Abstract
We have successfully used mutagenesis to engineer Taxol (paclitaxel) binding activity in Saccharomyces cerevisiae tubulin. Taxol, a successful antitumor agent, acts by promoting tubulin assembly and stabilizing microtubules. Several structurally diverse antimitotic compounds, including the epothilones, compete with Taxol for binding to mammalian microtubules, suggesting that Taxol and these compounds share an overlapping binding site. However, Taxol has no effect on tubulin or microtubules from S. cerevisiae, whereas epothilone does. After considering data on Taxol binding to mammalian tubulin and recent modeling studies, we have hypothesized that differences in five key amino acids are responsible for the lack of Taxol binding to yeast tubulin. After changing these amino acids to those found in mammalian brain tubulin, we observed Taxol-related activity in yeast tubulin comparable to that in mammalian tubulin. Importantly, this experimental system can be used to reveal tubulin interactions with Taxol, the epothilones, and other Taxol-like compounds.
Taxol (paclitaxel) is a mitotic inhibitor that has been successfully used in the treatment of breast, ovarian, and lung carcinomas. The powerful antitumor activity of Taxol results from its ability to promote the assembly and stabilization of microtubules (1). Microtubules are structures composed of polymerized tubulin heterodimers and play fundamental roles in vital cell processes such as chromosome segregation and intracellular transport. Extensive research has been directed toward understanding the tubulin–Taxol interaction and specifically, the pharmacophore of the Taxol molecule (reviewed in ref. 2). Photoaffinity crosslinking studies have localized Taxol binding to the β-subunit of tubulin (3–7) and, specifically, to peptides 1–31 (5), 217–231 (6), and 277–293 (7). Structure–activity relationship studies have defined the contribution of various substituents in the Taxol molecule (reviewed in ref. 8), whereas studies with Taxol-resistant mammalian cell lines containing mutations in β-tubulin have identified specific amino acids that may be involved in Taxol binding (9–12). These results, together with the 3.7-Å electron crystal structure of a tubulin polymer showing the electron density of bound Taxol (13), have been used to propose sites of interaction and conformations for bound Taxol (9, 14–17). However, there has been no direct experimental evidence supporting any of the putative binding interactions. Other recently discovered, potent, microtubule-stabilizing compounds such as the epothilones, discodermolide, and eleutherobins compete with Taxol for binding to mammalian microtubules (18–20). Therefore, despite a lack of structural similarity, these compounds seem to share an overlapping binding site with Taxol. Several of these compounds are noteworthy because they are effective against some multidrug-resistant tumor cell lines that show Taxol resistance and they are more water-soluble than Taxol. Thus, an understanding of the binding site shared by these compounds could be exploited for the rational design of novel and more effective chemotherapeutic agents for the treatment of cancer.
Tubulin from the budding yeast Saccharomyces cerevisiae shares 75% amino acid identity with mammalian brain tubulin and can coassemble with brain tubulin into microtubules (21). Despite these similarities, yeast tubulin is unaffected by most of the antimitotic compounds that bind strongly to mammalian tubulin or microtubules, including Taxol (21, 22). As a result, yeast tubulin has not been used in the investigation of tubulin–Taxol interactions. However, we recently showed that the epothilones do promote the assembly and stabilization of yeast microtubules (22). Thus, there are important differences in the sequences and structures of yeast and mammalian tubulin that permit strong binding of the epothilones but not of Taxol. Because yeast contains only one β-tubulin gene and site-directed mutations are easily accomplished in this organism, yeast is an excellent system for the investigation of the differences in Taxol and epothilone binding to tubulin.
We considered data from photoaffinity labeling studies (5, 6, 7), Taxol-resistant cell lines with mutations in β-tubulin (9–12), recent molecular modeling studies (9, 15–17), and the amino acid sequences of yeast and mammalian brain tubulin to identify five amino acid differences in yeast tubulin, relative to mammalian tubulin, that could weaken Taxol binding (22). We hypothesized that these differences were responsible for the lack of key interactions between Taxol in the T conformation (T-Taxol) (16) and yeast tubulin (22). The interactions are with the C3′ benzamido group of Taxol (Fig. 1), which is known to contribute significantly to the biological activity of Taxol with mammalian tubulin (23, 24) and with the diterpene ring.
Fig. 1.
Structure of Taxol (paclitaxel).
We mutated the single β-tubulin gene in S. cerevisiae and purified and studied the resultant protein in vitro. Five amino acids in yeast tubulin (A19, T23, G26, N227, and Y270) were changed to the respective residues found in mammalian brain tubulin (K19, V23, D26, H227, and F270). These mutations effectively created a Taxol binding site on yeast tubulin.
Materials and Methods
Tubulin Mutation and Purification. The five mutations were created in the β-tubulin gene of the S. cerevisiae haploid strain FY41 (25) to produce strain MGY1-tax (genotype: MATa, leu2Δ1, trp1Δ63, his4-917, URA3/ura3-52, tub2-His6-A19K-T23V-G26D-N227H-Y270F) as described (25). Mutations were verified by DNA sequencing done at the Biochemical Research Services Laboratory at the University of Kansas. The mutated strain had a doubling time equivalent to the corresponding wild-type strain MGY1. To rule out the possibility that the mutations were lethal and the lethality was masked by a secondary mutation, the mutated gene was used to create the heterozygous diploid strain MAY1210-tax (genotype: MATa/MATα, ADE2/ade2, his3/his3, leu2/leu2, LYS2/lys2, URA3/ura3/ura3, TUB2/tub2-His6-A19K-T23V-G26D-N227H-Y270F). This strain was then sporulated and the tetrads were dissected as described (25). The four resultant spores were viable with proper segregation of the metabolic markers, indicating that the mutations were not lethal. If the mutations were lethal only the two spores carrying the wild-type β-tubulin gene would have been viable.
Yeast tubulin was purified from the haploid strains MGY1 and MGY1-tax, both of which contain a His6 tag on the C terminus of β-tubulin, to apparent homogeneity by using a His6-tag-based affinity purification procedure (26). Our published procedure has been modified slightly to include 100 mM NaCl in the DE52 absorption and washing steps instead of 160 mM, and 250 mM imidazole instead of 300 mM to elute the protein from the Ni-affinity column. This procedure produces ≈5 mg of tubulin from 500 g of packed wet cells. Bovine brain tubulin was purified by two cycles of temperature-dependent polymerization (27) followed by phosphocellulose chromatography (28).
In Vitro Tubulin Assembly. Microtubule assembly reactions (typically 40–50 μl) containing 5 μM freshly cycled tubulin were performed under conditions that promote spontaneous assembly (100 mM Pipes/1 mM EGTA/1 mM MgSO4/0.5 mM GTP, pH 6.9) and under conditions that require Taxol or epothilone B for assembly (30 mM Pipes/1 mM EGTA/1 mM MgSO4/0.5 mM GTP, pH 6.9). Reactions were incubated for 30 min at 30°C, and the amount of polymerized tubulin was determined by sedimentation assay as described (22). Before sedimentation, 5-μl samples were fixed in 0.25% glutaraldehyde and negatively stained for electron microscopy. A small amount of aggregation of yeast tubulin occurred under nonassembly conditions in the absence of Taxol or epothilone B; however, microtubules were not observed. The length of individual microtubules assembled in the presence of 3 μM Taxol (n = 340) or epothilone B (n = 479) was determined on electron micrographs.
Taxol Binding to Yeast Tubulin. The stoichiometry of Taxol binding was determined by using [3H]Taxol (318–420 μCi/μmol; 1 μCi = 37 kBq) to induce microtubule assembly. After sedimentation, the polymer was washed by gently layering 100 μl of nonassembly buffer (30 mM Pipes/1 mM EGTA/1 mM MgSO4, pH 6.9) into the tube and then aspirating it, taking care not to disturb the pellet, before the amount of radioactivity associated with the polymerized tubulin was measured. Inhibition of Taxol binding by epothilone B was performed in the same manner by using 3 μM [3H]Taxol and increasing concentrations of epothilone B. Taxol binding to preformed microtubules was examined under spontaneous assembly conditions (100 mM Pipes) using 7.5 μM tubulin. After reaching steady state, [3H]Taxol was added to a final concentration of 7.5 μM. Reactions were incubated another 5 min, and the radioactivity associated with the polymerized microtubules was determined. To correct for any [3H]Taxol that may have become trapped in the microtubule pellets, identical reactions were carried out with unlabeled Taxol in the presence of [3H]H2O. After accounting for the amount of 3H incorporated into the exchangeable hydrogens on tubulin (<1% of the total 3H in the pellet in each case), the amount of trapped [3H]H2O was used to calculate the amount of [3H]Taxol trapped in the microtubule pellets (usually <1% and always <5% of the total [3H]Taxol found in the pellets).
Results
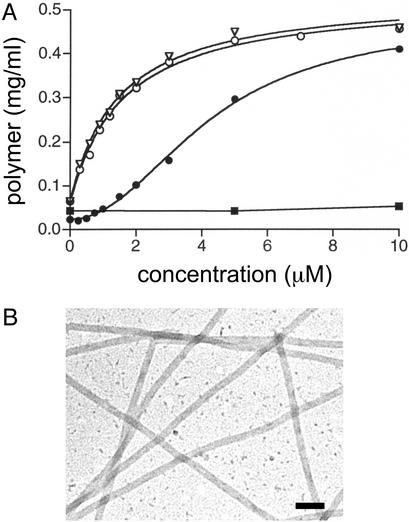
Under conditions that do not support the spontaneous assembly of 5 μM yeast tubulin, Taxol effectively promoted assembly of the mutated tubulin (Fig. 2A). The EC50 value for the stimulation of mutated yeast tubulin assembly by Taxol was 1.55 ± 0.16 μM. Electron microscopy was used to verify that the product of this Taxol-induced polymerization reaction was microtubules (Fig. 2B). Wild-type yeast tubulin did not assemble at Taxol concentrations up to 25 μM (Fig. 2 A and ref. 22). Baccatin III (Taxol lacking the C13 N-benzoylphenylisoserine side chain) is much less active than Taxol in promoting brain tubulin assembly, and 2-debenzoyl-2-(m-azidobenzoyl)baccatin III (2-m-azido baccatin III) (17) displays moderate activity. Therefore, as expected, neither of these compounds promoted the assembly of wild-type yeast tubulin at concentrations as high as 25 μM. However, at a concentration of 10 μM, baccatin III and 2-m-azido baccatin III were 15% and 28% as effective, respectively, as Taxol in promoting the assembly of mutated yeast tubulin. At a concentration of 50 μM, baccatin III and 2-m-azido baccatin III were 72% and 87% as effective, respectively, as 10 μM Taxol in promoting assembly of the mutated tubulin. This result is similar to observations made with mammalian tubulin that 2-m-azido baccatin III is more effective in promoting microtubule assembly than the relatively inactive baccatin III but is less effective than Taxol (17). The formation of microtubules induced by baccatin III and 2-m-azido baccatin III was confirmed by electron microscopy. Under the conditions used, Taxol-driven assembly of mutated yeast tubulin was more robust than with bovine tubulin (Fig. 2 A). This observation undoubtedly reflects the much lower critical concentration for yeast tubulin, which is ≈5–10% of that for mammalian brain tubulin (29, 30). The lower critical concentration allows yeast tubulin to assemble more efficiently than bovine brain tubulin at a concentration of 0.5 mg/ml at 30°C and in a low ionic-strength buffer.
Fig. 2.
Mutated yeast tubulin displays Taxol and epothilone-related activity. (A) Assembly of mutated yeast tubulin in the presence of Taxol (○) and epothilone B (▿) and of wild-type yeast (▪) and bovine brain (•) tubulin in the presence of Taxol. The reactions contained 5 μM (0.5 mg/ml) mutated tubulin from the yeast strain MGY1-tax, wild-type yeast tubulin, or bovine brain tubulin and 2.5% DMSO (see Materials and Methods). They were incubated for 30 min at 30°C under conditions that do not promote assembly in the absence of Taxol or epothilone B. The amount of polymer was determined by sedimentation assay. (B) Electron micrograph of negatively stained microtubules formed from mutated yeast tubulin in the presence of 3 μM Taxol. Reaction conditions were as described for A. (Bar = 100 nm.)
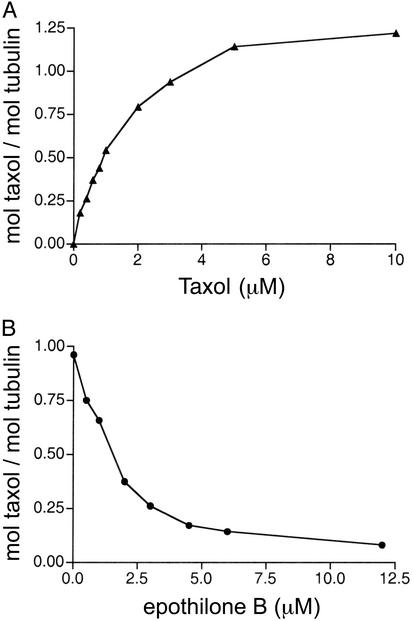
Using radiolabeled Taxol to promote tubulin assembly, we have observed the binding of close to one Taxol molecule per polymerized tubulin heterodimer (Fig. 3A), as has been observed with mammalian brain tubulin (31). When radiolabeled Taxol was added to preformed yeast microtubules (prepared under assembly-promoting conditions) at a Taxol/tubulin molar ratio of 1, the Taxol/tubulin molar binding ratio was 0.13 ± 0.01 and 1.01 ± 0.02 for microtubules assembled from wild-type and mutated tubulin, respectively.
Fig. 3.
Stoichiometry and competition by epothilone B of Taxol binding to polymerized, mutated yeast tubulin. (A) Stoichiometry of Taxol binding. (B) Inhibition of Taxol binding by epothilone B. Mutated yeast tubulin was purified from the yeast strain MGY1-tax. The reactions were performed as described for Fig. 2 A. The ratio of Taxol to tubulin was determined from the amount of [3H]Taxol bound to the tubulin polymer after sedimentation. For B, the reactions contained 3 μM [3H]Taxol, with a total of 5% DMSO.
The five mutations did not alter the interaction between yeast tubulin and epothilone B (Fig. 2 A). The EC50 value for epothilone B-induced assembly of the mutated tubulin was 1.45 ± 0.17 μM, which is in excellent agreement with values reported for assembly of both wild-type yeast and mammalian brain tubulin (22). The average length of mutated microtubules assembled in the presence of 3 μM Taxol or epothilone B was 1.54 ± 0.93 and 1.06 ± 0.68 μm, respectively (P < 0.001). As with mammalian brain microtubules, epothilone B effectively inhibited the binding of Taxol to the mutated yeast tubulin (Fig. 3B).
Discussion
Molecular modeling studies from several laboratories (9, 15–17) have provided information about the amino acid residues that may be in close contact with tubulin-bound Taxol. For example, Li et al. (15) have identified 27 residues in β-tubulin that are likely to be within 4 Å of the Taxol molecule. Snyder et al. (16) have proposed interactions between specific amino acids in brain tubulin and a “T” model of bound Taxol. We mutated five of the residues that were different in yeast tubulin to those that occur in brain tubulin and created Taxol binding in yeast tubulin. Fig. 4 illustrates the position of these five amino acids relative to tubulin-bound T-Taxol. Specifically, four of the changes restore interactions with the C3′ benzamido group of Taxol. In the T-Taxol model, the C3′ benzamido phenyl ring of Taxol is adjacent to the isopropyl group of V23 in brain tubulin. The more polar threonine at position 23 in yeast tubulin is likely to distort the positioning of the phenyl ring. According to the model, the C3′ benzamido phenyl ring is also stabilized by short contacts with methylene groups from K19, E22, and D26. In yeast tubulin, A19 and G26 would significantly reduce the number of methylene groups available to interact with the phenyl ring. The imidazole ring of H227 lies between the C2 benzoyl phenyl and C3′ benzamido phenyl rings in the T-Taxol model (16), interfering with the formation of the hydrophobically collapsed conformation of Taxol (32). H227 is also positioned between the C2 benzoyl phenyl and C3′ benzamido phenyl rings in a model of Taxol binding proposed by Li et al. (15). In addition, in a third model proposed by He et al. (17), H227 was found to be near the C2 benzoyl group. In yeast tubulin, asparagine occupies position 227 and eliminates the opportunity for π electron stacking between the imidazole and the C2 benzoyl and C3′ benzamido phenyl rings. Poor binding of the C3′ benzamido group could certainly result in decreased Taxol binding affinity. In fact, removing the benzamido group from Taxol results in a 94% loss of microtubule-promoting activity with mammalian tubulin (23, 24), even though the baccatin III portion of the molecule may provide as much as 75% of the free energy change for Taxol binding (33).
Fig. 4.
Taxol binding site on mammalian β-tubulin. The location of residues Lys-19, Val-23, Asp-26, His-227, and Phe-270 are indicated and are shown in dark gray. Labels on Taxol (gray) denote the following: I, C3′ phenyl ring; II, C3′ benzamido phenyl ring; and III, C2 benzoyl phenyl ring. Specific regions of β-tubulin that form the binding pocket are labeled, including α-helices H1, H7, H9, and H10, the β-strands B7–B10, and the B7–H9 M-loop. The structure was drawn with the modeling programs molscript (34) and raster3d (35) by using the coordinates (PDB ID code 1JFF) determined by Snyder et al. (16) for T-Taxol bound to the refined model of bovine brain tubulin (36).
The five mutations also created baccatin III and 2-m-azido baccatin III binding activity in yeast tubulin. This finding implies that the mutations increased the binding affinity of the baccatin III portion of Taxol for yeast tubulin. Of the five mutations made, two are most likely to affect binding of the baccatin III portion, N227H and Y270F. As stated previously, the imidazole ring of H227 is proposed to stack with the C2 benzoyl phenyl ring of Taxol. In addition, F270 comprises part of a hydrophobic basin in brain tubulin that is proposed to cradle the C4 acetoxy group of Taxol (16). The importance of this amino acid is evident in the fact that one Taxol-resistant cell line has been shown to contain a mutation at position 270 (10). The Y270F mutation removes the more polar tyrosine residue that likely disturbs interactions between the hydrophobic basin and the C4 acetoxy group.
Deciphering the tubulin–Taxol interactions is prerequisite to understanding the molecular mechanism that makes Taxol a successful antimitotic agent. Furthermore, knowledge of the binding site shared by Taxol and other structurally diverse antimitotic compounds that bind at the Taxol site holds great potential for the rational design of antitumor agents that exploit this region of the tubulin molecule. Despite significant effort, the orientation and conformation of Taxol bound to tubulin has remained unsolved. For example, in several recently published studies different conformations that can satisfy most of the available biochemical and structure–activity relationship data were proposed (9, 14–17). After changing 5 of the 124 amino acids that are different between yeast and mammalian brain β-tubulin, the mutated yeast tubulin displayed Taxol-related biological activity similar to that seen with mammalian brain tubulin, whereas wild-type yeast tubulin displayed none. This result provides direct experimental support for the involvement of specific amino acids in Taxol binding and activity. Although Taxol and the epothilones compete for binding to microtubules, and attempts have been made to find a common pharmacophore for the two compounds (9, 17), the epothilones bind equally well to wild-type and the mutated yeast tubulin. Thus, the epothilones can tolerate the changes made at the five positions in β-tubulin. This result is interesting because in two models for the interaction of epothilone with tubulin, F270 (9) and H227 (17) are positioned near the thiazole ring of epothilone.
Further mutagenesis studies are necessary to determine the relative importance of each of the five mutated residues in Taxol binding. In conjunction with molecular modeling, mutagenesis can also be used to identify other residues that make important contacts with Taxol. In addition, a more comprehensive mutational analysis of the Taxol binding region will provide a thorough understanding of the interactions required for Taxol, the epothilones, and other compounds to bind to this region of β-tubulin. Therefore, this engineered yeast tubulin represents a critical advance in understanding the fundamental protein–drug interactions at the Taxol binding site. The mutated tubulin, containing only a single β-isotype and possessing full Taxol binding activity, provides an ideal system in which to determine the importance of all of the putative interactions between tubulin and Taxol, and between tubulin and other agents that bind at the Taxol site.
Acknowledgments
We thank Dr. C. A. Dougherty for her generous help with tetrad dissection and analysis, and Brandon Turunen for the synthesis of 2-debenzoyl-2-(m-azidobenzoyl)baccatin III. This work was supported by the University of Kansas and National Institutes of Health Grants CA79641 and CA82801. C.J.B. was a recipient of National Institutes of Health Predoctoral Traineeship GM08545.
Abbreviations: T-Taxol, Taxol in the T-conformation; baccatin III, Taxol lacking the C13 N-benzoylphenylisoserine side chain; 2-m-azido baccatin III, 2-debenzoyl-2-(m-azidobenzoyl)baccatin III.
References
- 1.Jordan, M. A. (2002) Curr. Med. Chem. Anti-Cancer Agents 2, 1–17. [DOI] [PubMed] [Google Scholar]
- 2.Jiménez-Barbero, J., Amat-Guerri, F. & Snyder, J. P. (2002) Curr. Med. Chem. Anti-Cancer Agents 2, 91–122. [DOI] [PubMed] [Google Scholar]
- 3.Dasgupta, D., Park, H., Harriman, G. C., Georg, G. I. & Himes, R. H. (1994) J. Med. Chem. 37, 2976–2980. [DOI] [PubMed] [Google Scholar]
- 4.Combeau, C., Commerçon, A., Mioskowski, C., Rousseau, B., Aubert, F. & Goeldner, M. (1994) Biochemistry 33, 6676–6683. [DOI] [PubMed] [Google Scholar]
- 5.Rao, S., Krauss, N. E., Heerding, J. M., Swindell, C. S., Ringel, I., Orr, G. A. & Horwitz, S. B. (1994) J. Biol. Chem. 269, 3132–3134. [PubMed] [Google Scholar]
- 6.Rao, S., Orr, G. A., Chaudhary, A. G., Kingston, D. G. & Horwitz, S. B. (1995) J. Biol. Chem. 270, 20235–20238. [DOI] [PubMed] [Google Scholar]
- 7.Rao, S., He, L., Chakravarty, S., Ojima, I., Orr, G. A. & Horwitz, S. B. (1999) J. Biol. Chem. 274, 37990–37994. [DOI] [PubMed] [Google Scholar]
- 8.Kingston, D. G., Jagtap, P. G., Yuan, H. & Samala, L. (2002) Prog. Chem. Org. Nat. Prod. 84, 53–225. [DOI] [PubMed] [Google Scholar]
- 9.Giannakakou, P., Gussio, R., Nogales, E., Downing, K. H., Zaharevitz, D., Bollbuck, B., Poy, G., Sackett, D., Nicolaou, K. C. & Fojo, T. (2000) Proc. Natl. Acad. Sci. USA 97, 2904–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giannakakou, P., Sackett, D. L., Kang, Y. K., Zhan, Z., Buters, J. T., Fojo, T. & Poruchynsky, M. S. (1997) J. Biol. Chem. 272, 17118–17125. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-Garay, M. L., Chang, L., Blade, K., Menick, D. R. & Cabral, F. (1999) J. Biol. Chem. 274, 23875–23882. [DOI] [PubMed] [Google Scholar]
- 12.He, L., Yang, C.-H. H. & Horwitz, S. B. (2001) Mol. Cancer Ther. 1, 3–10. [PubMed] [Google Scholar]
- 13.Nogales, E., Wolf, S. G. & Downing, K. H. (1998) Nature 391, 199–203. [DOI] [PubMed] [Google Scholar]
- 14.Ojima, I., Chakravarty, S., Inoue, T., Lin, S., He, L., Horwitz, S. B., Kuduk, S. D. & Danishefsky, S. J. (1999) Proc. Natl. Acad. Sci. USA 96, 4256–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, Y., Poliks, B., Cegelski, L., Poliks, M., Gryczynski, Z., Piszczek, G., Jagtap, P. G., Studelska, D. R., Kingston, D. G., Schaefer, J. & Bane, S. (2000) Biochemistry 39, 281–291. [DOI] [PubMed] [Google Scholar]
- 16.Snyder, J. P., Nettles, J. H., Cornett, B., Downing, K. H. & Nogales, E. (2001) Proc. Natl. Acad. Sci. USA 98, 5312–5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, L., Jagtap, P. G., Kingston, D. G., Shen, H. J., Orr, G. A. & Horwitz, S. B. (2000) Biochemistry 39, 3972–3978. [DOI] [PubMed] [Google Scholar]
- 18.Hamel, E., Sackett, D. L., Vourloumis, D. & Nicolaou, K. C. (1999) Biochemistry 38, 5490–5498. [DOI] [PubMed] [Google Scholar]
- 19.Kowalski, R. J., Giannakakou, P., Gunasekera, S. P., Longley, R. E., Day, B. W. & Hamel, E. (1997) Mol. Pharmacol. 52, 613–622. [PubMed] [Google Scholar]
- 20.Kowalski, R. J., Giannakakou, P. & Hamel, E. (1997) J. Biol. Chem. 272, 2534–2541. [DOI] [PubMed] [Google Scholar]
- 21.Barnes, G., Louie, K. A. & Botstein, D. (1992) Mol. Biol. Cell 3, 29–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bode, C. J., Gupta, M. L., Jr., Reiff, E. A., Suprenant, K. A., Georg, G. I. & Himes, R. H. (2002) Biochemistry 41, 3870–3874. [DOI] [PubMed] [Google Scholar]
- 23.Gueritte-Voegelein, F., Guenard, D., Lavelle, F., Le Goff, M. T., Mangatal, L. & Potier, P. (1991) J. Med. Chem. 34, 992–998. [DOI] [PubMed] [Google Scholar]
- 24.Swindell, C. S., Krauss, N. E., Horwitz, S. B. & Ringel, I. (1991) J. Med. Chem. 34, 1176–1184. [DOI] [PubMed] [Google Scholar]
- 25.Gupta, M. L., Jr., Bode, C. J., Dougherty, C. A., Marquez, R. T. & Himes, R. H. (2001) Cell Motil. Cytoskeleton 49, 67–77. [DOI] [PubMed] [Google Scholar]
- 26.Gupta, M. L., Jr., Bode, C. J., Thrower, D. A., Pearson, C. G., Suprenant, K. A., Bloom, K. S. & Himes, R. H. (2002) Mol. Biol. Cell 13, 2919–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tiwari, S. C. & Suprenant, K. A. (1993) Anal. Biochem. 215, 96–103. [DOI] [PubMed] [Google Scholar]
- 28.Algaier, J. & Himes, R. H. (1988) Biochim. Biophys. Acta 954, 235–243. [DOI] [PubMed] [Google Scholar]
- 29.Davis, A., Sage, C. R., Wilson, L. & Farrell, K. W. (1993) Biochemistry 32, 8823–8835. [DOI] [PubMed] [Google Scholar]
- 30.Bode, C. J., Gupta, M. L., Suprenant, K. A. & Himes, R. H. (2003) EMBO Rep. 4, 94–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parness, J. & Horwitz, S. B. (1981) J. Cell Biol. 91, 479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vander Velde, D. G., Georg, G. I., Grunewald, G. L., Gunn, K. & Mitscher, L. A. (1993) J. Am. Chem. Soc. 115, 11650–11651. [Google Scholar]
- 33.Andreu, J. M. & Barasoain, I. (2001) Biochemistry 40, 11975–11984. [DOI] [PubMed] [Google Scholar]
- 34.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 35.Merrit, E. A. & Murphy, M. E. P. (1994) Acta Crystallogr. D 50, 869–873. [DOI] [PubMed] [Google Scholar]
- 36.Lowe, J., Li, H., Downing, K. H. & Nogales, E. (2001) J. Mol. Biol. 313, 1045–1057. [DOI] [PubMed] [Google Scholar]