Abstract
The atomic resolution structure of fd coat protein determined by solid-state NMR spectroscopy of magnetically aligned filamentous bacteriophage particles differs from that previously determined by x-ray fiber diffraction. Most notably, the 50-residue protein is not a single curved helix, but rather is a nearly ideal straight helix between residues 7 and 38, where there is a distinct kink, and then a straight helix with a different orientation between residues 39 and 49. Residues 1–5 have been shown to be mobile and unstructured, and proline 6 terminates the helix. The structure of the coat protein in virus particles, in combination with the structure of the membrane-bound form of the same protein in bilayers, also recently determined by solid-state NMR spectroscopy, provides insight into the viral assembly process. In addition to their roles in molecular biology and biotechnology, the filamentous bacteriophages continue to serve as model systems for the development of experimental methods for determining the structures of proteins in biological supramolecular assemblies. New NMR results include the complete sequential assignment of the two-dimensional polarization inversion spin-exchange at the magic angle spectrum of a uniformly 15N-labeled 50-residue protein in a 1.6 × 107 Da particle in solution, and the calculation of the three-dimensional structure of the protein from orientational restraints with an accuracy equivalent to an rms deviation of ≈1Å.
Keywords: protein structure, virus structure, polarity index slant angle wheel, dipolar wave, supramolecular assembly
Filamentous bacteriophages have been studied in the context of a wide range of prokaryotic biology (1, 2, 3), and they have essential roles in many laboratory procedures of molecular biology and biotechnology, including the cloning and sequencing of DNA, and the generation and screening of peptide libraries (1, 4). fd, M13, and f1 are very similar inoviruses with their single-stranded circular DNA packaged inside a cylinder that consists almost entirely of ≈2,700 copies of structurally identical coat protein subunits. Although filamentous bacteriophages do not have a membrane, the major coat protein (pVIII) is a membrane protein in the course of the viral lifecycle because it is stored in the bacterial membrane after synthesis and processing before the assembly of new virus particles. Filamentous bacteriophage coat proteins are popular systems for the development of NMR methods applicable to helical membrane proteins in micelles (5–10). The three-dimensional structure of the membrane-bound form of fd coat protein, which has been determined in micelles by solution NMR (11), and in bilayers by solid-state NMR (12), is that of a typical membrane protein with a hydrophobic transmembrane helix connected by a short loop to an amphipathic in-plane helix that interacts with the lipid head groups. During the viral assembly process, the tertiary structure of the protein changes dramatically as it undergoes the transition from a membrane protein to the principal structural (and DNA-binding) protein of bacteriophage particles. There have been numerous x-ray fiber diffraction studies of filamentous bacteriophages (3, 13–17), and although the resulting protein structures have relatively low resolution, these studies provide crucial information about the symmetry and packing of the coat protein subunits (15, 18). Other spectroscopic investigations (19–21), as well as our earlier solid-state NMR studies (22–26), demonstrate that the coat protein is nearly all α-helix, its positively charged C-terminal residues are buried on the interior of the particle where they interact with the DNA, and the N-terminal residues are mobile (26) and exposed on the exterior of the virus particles.
As is the case for most proteins associated with biological supramolecular assemblies, determinations of the structures of fd coat protein in virus particles and in bilayers are problematic for the x-ray crystallography and solution NMR methods that work well with globular proteins. Fortunately, NMR studies of proteins in large assemblies are formidable only because of the correlation time problem, because crystals are not necessary, and no other chemical or physical properties of the polypeptides interfere with NMR experiments. The experimental methods, instruments, and theories of solid-state NMR developed for crystalline and amorphous solids are applicable to samples where the molecules of interest are immobile on the millisecond time scales of the operative chemical-shift and dipole–dipole coupling spin interactions. With the exception of a few residues at the N terminus, which have narrow isotropic resonances, there is no evidence of motional averaging of backbone sites (27), although many of the side chains undergo large amplitude motions, in fd coat protein in bacteriophage particles (26, 28).
Because solid-state NMR procedures can be applied in ways that lead to the selective averaging and temporal separation of the evolution of the anisotropic spin interactions, spectra can be obtained that provide directly interpretable orientational restraints as input for structure determination (29). The earliest NMR results on single crystals (30, 31) demonstrated that high spectral resolution can be achieved through a combination of radio frequency irradiations and sample alignment, and that the observable spectral parameters reflect the orientations of molecular sites with respect to the direction of the applied magnetic field. Structure determination of proteins in aligned samples by solid-state NMR (24) takes advantage of these spectroscopic principles, as initially demonstrated with uniaxially oriented polymer fibers (32), and subsequently with magnetically aligned filamentous bacteriophage particles (25), mechanically aligned membrane proteins in bilayers (33), and magnetically aligned membrane proteins in bicelles (34). The structures of several membrane-associated peptides (35) and proteins (12, 36, 37) have been determined by solid-state NMR of aligned bilayer samples. In this article, we describe the structure determination of fd coat protein in magnetically aligned filamentous bacteriophage particles in solution by solid-state NMR spectroscopy.
Materials and Methods
Sample Preparation. Uniformly 15N-labeled samples of Y21M fd bacteriophage were obtained by infecting Escherichia coli cells grown in M9 media containing 15N-enriched ammonium sulfate (Cambridge Isotope Laboratories, Cambridge, MA). Cells were inoculated at mid-logarithmic phase (OD550 = 0.3) with virus stock at 10:1 multiplicity, based on infectivity determined by plaque assay. Selectively labeled samples were obtained by using M9 media supplemented with one 15N-labeled amino acid (Cambridge Isotopes Laboratories) and the other amino acids, but not proline, in natural isotopic abundance (Sigma). The isotopic labeling was verified with solution NMR spectra of the coat proteins in micelles. Virus particles were isolated from the growth media, after removal of bacteria by precipitation with polyethylene glycol, and then purified by cesium chloride gradient ultracentrifugation. The samples for the solid-state NMR experiments were 5-mm o.d. thin-wall glass tubes containing 180 μl of 60 mg/ml solutions of bacteriophage in 5 mM sodium borate buffer, pH 8.0, and 0.1 mM sodium azide. Under these conditions, the bacteriophage particles align with their long axis parallel to the direction of the applied magnetic field.
NMR Spectroscopy. Two-dimensional polarization inversion spin-exchange at the magic angle (PISEMA) spectra (38, 39) were obtained on a home-built spectrometer with a wide-bore Magnex (Oxford) 550/89 magnet with a field strength corresponding to a 1H-resonance frequency of 550 MHz and a 15N-resonance frequency of 55.7 MHz. The sample tubes containing the isotopically labeled bacteriophage particles were placed horizontally within the solenoid coil of a home-built probe double-tuned for the 1H- and 15N-resonance frequencies. The temperature of the sample was controlled at 65°C. Typically, 256 transients were acquired for 64 t1 points in the indirect dimension in these experiments. The initial spin-lock cross-polarization time was 1 ms. Decoupling field strengths were between 60 and 70 kHz. The data were processed with the program FELIX (Accelrys, San Diego) using sine bell apodization and zero filling in the chemical-shift dimension and linear prediction followed by sine bell apodization in the indirect heteronuclear dipolar-coupling dimension.
Structure Determination. The resonances were assigned by using a modified version of the shotgun NMR approach (12). The two-dimensional PISEMA spectra from one uniformly 15N-labeled and several selectively 15N-labeled samples were analyzed with a combination of polarity index slant angle (PISA) wheels (40, 41) and dipolar waves (42). A sinusoid with a period of 3.6 residues was fit to the experimental 1H–15N dipolar couplings as a function of residue number to characterize the length and orientation of each helical segment relative to the direction of the applied magnetic field (42). Based on the assumption that the H–N bond vectors in an ideal α-helix are tilted 15.8° away from the long axis of the helix, the fit of the properly parameterized expression yields values for the tilt and rotation of the helix and serves as an index of the regularity of the helix.
The atomic resolution structure was calculated through structural fitting (43) of the 1H–15N dipolar coupling and 15N-chemical-shift frequencies measured from the spectrum in Fig. 1 and listed in Table 1, which is equivalent to the direct calculation of the structure because all of the resonances are assigned to specific residues (43). The accuracy of the structure was evaluated with a statistical analysis that takes into account both experimental errors in the measurement of the dipolar coupling and chemical-shift frequencies and uncertainties in the magnitudes of the principal values of the chemical-shift tensors. For every calculation of the torsion angles between two residues based on the pairs of frequencies associated with their resonances, the chemical-shift tensor components were allowed to vary within ±5 ppm relative to the canonical values (σ11 = 76 ppm, σ22 = 89 ppm, and σ33 = 229 ppm, relative to solid [15N]ammonium sulfate at 26.8 ppm). The experimental accuracy for the determination of the frequencies was conservatively estimated to be ±100 Hz in both frequency dimensions. To pick up all torsion angles consistent with the resulting ranges in frequencies and principal values, during the fitting the initial values for the angles φ and Ψ were randomized within ±10° relative to their ideal values, φ0 = –65o, Ψ0 = –40°. The mean rms deviation of the 100 calculated structures was 0.98 Å relative to their average structure.
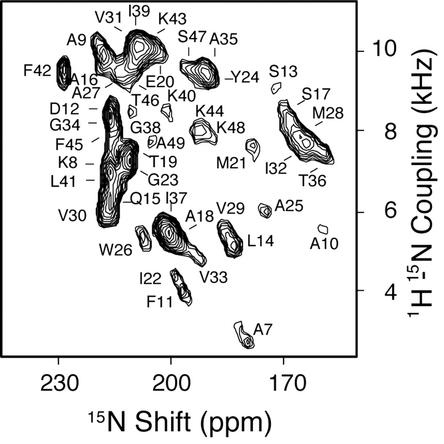
Fig. 1.
Experimental two-dimensional 1H/15N PISEMA spectrum of uniformly 15N-labeled Y21M fd bacteriophage particles aligned in the magnetic field of the NMR spectrometer. The assignments of the amide resonances are indicated. Each resonance is characterized by the orientationally dependent frequencies associated with the 1H–15N heteronuclear dipolar coupling and 15N-chemical-shift interactions that are listed in Table 1.
Table 1. Experimental frequencies measured from the PISEMA spectrum in Fig. 1.
| Residue no. | 15N shift, ppm | 1H-15N coupling, kHz |
|---|---|---|
| 7 | 179.4 | 2.7 |
| 8 | 217.0 | 7.1 |
| 9 | 219.6 | 10.1 |
| 10 | 159.5 | 5.6 |
| 11 | 197.2 | 4.0 |
| 12 | 216.7 | 8.6 |
| 13 | 172.3 | 9.1 |
| 14 | 183.5 | 5.1 |
| 15 | 216.5 | 6.3 |
| 16 | 217.9 | 9.7 |
| 17 | 169.8 | 8.3 |
| 18 | 200.0 | 5.3 |
| 19 | 211.5 | 7.5 |
| 20 | 203.8 | 9.9 |
| 21 | 179.0 | 7.6 |
| 22 | 199.0 | 4.3 |
| 23 | 211.8 | 7.1 |
| 24 | 190.4 | 9.3 |
| 25 | 175.0 | 6.0 |
| 26 | 207.7 | 5.4 |
| 27 | 214.6 | 9.5 |
| 28 | 166.2 | 7.9 |
| 29 | 185.3 | 5.4 |
| 30 | 217.0 | 6.0 |
| 31 | 210.0 | 9.9 |
| 32 | 164.0 | 7.7 |
| 33 | 193.2 | 4.9 |
| 34 | 216.0 | 8.4 |
| 35 | 192.0 | 9.6 |
| 36 | 159.6 | 7.5 |
| 37 | 202.0 | 5.6 |
| 38 | 210.8 | 8.5 |
| 39 | 209.2 | 10.3 |
| 40 | 201.5 | 8.5 |
| 41 | 216.6 | 6.9 |
| 42 | 229.0 | 9.5 |
| 43 | 206.3 | 10.0 |
| 44 | 192.5 | 8.0 |
| 45 | 215.7 | 8.0 |
| 46 | 210.5 | 9.6 |
| 47 | 196.2 | 9.6 |
| 48 | 190.2 | 7.9 |
| 49 | 205.4 | 7.7 |
Capsid Model. A model of the virus capsid was constructed by using the structure of the coat protein obtained from the NMR data and the known symmetry of the particles (16, 17). Because the NMR data are ambiguous with respect to the rotation of the protein about the alignment axis, initial backbone models of the virus capsid were built with several different rotations of the molecule about the z axis, consistent with the C terminus lysine side chains on the inside pointing toward the DNA and the N terminus pointing away from the particle. Side chains were added to the backbone structure, using the program SCRWL (side-chain placement with a rotamer library; ref. 44) and the capsid model was energy minimized. The minimum energy configuration obtained by using this algorithm was selected as the best model because the vast majority of models produced clashes that were not removable.
Results
The coat protein subunits are immobilized by, and aligned along with, the filamentous bacteriophage particles parallel to the direction of the applied magnetic field. Because all of the coat protein subunits have the identical structure and are arranged symmetrically in the virus particles, only by rotation and translation, each amide site in the polypeptide backbone contributes one resonance to the NMR spectra of 15N-labeled samples (25, 32). The two-dimensional PISEMA spectrum of a uniformly 15N-labeled sample of Y21M fd bacteriophage shown in Fig. 1 contains resonances from all of the structured amide sites of the coat protein. Complementary data from selectively 15N-labeled samples (Fig. 2) provide assurances about the attributions of partially overlapped resonances, and the information needed for simultaneously assigning resonances and determining the protein structure through the shotgun NMR (12) and structural fitting (43) approaches.
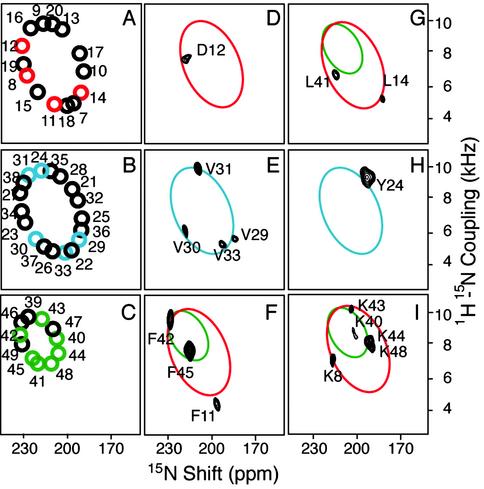
Fig. 2.
(A–C) Representations of ideal PISA wheels for three regions of the coat protein. The colored circles represent the resonances present in the experimental spectra of selectively labeled fd virions in D–I with the labeled amino acids identified by the single-letter code. Superimposed on the experimental data are colored ellipses corresponding to the ideal wheels.
Samples of Y21M fd bacteriophage give better resolved solid-state NMR spectra than those of wild-type fd (45). The association of one resonance with each amide site is unambiguous in the PISEMA spectra of the selectively 15N-labeled samples in Fig. 2. Each resonance is characterized by orientationally dependent frequencies from the 1H–15N heteronuclear dipole–dipole coupling and 15N-chemical-shift interactions, which, in combination with the magnitudes and orientations of the spin-interaction tensors in the molecular frame of the peptide bond (38, 46), provide input for structure determination (24). In aligned samples, the orientations of the spin-interaction tensors effect the mapping of the protein structure onto the spectra. Simple patterns are observed in the spectra for regular secondary structures of α-helix (40, 41) and β-sheet (47). The spectrum in Fig. 1 has a distinct “wheel-like” pattern of resonances because the helix axes are aligned approximately parallel to the direction of the applied magnetic field.
PISA wheel patterns are sensitive indicators of the tilt (slant angle) of the helix because the dipolar coupling and chemical-shift frequencies reflect the orientations of the amide groups relative to the direction of the magnetic field. However, spectra for different rotations (polarities) of the helix vary only in their resonance assignments, whose patterns mirror exactly the polarity of the corresponding helical wheel. When a wheel-like pattern of resonances is observed in a PISEMA spectrum, no resonance assignments are needed to determine the tilt of a helix, and only a single resonance assignment or the patterns of resonances from several labeled sites, as obtained from a selectively 15N-labeled sample, is sufficient to determine helix polarity because sequential resonance assignments are intimately linked with the spectral features of PISA wheels (12, 40, 48, 49). Fig. 2 illustrates how PISA wheels simplify and accelerate the resonance assignment and structure determination process. Fig. 2 A–C represent ideal PISA wheels for three segments of the polypeptide. Ellipses that match the color and tilt angles of the PISA wheels for the protein segments are superimposed on the experimental data and demonstrate that all of the resonances fall on, or close to, the wheels. The asymmetric distribution of the four valine resonances on the wheel in Fig. 2E is sufficient to index the wheel, determine the helix rotation, and assign resonances from other residues in the helix, including those present only in the spectrum of the uniformly 15N-labeled sample. These conclusions are reiterated in the spectra from other selectively 15N-labeled samples. Representative one-dimensional spectral slices from the two-dimensional experimental PISEMA spectra of uniformly and selectively 15N-labeled Y21M fd bacteriophage particles shown in Figs. 1 and 2 are shown in Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org.
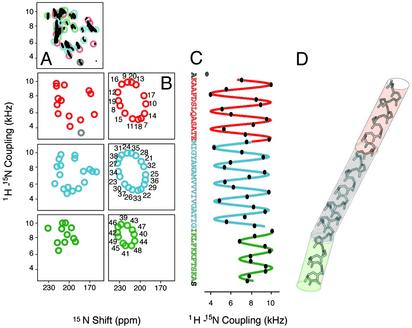
Fig. 3 summarizes the structural analysis of the frequencies measured from the spectrum in Fig. 1 and are listed in Table 1. Although multiple samples are used in the qualitative assignment process, all of the frequencies actually used for structure determination are measured from a single spectrum, avoiding potential problems due to variations among samples and conditions. For clarity of presentation, and to focus on the principal structural features of the protein, its sequence is divided into four segments: residues 1–6 are not shown, because residues 1–5 are mobile (26) and proline 6 terminates the helix; residues 7–20 (red) constitute the amphipathic helix that lies on top of bilayers in the membrane-bound form of the protein; residues 21–38 (blue) are hydrophobic and are part of the transmembrane helix in the membrane-bound form of the protein; and the side chains of residues 39–49 (green) provide the positive charges for DNA interactions in the virus particles. Fig. 3A Left sorts the experimental PISA wheels by segment for comparison to the corresponding ideal PISA wheels in the adjacent column (Fig. 3B). The residues at the ends of the helix, Ala-7 and Ser-50, are gray. The dipolar coupling frequency measured for Ala-7 does not fit well to a wave, indicating some distortion at the start of the helix, and the resonance for the C-terminal Ser residue has not been identified in the spectrum.
Fig. 3.
(A) Representations of the experimental PISEMA spectrum of uniformly and selectively 15N-labeled Y21M fd bacteriophage particles. The midpoint of each circle represents the pair of frequencies associated with a resonance in the spectrum shown in Fig. 1 and the top of column, and listed in Table 1. (B) Ideal PISA wheels corresponding to the experimental data in A. (C) Dipolar waves fit to the experimental dipolar coupling frequencies as a function of residue number. The frequencies are measured from the resonances in the spectrum in Fig. 1, listed in Table 1, and represented by the circles in A. (D) Three-dimensional structure of Y21M fd coat protein in bacteriophage particles, whose coordinates are deposited as PDB ID code 1NH4, superimposed on colored tubes with orientations determined from the fits to the dipolar waves in C.
Dipolar waves for residues 8–49 are shown in Fig. 3C, where sine waves with the 3.6-residues-per-turn periodicity characteristic of a α-helix are fit to the experimental dipolar-coupling frequencies as a function of residue number (42). A break in the helix was anticipated near residue 21, the site of the turn connecting the hydrophobic and amphipathic helices in the membrane-bound form of the protein (11, 12). Remarkably, both the PISA wheels (Fig. 3B), and especially the dipolar waves (Fig. 3C), indicate that residues 8–38 are in a single continuous helix, with no evidence of a kink, curvature, or other distortion. In contrast, there is an unexpected kink near residue 39. The ideal and experimental PISA wheels corresponding to residues 21–38 and 39–49 reflect helices with different tilt angles, and the dipolar waves demonstrate that the helix abruptly changes direction near residue 39. Both the qualitative PISA wheels and the quantitative dipolar waves indicate that all segments of the protein form nearly ideal α-helices. The discrepancies between the experimental and ideal PISA wheels in Fig. 3 are typical of those due to local variations of chemical-shift tensors. The R factors for the fits of the experimental data to sine waves are less than the experimental errors for the measurements of the dipolar-coupling frequencies.
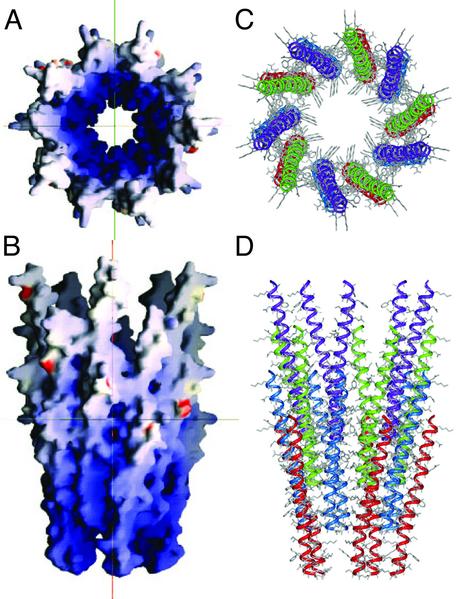
The backbone structure of the coat protein obtained by structural fitting (43) to the data in Fig. 1 and Table 1 is shown in Fig. 3D. The colored cylinders represent the orientations of the three segments of the molecule obtained from the dipolar waves. A conservative analysis of the accuracy of the structure determination, as described in Materials and Methods, indicates that it is equivalent to an rms deviation of ≈1 Å. A model of the virus capsid was constructed by placing the coat protein monomers in a five-start helix with a two-fold screw axis with a 16-Å pitch and –33.23° rotation (3, 16, 17), and is presented in Fig. 4. Sections of the capsid were built containing four interleaved layers of the coat protein with rotations about the z axis varying over a range of 180° (in 1° increments), which is consistent with the C terminus on the inside of the capsid, and with a diameter varying by a few angstroms about the literature value of 65 Å. Using the program SCRWL (44), side chains were added to the models with the backbone atoms of the capsid-held fix. This method begins by placing side chains into the model in their most favorable rotamer and then relieving sterical clashes by varying the preferred rotameric configurations of those side chains to lower the overall energy of the model. The algorithm then performs a brief energy minimization by using a hard repulsive potential to remove steric clashes. Using the parameters of the AMBER 4.1 potential, the final energy of the models are compared and the lowest energy configuration was chosen as the final model of the capsid. This calculation converges to a single orientation of the protein molecule. A section of the capsid is shown in Fig. 4. In Fig. 4 A and B, the molecular surface of the capsid is colored according to the electrostatic potential calculated with the program GRASP (50). Fig. 4A is a view from the top along the virion axis and Fig. 4B is a side view. Figs. 4 C and D present the same views of the capsid as rendered by the program INSIGHT II (Accelrys).
Fig. 4.
Model of a section of the Y21M fd filamentous bacteriophage capsid built from the coat protein subunit structure, which was determined by solid-state NMR spectroscopy. The symmetry was derived from fiber diffraction studies. (A and B) Representations of the electrostatic potential on the molecular surface of the virus (A is a bottom view and B is a side view along the virus axis) obtained by using the program grasp (57). (C and D) Views of the capsid structure showing the arrangement of the coat proteins in pentamers and further assembly of the 2-fold helical structure obtained by using the program insight ii (Accelrys).
Discussion
The structure of fd coat protein shown in Figs. 3D and 5A results from a direct fit to the experimental data. Protein structures based on ideal α-helices represented by tubes (Fig. 3D) and ribbons are also consistent with the experimental PISA wheels and well fit dipolar waves. In contrast, the fiber-diffraction structure results from model fitting of data with a resolution of ≈7 Å (16, 17). However, the differences between the protein structures determined by NMR spectroscopy and x-ray fiber diffraction cannot be explained on the basis of differences in resolution alone. The structure of the coat protein determined by x-ray fiber diffraction has been described as a single gently curving α-helix (16), a gently curved α-helix (17) oriented roughly parallel to the virion axis (14, 15), and as having a 20° tilt with respect to the same axis (16). Raman spectroscopy data suggest an average tilt of the helix between 13° and 20° (51). The solid-state NMR data give very accurate values based on dipolar couplings for the tilt of the helical regions of the coat protein, with residues 8–38 having a 23° tilt and the C-terminal region, starting at residue 40, having a somewhat smaller tilt, of 18° with respect to the filament (and magnetic field) axis. The difference is patent in the dipolar waves in Fig. 3D, from which the tilt angles are calculated. The PISA wheel analysis and the structural fitting of the spectra confirm these findings.
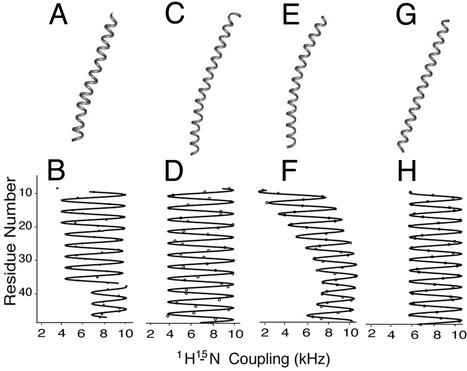
Fig. 5.
(A) Ribbon rendering of the structure of fd coat protein determined by NMR spectroscopy and shown in Fig. 3D. (C) Ribbon rendering of the x-ray fiber diffraction structure of fd coat protein (PDB ID code 1IFI). (E) Ideal gently curved helix. (G) Ideal straight tilted helix. (B, D, F, and H) The corresponding dipolar waves. (B) The fit to the experimental NMR data in Fig. 4C. (D) Calculated from the x-ray fiber diffraction structure. (F and H) Simulations for the corresponding ideal helices.
In comparisons between the solid-state NMR structure (PDB ID code 1NH4) and the fiber-diffraction structure of fd (PDB ID code 1IFI), a small difference in the length of the individual coat protein subunits is apparent. Ideal α-helices have a periodicity very close to 3.6 residues per turn (52). The fiber diffraction structure has a periodicity of 3.55 residues per turn, whereas the solid-state NMR data have 3.68 residues per turn. These small variations in periodicity might reflect slight differences in the peptide plane geometries and bond lengths used to calculate the helical structures, and result in the overall differences in length of the protein. A simulated dipolar wave fit to 1IFI is shown in Fig. 5D, and compared with dipolar waves corresponding to the NMR structure in Fig. 5B, and to ideal helices: in Fig. 5F, a straight, 45-residue helix tilted 20° with respect to the z axis; and in Fig. 5H, a slightly curved 45-residue helix. The ribbon representation for each structure is shown in Fig. 5 A, C, E, and G. The dipolar waves used to determine the structure by NMR (Fig. 5B) are clearly different from that derived from the fiber diffraction structure [Fig. 5D, because the coordinates from 1IFI give rise to a dipolar wave that appears to correspond to a straight helix with a tilt of >23o, and neither is consistent with the dipolar wave for a curved helix (Fig. 5F) or a straight helix with a 20° tilt (Fig. 5H)].
The N-terminal amphipathic helix (residues 7–20), which lies on the surface of lipid bilayers (16, 11, 12), undergoes a large rearrangement during the assembly process, which is equivalent to a rigid body rotation of ≈90°, after which it is seamlessly integrated, along with the residues that act as a hinge, into a helix that starts immediately after Pro 6 and continues through the C terminus in the bacteriophage particles. The helix is straight, except for a kink near residue 39, which enables the C-terminal residues (39–49) to have a somewhat different orientation than residues 8–38. The change in orientation at residue 39 may be necessary to optimize interactions of the positive charges of the side chains of the C-terminal residues with the DNA packed inside. Alternatively, the length of the C-terminal region is compatible with the rise of ≈16 Å per turn of the 2-fold screw axis symmetry observed for the virus capsid, considering the α-helical rise of 5.4 Å per turn, or 16.5 Å for 11 residues (52). The same kink is present in the structures of the membrane-bound form of the coat protein determined in micelles (11) and bilayers (12), therefore it may satisfy structural requirements of the membrane-bound form of the protein unrelated to its roles in the bacteriophage particles.
Depending on the extent of hydration, the diameter of the capsid as determined by x-ray fiber diffraction varies between 55 Å and 65 Å (53), and most studies report external diameters of 60 Å (17) or 65 Å (16). In our model-building approach to the virus particles based on the atomic resolution structure of the coat protein subunits, an external diameter of 67 Å was the most energetically favorable after the side chains were added. The model of the virus capsid shown in Fig. 4, as well as those derived from the x-ray fiber diffraction results, have similar side-chain interactions among the coat protein subunits and between the C-terminal residues and the DNA.
Recent mutational analysis (54) has shown that only a small group of residues is absolutely required for virus viability, including residues Ile-39, Leu-41, Phe-42, and Phe-45. This finding is supported by hydrogen-exchange experiments (data not shown) where two-dimensional PISEMA spectra were obtained after resuspension of uniformly 15N-labeled bacteriophage particles in D2O at pH 8.0. The resonances corresponding to these four residues are present in spectra obtained on samples exposed to D2O for as long as 6 months, whereas nearly all other resonances disappear within a few hours of resuspension of the sample in D2O. This result suggests that the inter-subunit interactions involving these residues are very stable, and reflect the resistance of the virus particles to extremes of temperature, pH, and ionic strength (55). Three of the C-terminal lysine residues (40, 43, and 44) are also highly conserved, and their side chains point toward the center of the capsid and interact with the DNA. The electrostatic surface of the capsid in Fig. 4 shows the prevalence of positively charged residues on the interior and the location of acidic residues on the exterior.
Solid-state NMR spectroscopy of aligned samples yields atomic resolution structures of proteins in assemblies that cannot be crystallized and are too large to reorient rapidly in solution. Previous examples were membrane proteins in bilayers aligned between glass plates (12, 35–37), and here we present the structure of the coat protein in virus particles. The structure of the membrane-bound form of fd coat protein was determined in fully hydrated lipid bilayers (12) and the solutions of filamentous bacteriophage particles are infectious, thus the biological relevance of the samples is very high. Only two-dimensional NMR experiments were needed to determine the structure of the 50-residue coat protein; however, three-dimensional NMR spectra with much greater spectral resolution have been obtained (56), indicating that it is feasible to determine the structures of substantially larger polypeptides with this approach. Further, it has been shown that it is possible to significantly enhance the sensitivity of these experiments by performing them on bacteriophage samples at low temperatures and with special techniques like dynamic nuclear polarization (57). There are substantial benefits to performing the experiments at the high magnetic fields that are now becoming available. As a result, solid-state NMR spectroscopy has the potential to extend structural biology to a much broader range of proteins than is currently feasible with the most widely used methods of structure determination.
Supplementary Material
Acknowledgments
We thank F. Marassi, D. Thiriot, and L. Zagyanskiy for helpful discussions and assistance with the sample preparation; C. Grant and C. H. Wu for assistance with the instrumentation; and R. N. Perham for providing the Y21M mutant strain of fd. This research was supported by National Institutes of Health Grants R37GM24266 and RO1GM29754 (to S.J.O.) and utilized the Resource for Solid-State NMR of Proteins supported by National Institutes of Health Grant P41EB002031.
Abbreviations: PISEMA, polarization inversion spin-exchange at the magic angle; PISA, polarity index slant angle.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1NH4).
References
- 1.Webster, R. (2001) in Phage Display: A Laboratory Manual, ed. Burton, D. R. (Cold Spring Harbor Lab. Press, Plainview, NY).
- 2.Marvin, D. A. & Hohn, B. (1969) Bacteriol. Rev. 33, 172–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marvin, D. A. (1998) Curr. Opin. Struct. Biol. 8, 150–158. [DOI] [PubMed] [Google Scholar]
- 4.Sambrook, J. & Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 5.Cross, T. A. & Opella, S. J. (1980) Biochem. Biophys. Res. Commun. 92, 478–484. [DOI] [PubMed] [Google Scholar]
- 6.McDonnell, P. A., Shon, K., Kim, Y. & Opella, S. J. (1993) J. Mol. Biol. 233, 447–463. [DOI] [PubMed] [Google Scholar]
- 7.Henry, G. D. & Sykes, B. D. (1992) Biochemistry 31, 5284–5297. [DOI] [PubMed] [Google Scholar]
- 8.van de Ven, F. J., van Os, J. W., Aelen, J. M., Wymenga, S. S., Remerowski, M. L., Konings, R. N. & Hilbers, C. W. (1993) Biochemistry 32, 8322–8328. [DOI] [PubMed] [Google Scholar]
- 9.Williams, K. A., Farrow, N. A., Deber, C. M. & Kay, L. E. (1996) Biochemistry 35, 5145–5157. [DOI] [PubMed] [Google Scholar]
- 10.Papavoine, C. H., Christiaans, B. E., Folmer, R. H., Konings, R. N. & Hilbers, C. W. (1998) J. Mol. Biol. 282, 401–419. [DOI] [PubMed] [Google Scholar]
- 11.Almeida, F. C. & Opella, S. J. (1997) J. Mol. Biol. 270, 481–495. [DOI] [PubMed] [Google Scholar]
- 12.Marassi, F. M. & Opella, S. J. (2003) Protein Sci. 12, 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marvin, D. A. (1966) J. Mol. Biol. 15, 8–17. [DOI] [PubMed] [Google Scholar]
- 14.Marvin, D. A., Pigram, W. J., Wiseman, R. L., Wachtel, E. J. & Marvin, F. J. (1974) J. Mol. Biol. 88, 581–598. [DOI] [PubMed] [Google Scholar]
- 15.Banner, D. W., Nave, C. & Marvin, D. A. (1981) Nature 289, 814–816. [DOI] [PubMed] [Google Scholar]
- 16.Glucksman, M. J., Bhattacharjee, S. & Makowski, L. (1992) J. Mol. Biol. 226, 455–470. [DOI] [PubMed] [Google Scholar]
- 17.Marvin, D. A., Hale, R. D., Nave, C. & Helmer-Citterich, M. (1994) J. Mol. Biol. 235, 260–286. [DOI] [PubMed] [Google Scholar]
- 18.Caspar, D. L. & Makowski, L. (1981) J. Mol. Biol. 145, 611–617. [DOI] [PubMed] [Google Scholar]
- 19.Thomas, G. J., Jr., Prescott, B. & Day, L. A. (1983) J. Mol. Biol. 165, 321–356. [DOI] [PubMed] [Google Scholar]
- 20.Clack, B. A. & Gray, D. M. (1989) Biopolymers 28, 1861–1873. [DOI] [PubMed] [Google Scholar]
- 21.Williams, R. W., Dunker, A. K. & Peticolas, W. L. (1984) Biochim. Biophys. Acta 791, 131–144. [DOI] [PubMed] [Google Scholar]
- 22.Cross, T. A. & Opella, S. J. (1985) J. Mol. Biol. 182, 367–381. [DOI] [PubMed] [Google Scholar]
- 23.Cross, T. A., Tsang, P. & Opella, S. J. (1983) Biochemistry 22, 721–726. [DOI] [PubMed] [Google Scholar]
- 24.Opella, S. J., Stewart, P. L. & Valentine, K. G. (1987) Q. Rev. Biophys. 19, 7–49. [DOI] [PubMed] [Google Scholar]
- 25.Cross, T. A. & Opella, S. J. (1983) J. Am. Chem. Soc. 105, 306–308. [Google Scholar]
- 26.Colnago, L. A., Valentine, K. G. & Opella, S. J. (1987) Biochemistry 26, 847–854. [DOI] [PubMed] [Google Scholar]
- 27.Cross, T. A. & Opella, S. J. (1982) J. Mol. Biol. 159, 543–549. [DOI] [PubMed] [Google Scholar]
- 28.Gall, C. M., Cross, T. A., DiVerdi, J. A. & Opella, S. J. (1982) Proc. Natl. Acad. Sci. USA 79, 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waugh, J. S. (1976) Proc. Natl. Acad. Sci. USA 73, 1394–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pausak, S., Pines, A. & Waugh, J. S. (1973) J. Chem. Phys. 59, 591–595. [Google Scholar]
- 31.Hester, R., Ackerman, J. L., Neff, B. L. & Waugh, J. S. (1976) Phys. Rev. Lett. 36, 1081–1083. [Google Scholar]
- 32.Opella, S. J. & Waugh, J. S. (1977) J. Chem. Phys. 66, 4919–4924. [Google Scholar]
- 33.Opella, S. J., Kim, Y. & McDonnell, P. (1994) Methods Enzymol. 239, 536–560. [DOI] [PubMed] [Google Scholar]
- 34.Howard, K. P. & Opella, S. J. (1996) J. Magn. Reson. 112, 91–94. [DOI] [PubMed] [Google Scholar]
- 35.Ketchem, R. R., Hu, W. & Cross, T. A. (1993) Science 261, 1457–1460. [DOI] [PubMed] [Google Scholar]
- 36.Opella, S. J., Marassi, F. M., Gesell, J. J., Valente, A. P., Kim, Y., Oblatt-Montal, M. & Montal, M. (1999) Nat. Struct. Biol. 6, 374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, J., Kim, S., Kovacs, F. & Cross, T. A. (2001) Protein Sci. 10, 2241–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, C. H., Ramamoorthy, A. & Opella, S. J. (1994) J. Magn. Reson. 109, 270–272. [Google Scholar]
- 39.Ramamoorthy, A., Wu, C. H. & Opella, S. J. (1999) J. Magn. Reson. 140, 131–140. [DOI] [PubMed] [Google Scholar]
- 40.Marassi, F. M. & Opella, S. J. (2000) J. Magn. Reson. 144, 150–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, J., Denny, J., Tian, C., Kim, S., Mo, Y., Kovacs, F., Song, Z., Nishimura, K., Gan, Z., Fu, R., et al. (2000) J. Magn. Reson. 144, 162–167. [DOI] [PubMed] [Google Scholar]
- 42.Mesleh, M. F., Veglia, G., DeSilva, T. M., Marassi, F. M. & Opella, S. J. (2002) J. Am. Chem. Soc. 124, 4206–4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nevzorov, A. & Opella, S. J. (2003) J. Magn. Reson. 160, 33–39. [DOI] [PubMed] [Google Scholar]
- 44.Bower, M. J., Cohen, F. E. & Dunbrack, R. L., Jr. (1997) J. Mol. Biol. 267, 1268–1282. [DOI] [PubMed] [Google Scholar]
- 45.Tan, W. M., Jelinek, R., Opella, S. J., Malik, P., Terry, T. D. & Perham, R. N. (1999) J. Mol. Biol. 286, 787–796. [DOI] [PubMed] [Google Scholar]
- 46.Oas, T. G., Hartzell, C. J., Dahlquist, F. W. & Drobny, G. P. (1987) J. Am. Chem. Soc. 109, 5962–5966. [Google Scholar]
- 47.Marassi, F. M. (2001) Biophys. J. 80, 994–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marassi, F. M. & Opella, S. J. (2002) J. Biomol. NMR 23, 239–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Opella, S. J., Nevzorov, A., Mesleh, M. F. & Marassi, F. M. (2002) Biochem. Cell Biol. 80, 597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281–296. [DOI] [PubMed] [Google Scholar]
- 51.Overman, S. A., Tsuboi, M. & Thomas, G. J., Jr. (1996) J. Mol. Biol. 259, 331–336. [DOI] [PubMed] [Google Scholar]
- 52.Voet, D., & Voet, J. G. (1995) Biochemistry (Wiley, New York), 2nd Ed.
- 53.Wachtel, E. J., Wiseman, R. L., Pigram, W. J. & Marvin, D. A. (1974) J. Mol. Biol. 88, 601–618. [DOI] [PubMed] [Google Scholar]
- 54.Roth, T. A., Weiss, G. A., Eigenbrot, C. & Sidhu, S. S. (2002) J. Mol. Biol. 322, 357–367. [DOI] [PubMed] [Google Scholar]
- 55.Marvin, D. A. & Wachtel, E. J. (1976) Philos. Trans. R. Soc. London B 276, 81–98. [DOI] [PubMed] [Google Scholar]
- 56.Jelinek, R., Ramamoorthy, A. & Opella, S. J. (1995) J. Am. Chem. Soc. 117, 12348–12349. [Google Scholar]
- 57.Rosay, M., Zeri, A. C., Astrof, N. S., Opella, S. J., Herzfeld, J. & Griffin, R. G. (2001) J. Am. Chem. Soc. 123, 1010–1011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.