF-Tu (32–34), may share a similar sensitivity to NO and display enhanced GNE and, hence, increased GTP loading. Furthermore, other redox active agents may show similar affects. The mechanisms by which NO and other reactive oxygen and nitrogen species modulate the activities of Ras and other GTPases warrant further investigation.
Acknowledgments
We thank Min-qi Lu for excellent technical assistance and Kirk Prutzman for processing the NOE data. We also thank Michael Clarkson and Sean Palmer for their experimental assistance with this work. The Lineberger Comprehensive Cancer Center's Cancer Cell Biology Training Program supported J.G.W. This work was funded by American Cancer Society Grant RDG-99-060-01-CNE (to S.L.C.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Cys-NO, S-nitroso-cysteine; GNE, guanine nucleotide exchange; GEF, GNE factor; GS-NO, S-nitroso-glutathione; GTPase, guanosine triphosphatase; HSQC, heteronuclear single quantum coherence; NOE, nuclear Overhauser enhancement; Raf-RBD, Ras-binding domain of Raf-1; mSOS, murine Son of Sevenless GEF.
References
- 1.Ignarro, L. J., Cirino, G., Casini, A. & Nopoli, C. (1999) J. Cardiovasc. Pharmacol. 34, 879–886. [DOI] [PubMed] [Google Scholar]
- 2.Groves, J. T. & Wang, C. C. (2000) Curr. Opin. Chem. Biol. 4, 687–695. [DOI] [PubMed] [Google Scholar]
- 3.Alderton, W. K., Cooper, C. E. & Knowles, R. G. (2001) Biochem. J. 357, 593–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ford, P. C. & Lorkovick, I. M. (2002) Chem. Rev. 102, 993–1018. [DOI] [PubMed] [Google Scholar]
- 5.Gow, A. J. & Stamler, J. S. (1998) Nature 391, 169–173. [DOI] [PubMed] [Google Scholar]
- 6.Stamler, J. S. (1994) Cell 78, 931–936. [DOI] [PubMed] [Google Scholar]
- 7.Lander, H. M., Ogiste, J. S., Pearce, S. F., Levi, R. & Novogrodsky, A. (1995) J. Biol. Chem. 270, 7017–7020. [DOI] [PubMed] [Google Scholar]
- 8.Wittinghofer, A. (1998) Biol. Chem. 379, 933–937. [PubMed] [Google Scholar]
- 9.Shields, J. M., Pruitt, K., McFall, A., Shaub, A. & Der, C. J. (2000) Trends Cell Biol. 10, 147–154. [DOI] [PubMed] [Google Scholar]
- 10.Campbell, S. L., Khosravi-Far, R., Rossman, K. L., Clark, G. J. & Der, C. J. (1998) Oncogene 17, 1395–1413. [DOI] [PubMed] [Google Scholar]
- 11.Geyer, M. & Wittinghofer, A. (1997) Curr. Opin. Struct. Biol. 7, 786–792. [DOI] [PubMed] [Google Scholar]
- 12.Cherfils, J. & Chardin, P. (1999) Trends Biochem. Sci. 24, 306–311. [DOI] [PubMed] [Google Scholar]
- 13.McCormick, F. (1998) Curr. Biol. 8, R673–R674. [DOI] [PubMed] [Google Scholar]
- 14.Lander, H. M., Hajjar, D. P., Hempstead, B. L., Mirza, U. A., Chait, B. T., Campbell, S. & Quilliam, L. A. (1997) J. Biol. Chem. 272, 4323–4326. [DOI] [PubMed] [Google Scholar]
- 15.Yun, H. Y., Gonzalez-Zulueta, M., Dawson, V. L. & Dawson, T. M. (1998) Proc. Natl. Acad. Sci. USA 95, 5773–5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mott, H. R., Carpenter, J. W. & Campbell, S. L. (1997) Biochemistry 36, 3640–3644. [DOI] [PubMed] [Google Scholar]
- 17.Wittinghofer, F., Krengel, U., John, J., Kabsch, W., Kabsch, W. & Pai, E. F. (1991) Environ. Health Perspect. 93, 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunger, A. T., Milburn, M. V., Tong, L., deVos, A. M., Jancarik, J., Yamaizumi, Z., Nishimura, S., Ohtsuka, E. & Kim, S. H. (1990) Proc. Natl. Acad. Sci. USA 87, 4849–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell-Burk, S. L. & Carpenter, J. W. (1995) Methods Enzymol. 255, 3–13. [DOI] [PubMed] [Google Scholar]
- 20.Zhang, Y. Y., Xu, A. M., Nomen, M., Walsh, M., Keaney, J. F., Jr., & Loscalzo, J. (1996) J. Biol. Chem. 271, 14271–14279. [PubMed] [Google Scholar]
- 21.Grzesiek, S. & Bax, A. (1993) J. Biomol. NMR 3, 627–638. [DOI] [PubMed] [Google Scholar]
- 22.Kay, L., Keifer, P. & Saarinen, T. (1992) J. Am. Chem. Soc. 114, 10663–10665. [Google Scholar]
- 23.Zhang O. W., Kay L. E., Olivier J. P. & Forman-Kay, J. D. (1994) J. Biomol. NMR 4, 845–858. [DOI] [PubMed] [Google Scholar]
- 24.Deora, A. A., Hajjar, D. P. & Lander, H. M. (2000) Biochemistry 39, 9901–9908. [DOI] [PubMed] [Google Scholar]
- 25.Deora, A. A., Win, T., Vanhaesebroeck, B. & Lander, H. M. (1998) J. Biol. Chem. 273, 29923–29928. [DOI] [PubMed] [Google Scholar]
- 26.Nassar, N., Horn, G., Herrmann, C., Scherer, A., McCormick, F. & Wittinghofer, A. (1995) Nature 375, 554–560. [DOI] [PubMed] [Google Scholar]
- 27.Teng, K. K., Esposito, D. K., Schwartz, G. D., Lander, H. M. & Hempstead, B. L. (1999) J. Biol. Chem. 274, 37315–37320. [DOI] [PubMed] [Google Scholar]
- 28.Lander, H. M., Ogiste, J. S., Teng, K. K. & Novogrodsky, A. (1995) J. Biol. Chem. 270, 21195–21198. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez-Zulueta, M., Feldman, A. B., Klesse, L. J., Kalb, R. G., Dillman, J. F., Parada, L. F., Dawson, T. M. & Dawson, V. L. (2000) Proc. Natl. Acad. Sci. USA 97, 436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wittinghofer, A. & Nassar, N. (1996) Trends Biochem. Sci. 21, 488–491. [DOI] [PubMed] [Google Scholar]
- 31.Andrew, P. J. & Mayer, B. (1999) Cardiovasc. Res. 43, 521–531. [DOI] [PubMed] [Google Scholar]
- 32.Bos, J. L. (1998) EMBO J. 17, 6776–6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stenmark, H. & Olkkonen, V. M. (2001) Genome Biol. 2, 3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takai, Y., Sasaki, T., Sasaki, T. & Matozaki, T. (2001) Physiol. Rev. 81, 153–208. [DOI] [PubMed] [Google Scholar]