Abstract
How myosin II localizes to the cleavage furrow of dividing cells is largely unknown. We show here that a 283-residue protein, assembly domain (AD)1, corresponding to the AD in the tail of Dictyostelium myosin II assembles into bundles of long tubules when expressed in myosin II-null cells and localizes to the cleavage furrow of dividing cells. AD1 mutants that do not polymerize in vitro do not go to the cleavage furrow in vivo. An assembly-competent polypeptide corresponding to the C-terminal 256 residues of Acanthamoeba myosin II also goes to the cleavage furrow of Dictyostelium myosin II-null cells. When overexpressed in wild-type cells, AD1 colocalizes with endogenous myosin II (possibly as a copolymer) in interphase, motile, and dividing cells and under caps of Con A receptors but has no effect on myosin II-dependent functions. These results suggest that neither a specific sequence, other than that required for polymerization, nor interaction with other proteins is required for localization of myosin II to the cleavage furrow.
Efficient cytokinesis generally depends on the constriction of a contractile ring driven by the motor activity of actomyosin II (1), but the mechanisms by which actin and myosin II localize at the cleavage furrow (CF) of dividing cells are largely unknown. Dictyostelium myosin II heavy chain-null cells, which cannot divide in suspension culture but do form a CF when grown on a substrate (2–4), provide an excellent experimental system for determining the domains of myosin II that are required for localization to the CF and growth of cells in suspension culture. Myosin must be able to polymerize to go to the CF (3, 5, 6), but neither the ATPase nor actin-binding activity of the motor domain is required; a polymerization-competent myosin tail localizes to the CF when expressed in myosin II-null cells (7). Moreover, it has been inferred (8) from experiments with heavy chains with different tail regions deleted (8–11) that the region between tail residues 1465 and 1819 may be sufficient for localization of myosin to the CF. This region includes the assembly domain (AD), residues 1533–1819, the shortest region of the tail that had been shown to be capable of polymerization in vitro (12). Furthermore, chimeric myosins consisting of the motor domain of Dictyostelium myosin II and the tails of either Acanthamoeba (13) or chicken smooth (13) or skeletal (8) muscle myosin II localize to the CF of myosin-null cells. Interestingly, the only region of the Dictyostelium myosin II tail that has significant sequence similarity to the tails of the three other myosins lies within the AD domain, residues 1695–1741 (13), with 19% sequence identity among the homology regions of the four myosins.
These results are consistent with the hypothesis that tail residues 1465–1819, and possibly just the AD, are sufficient for localization of Dictyostelium myosin II to the CF. However, all the deletion and substitution experiments were done within the context of a full-length myosin, the effect of deleting residues 1465–1819 has not been investigated, and the smallest constructs that have been shown to go to the CF are the full-length tails of Dictyostelium (7) and Acanthamoeba (13) myosin II. Therefore, to determine directly whether the AD contains all the information necessary for localization to the CF, we expressed N-terminal FLAG-tagged AD constructs in Dictyostelium myosin II heavy chain-null cells.
Methods
Plasmids. The actin 15 promoter DNA and AD1 DNA spanning bases 4666–5514 of Dictyostelium myosin II, with a FLAG peptide (DYKDDDDK) at the N-terminal end, were synthesized by PCR with pTIKLMyD as the template (14), and the two fragments were fused by overlapping PCR. An XbaI restriction site was introduced at the 5′ end of the promoter, and a stop codon and an MluI restriction site (TAACGCGT) were created downstream of base A5514. The PCR product was digested with MluI and XbaI and subcloned into the pGEM7 vector, resulting in pGEMAD1, and into pTKLMyD, which has an MluI site downstream of the stop codon, resulting in pTKLAD1. DNAs for AD2–AD6 and the Acanthamoeba myosin II assembly region (V1254–E1509) were synthesized by PCR, subcloned into pGEMAD1 by digestion with EcoRI and MluI, and then subcloned into pTIKLAD1 by using XbaI and MluI. All sequences obtained by PCR were confirmed by sequencing.
Cell Culture. pTIKLAD plasmids were introduced into Dictyostelium wild-type (AX3) and myosin II-null (HS1) cells (15) by electroporation (16). The transformants were selected and maintained on plates in the presence of 12 μg/ml G418 in HL5 medium containing 60 μg/ml each of penicillin and streptomycin at 22°C. For large-scale cultures and growth-rate assays, all transformants were grown in the same medium but without G418 in conical flasks on a rotary shaker at 145 rpm at 22°C (17). Cells were counted daily by using a hemocytometer.
Immunofluorescence. The cell cycle was partially synchronized by cold treatment (8). Cell motility was initiated by treatment with chemotaxing-inducing starvation conditions as described (18, 19). For indirect immunofluorescence of FLAG-tagged proteins, cells were fixed for 5 min in –20°C methanol containing 1% formalin and then incubated with 700-fold-diluted anti-FLAG M2 monoclonal antibody (Sigma) for 1 h at 37°C, washed, and incubated with 500-fold-diluted FITC-conjugated goat anti-mouse IgG (Molecular Probes). The same procedure was used for double staining for AD1 and myosin II by using as primary antibodies 700-fold-diluted anti-FLAG M2 antibody and 1,000-fold-diluted anti-Dictyostelium myosin II antibody (a kind gift of T. Q. P. Uyeda, National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan) and as secondary antibodies 150-fold-diluted Cy3-conjugated anti-mouse IgG (Sigma) and 200-fold-diluted FITC-conjugated goat anti-rabbit antibody (Molecular Probes).
To determine the time course of Con A capping, cells were incubated on glass coverslips with 50 μl of 30 μg/ml tetramethyl rhodamine isothiocyanate-conjugated Con A (Molecular Probes) as described (13, 20). Excess Con A was removed by rinsing the coverslips briefly in starvation buffer, and the cells were fixed and stained for AD1 and myosin II with the primary antibodies described above, the same secondary antibody for myosin II, and 1,000-fold-diluted Alexa Fluor 647-conjugated anti-mouse IgG (Molecular Probes) for AD1. Micrographs were taken on a Zeiss LSM 510 laser scanning fluorescence confocal microscope equipped with a Plan apo ×63 oil objective.
Development Assay. Small aliquots of amoebae were spotted on agar plates in starvation medium (10, 13), and phenotypes were observed under a dissection microscope over a 24-h period.
Electrophoresis and Western Blots. Cells were harvested by low-speed centrifugation, 50 μl of resuspended cells (2 × 106 cells) were added to 50 μlof2× SDS sample buffer in a boiling water bath, and equivalent aliquots were analyzed by 10% SDS/PAGE. Gels were either stained with Coomassie blue or transferred electrophoretically to nitrocellulose paper and blotted with either 10,000-fold-diluted polyclonal anti-Dictyostelium myosin II antibody or 1,000-fold-diluted anti-FLAG M2 monoclonal antibody and then incubated in either 2,000-fold-diluted horseradish peroxidase-labeled goat anti-rabbit (Bio-Rad) or 5,000-fold-diluted horseradish peroxidase-labeled goat anti-mouse (Amersham Biosciences) Ig, respectively.
Protein Purification. AX3 cells expressing each of the proteins were harvested by low-speed centrifugation and lysed in 300 mM NaCl/20 mM Hepes (pH 7.5)/1mMDTT/0.6% Triton X-100/0.1 mM PMSF/protease inhibitor mixture (Roche Diagnostics), and the lysates were centrifuged in a Beckman 70Ti rotor at 50,000 rpm for 1 h at 4°C. The supernatants were loaded onto anti-FLAG antibody-conjugated resin (Sigma), and the proteins were eluted with lysate buffer without Triton but containing 0.1 mg/ml FLAG-peptide.
Assembly Assay. The purified, soluble proteins were dialyzed overnight against assembly buffer (50 mM NaCl/10 mM Hepes, pH 7.5/1 mM DTT/10 mM MgCl2) unless otherwise specified and centrifuged for1hat 240,000 × g in a Beckman TL-100 centrifuge. The pellets were resuspended to the original volume in dialysis buffer, and equal aliquots of total sample, supernatant, and pellet were analyzed by SDS/PAGE.
Electron Microscopy. For negatively stained images, 5-μl samples were applied to formvar/carbon-coated grids and stained with 2% uranyl acetate. Cells were fixed with 2.5% glutaraldehyde/1% paraformaldehyde in 0.1 M cacodylate buffer for 1 h at room temperature and overnight at 4°C, postfixed in 1% OsO4, and stained with 2% uranyl acetate and 1% tannic acid before embedding in Epon. The thin sections were stained further with 2% uranyl acetate and Reynold's lead citrate solution. Micrographs were taken by using a JEOL EX II electron microscope at 80 kV.
Results
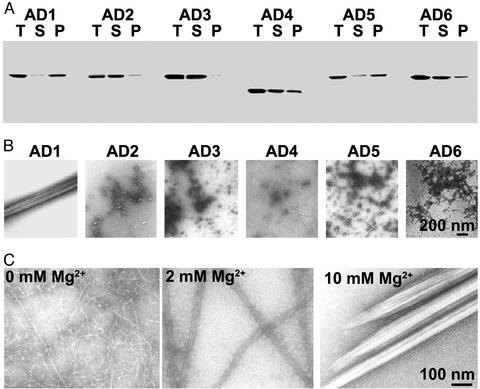
AD1 Assembles in Vitro, but Truncated and Substitution Mutants Do Not. Dictyostelium-expressed AD1 (tail residues 1533–1815; Fig. 1) differs only slightly from the two Escherichia coli-expressed ADs, the in vitro assembly properties of which were described previously: residues 1533–1819 (12) and 1531–1824 (22). When dialyzed against assembly buffer, >95% of AD1 was recovered in the high-speed pellet (Fig. 2A), as in the earlier studies with the slightly larger constructs (12, 22). AD1 formed short tubules 10–15 nm in width when the assembly buffer contained 25 mM NaCl and no MgCl2 (Fig. 2C), bundles of longer tubules when 2 mM MgCl2 was included in the buffer (Fig. 2C), and larger bundles and paracrystals in the presence of 10 mM MgCl2 (Fig. 2 B and C).
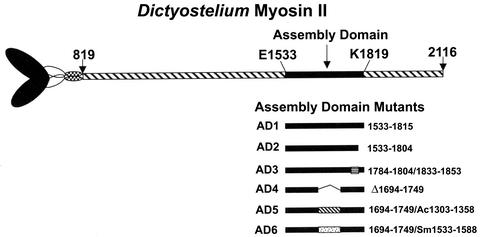
Fig. 1.
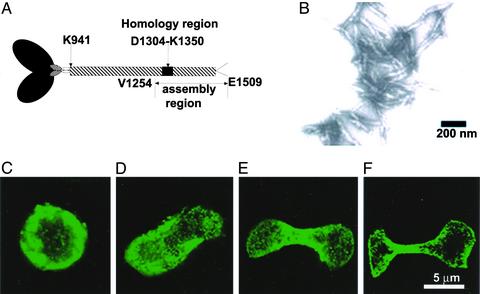
AD constructs. Schematic representation of Dictyostelium myosin II heavy chain showing the coiled-coil helical tail (residues 819–2116) and the location of the AD (residues E1533–K1819). The AD mutants discussed in this article are AD1 (residues 1533–1815), AD2 (residues 1533–1804), AD3 (residues 1784–1804 of AD1 replaced by Dictyostelium myosin II tail residues 1833–1853), AD4 (residues 1694–1749, the homology region, of AD1 deleted), AD5 (residues 1694–1749 of AD1 replaced by the homology region of Acanthamoeba myosin II, residues 1303–1358), and AD6 (residues 1694–1749 of AD1 replaced by the homology region of smooth muscle myosin II, residues 1533–1588). All constructs had an N-terminal FLAG tag and are predicted to be 100% coiled-coil (21).
Fig. 2.
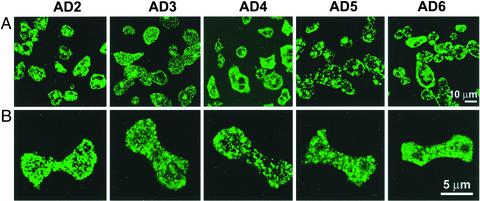
In vitro properties of AD1 and AD1 mutants. (A) Coomassie blue-stained SDS/PAGE of total mixture (T), supernatant (S), and pellet (P) after dialysis against 50 mM NaCl/10 mM MgCl2 and centrifugation. (B) Negatively stained electron-microscopic images of AD1–AD6 after dialysis against 50 mM NaCl/10 mM MgCl2.(C) Negatively stained images of AD1 after dialysis against 25 mM NaCl, 50 mM NaCl/2 mM MgCl2, or 50 mM NaCl/10 mM MgCl2.
Shoffner and De Lozanne (22) found that deletion of 28 residues (1531–1558) from the N-terminal end or 26 residues (1799–1824) from the C-terminal end made their AD construct unable to assemble in vitro. In agreement with their results, we found that assembly of AD1 was completely blocked by deletion of either residues 1533–1613 from the N-terminal end or residues 1783–1815 from the C-terminal end (data not shown).
When just 11 residues, 1805–1815, were deleted from the C-terminal end (Fig. 1, AD2), <10% of the polypeptide was recovered in the centrifugal pellet under assembly conditions (Fig. 2A), and only amorphous aggregates were seen by electron microscopy (Fig. 2B). AD3 (Fig. 1), in which AD1 residues 1784–1804 were replaced by residues 1833–1853 that lie immediately C-terminal to the AD in the myosin II tail, was completely soluble in assembly buffer (Fig. 2A). When AD1 residues 1694–1749, which include the homology region, were either deleted (Fig. 1, AD4) or replaced by a segment containing the homology region of either Acanthamoeba myosin II (Fig. 1, AD5) or smooth muscle myosin II (Fig. 1, AD6), 35%, 60%, and 20% of the polypeptides, respectively, were recovered in the pellets (Fig. 2A), but there was no evidence of assembly into filaments or tubules, only amorphous aggregates (Fig. 2B).
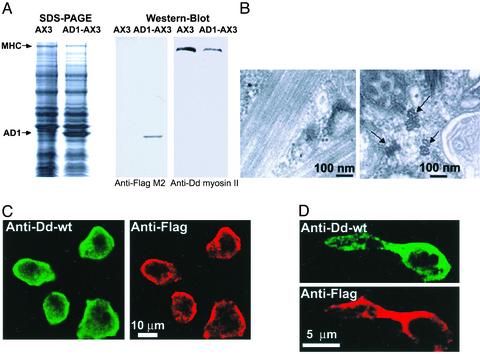
AD1 Assembles and Goes to the CF When Expressed in Dictyostelium Myosin II-Null (HS1) Cells, but Mutants AD2–AD6 Do Not. AD1–AD6 were heavily expressed in HS1 cells (Fig. 3). In ≈70% of the cells, AD1 had a predominantly cortical distribution similar to that of myosin II in wild-type AX3 cells, but in ≈30% of the cells AD1 formed a “bar” extending across the middle of the cell (Fig. 4A). In both locations, AD1 appeared to be filamentous by confocal microscopy. Thin-section electron microscopy (Fig. 4 B and C) showed both structures to consist of large assemblies of parallel arrays of very long tubules (o.d. ≈ 15 nm, i.d. ≈ 9 nm). Neither actin nor tubulin was associated with the AD1 in the cortex or bars (data not shown). Most importantly, AD1 was concentrated heavily at the CF of dividing cells (Fig. 4 D–F).
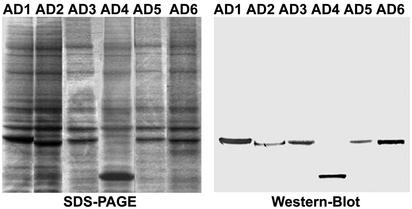
Fig. 3.
Overexpression of AD1 and AD1 mutants in Dictyostelium myosin II-null cells. Shown are SDS/PAGE and Western blot with anti-FLAG antibody of total cell extracts of HS1 cells overexpressing FLAG-tagged AD1–AD6.
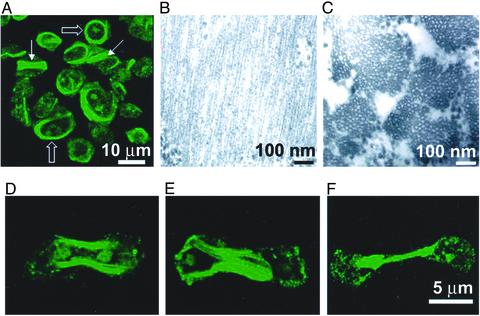
Fig. 4.
AD1 goes to the CF of Dictyostelium myosin II-null cells. (A) Immunostained interphase HS1 cells overexpressing FLAG-tagged AD1 showing predominantly cortical staining (wide arrow) and less frequent rods (thin arrow). (B and C) Thin-section electron micrographs showing longitudinal and cross-sectional views of bundles of tubules of AD1 in transfected interphase HS1 cells. (D–F) Immunostained dividing HS1 cells showing localization to the CF of overexpressed AD1.
None of the other constructs formed filamentous or tubular structures when expressed in HS1 cells (Fig. 5). In most interphase cells (Fig. 5A), AD2–AD6 were relatively uniformly distributed throughout the cytoplasm, although occasionally they appeared to be more concentrated at the periphery, and they remained uniformly distributed in the cytoplasm of dividing cells with no indication of any concentration at the CF (Fig. 5B).
Fig. 5.
Constructs AD2–AD6 do not assemble or go to the CF when expressed in Dictyostelium myosin II-null cells. Shown are immunostained interphase (A) and dividing (B) HS1 cells overexpressing AD2–AD6.
An Assembly-Competent Region of Acanthamoeba Myosin II Tail Goes to the CF of Dictyostelium Myosin II-Null Cells. An expressed protein corresponding to the C-terminal 256 residues (1254–1509) of Acanthamoeba myosin II (Fig. 6A), which includes the 47-residue homology region (residues 1304–1350), polymerized in vitro (Fig. 6B), forming aggregated bundles of short filaments indistinguishable from those formed by a corresponding fragment of native Acanthamoeba myosin II produced by proteolysis (23). When expressed in Dictyostelium myosin II-null cells, the assembly region localized to the cortex of interphase cells (Fig. 6C) and to the CF of dividing cells (Fig. 6 D–F); unlike AD1, the Acanthamoeba assembly region did not form bars in interphase cells. A shorter construct, residues 1373–1509, that began 23 residues C-terminal to the homology region did not polymerize in vitro (data not shown).
Fig. 6.
Assembly region of Acanthamoeba myosin II goes to the CF of Dictyostelium myosin II-null cells. (A) Schematic representation of Acanthamoeba myosin II showing the location of the homology region within the assembly region that spans the C-terminal half of the coiled-coil tail and the nonhelical tailpiece. (B) Negatively stained electron-microscopic image of filaments of the assembly region after dialysis against 50 mM NaCl/10 mM MgCl2. (C) Micrograph of immunostained HS1 interphase cell overexpressing FLAG-tagged assembly region. (D–F) Micrographs of immunostained HS1 dividing cells overexpressing FLAG-tagged assembly region at progressive stages of division.
AD1 Colocalizes with Myosin II When Expressed in Dictyostelium Wild-Type (AX3) Cells and Does Not Inhibit Myosin II Function. AD1 was greatly overexpressed in AX3 cells relative to the level of endogenous myosin II (Fig. 7A) and formed bundles of tubules (Fig. 7B) similar to but smaller than the bundles of tubules of AD1 expressed in HS1 cells (Fig. 4B). Although it is not apparent in the electron micrographs, the bundles probably also contained endogenous myosin II, because, as determined by confocal microscopy, myosin II and AD1 colocalized both in the cortex of interphase cells (Fig. 7C) and the CF of dividing cells (Fig. 7D).
Fig. 7.
Expressed AD1 colocalizes with myosin II in Dictyostelium wild-type cells. (A) SDS/PAGE and Western blot of total cell proteins of AX3 cells and AX3 cells expressing AD1. (B) Thin-section electron micrograph showing longitudinal and cross-sectional (arrows) views of bundles of tubules of overexpressed AD1 in transfected interphase AX3 cells. (C) Colocalization of endogenous wild-type myosin II (Anti-Dd-wt) and AD1 (Anti-Flag) in interphase AX3 cells overexpressing AD1. (D) Colocalization of endogenous wild-type myosin II (Anti-Dd-wt) and AD1 (Anti-Flag) in dividing AX3 cells overexpressing AD1.
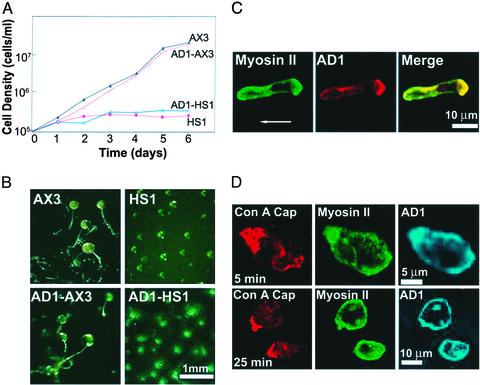
Surprisingly, despite the large excess of AD1 relative to myosin II heavy chain (Fig. 7A) [and in contrast to the dominant negative effect of full-length tail (residues 809–2116) and light meromyosin (residues 1528–2116); refs. 24 and 25], AD1 had no demonstrable effect on myosin II function in AX3 cells. AX3 cells expressing AD1 grew in suspension culture at the same rate as control AX3 cells (Fig. 8A) and completed normal development to fruiting bodies after 24 h in starvation medium (Fig. 8B), and myosin II localized normally at the rear of motile cells (Fig. 8C). Also, AX3 cells overexpressing AD1 capped Con A receptors within 5 min after exposure to Con A, with myosin II concentrated under the cap, and the myosin redistributed to the cortex within 25 min (Fig. 8D), as in control cells.
Fig. 8.
AD1 does not inhibit myosin II function in Dictyostelium wild-type cells. (A) Growth in suspension culture of HS1 (myosin-null) and AX3 (wild-type) cells and HS1 and AX3 cells overexpressing AD1, AD1-HS1, and AD1–AX3, respectively. (B) Development of cells after 24 h on starvation medium. (C) Colocalization of AD1 and myosin II at rear of a wild-type cell moving from right to left. (D) Co-capping of AD1 and myosin II under Con A caps at 5 min and redispersal of AD1 and myosin II by 25 min after capping in wild-type cells.
Even more surprisingly, overexpressed AD1 colocalized with myosin II at the rear of motile AX3 cells (Fig. 8C), concentrated under the Con A-receptor cap with myosin II (Fig. 8D), and AD1 redispersed from the cap on the same time scale as myosin II (Fig. 8D). AD1 did not localize to the rear of motile HS1 (myosin II-null) cells, and HS1 cells expressing AD1 did not form Con A-receptor caps (data not shown).
Discussion
The AD was originally defined by in vitro assembly studies as residues 1533–1819 (12). Truncation of the AD at residue 1798 blocked assembly in vitro (22), as assayed by insolubility, consistent with the earlier observation that deletion of residues 1784–1819 makes myosin II unable to assemble in vitro or to support cytokinesis or Con A capping in vivo (11). We have now found that the AD can be truncated at position 1815 and remain assembly-competent both in vitro and in vivo. However, truncation after residue 1804 blocks polymerization in vitro and in vivo, as does replacement of residues 1784–1804 with residues 1833–1853 even though the substitution maintains the heptad repeat and predicted coiled-coil structure (21) of the original construct.
The experiments described in this article establish unequivocally that the AD of Dictyostelium myosin II contains all the information necessary for localization to the CF. AD mutants that do not polymerize in vitro do not polymerize in vivo or relocate to the CF. Deletion of a 56-residue segment that includes the homology region from the middle of AD1, even when it is replaced by the homology region of Acanthamoeba or smooth muscle myosin II, makes AD1 incapable of polymerization in vitro and in vivo and unable to localize to the CF of myosin II-null cells. However, the 256-residue C-terminal region of Acanthamoeba myosin II that contains its homology region does polymerize in vitro and go to the CF when overexpressed in Dictyostelium myosin II-null cells. These results strongly suggest that species-specific sequences in the homology region are important for polymerization of myosin II but that a specific sequence is not required for localization of polymerized myosin to the CF.
We have shown that AD1 expressed in Dictyostelium wild-type cells colocalizes with endogenous myosin II not only in the cortex of interphase cells and at the CF of dividing cells but also when myosin II relocates to the rear of motile cells and under Con A-induced caps. These latter processes require myosin motor activity and the ability to interact with actin filaments (19), both of which AD1 lacks, suggesting that AD1 closely associates with polymerized myosin II in vivo, possibly as a copolymer that retains myosin II function. The further observation that AD1 dissociates from the Con A cap on the same time scale as myosin II is also suggestive of an AD1–myosin II copolymer, because dispersion of myosin II filaments from Con A caps requires regulated depolymerization mediated by phosphorylation of sites in the myosin tail (16) that are not present in AD1. Because no heavy chain was detectable on SDS/PAGE gels containing >10 times as much affinity-purified AD1 as the gels in Fig. 2, any copolymer would contain AD1 homodimers and full-length heavy chain homodimers but no heterodimers of AD1 and heavy chain (single-headed myosin).
That a large molar excess of AD1 has no effect on any of the myosin II-dependent activities of wild-type cells leads us to conclude that AD1 does not adversely compete with myosin II for any protein with which myosin II must interact in any of these processes. Moreover, that both AD1 and an assembly-competent region of Acanthamoeba myosin II heavy chain go to the CF when greatly overexpressed in Dictyostelium myosin II-null cells and that the large tubular structures formed by both polypeptides are very different from the bipolar filaments of myosin II strongly suggest that specific interactions with other proteins are not required for localization of myosin II to the CF.
Acknowledgments
We thank Myoung-Soon Cho for the electron microscopy.
Abbreviations: CF, cleavage furrow; AD, assembly domain.
References
- 1.Robinson, D. N. & Spudich, J. A. (2000) Trends Cell Biol. 14, 228–237. [DOI] [PubMed] [Google Scholar]
- 2.Knecht, D. A. & Loomis, W. F. (1987) Science 236, 1081–1086. [DOI] [PubMed] [Google Scholar]
- 3.De Lozanne, A. & Spudich, J. A. (1987) Science 236, 1086–1091. [DOI] [PubMed] [Google Scholar]
- 4.Manstein, D. J., Titus, M. A., De Lozanne, A. & Spudich, J. A. (1989) EMBO J. 8, 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukui, Y., De Lozanne, A. & Spudich, J. A. (1990) J. Cell Biol. 104, 367–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabry, J. H., Moores, S. L., Ryan, S., Zang, J. H. & Spudich, J. A. (1997) Mol. Biol. Cell 8, 2605–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zang, J. H. & Spudich, J. A. (1998) Proc. Natl. Acad. Sci. USA 95, 13652–13657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shu, S., Lee, R. J., LeBlanc-Straceski, J. M. & Uyeda, T. Q. P. (1999) J. Cell Sci. 112, 2195–2201. [DOI] [PubMed] [Google Scholar]
- 9.O'Halloran, T. J. & Spudich, J. A. (1990) Proc. Natl. Acad. Sci. USA 87, 8110–8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubalek, E. W., Uyeda, T. Q. P. & Spudich, J. A. (1992) Mol. Biol. Cell 3, 1455–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, R. J., Egelhoff, T. T. & Spudich, J. A. (1994) J. Cell Sci. 107, 2875–2886. [DOI] [PubMed] [Google Scholar]
- 12.O'Halloran, T., Ravid, S. & Spudich, J. A. (1990) J. Cell Biol. 110, 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shu, S., Liu, X., Parent, C. A., Uyeda, T. Q. P. & Korn, E. D. (2002) J. Cell Sci. 115, 4237–4249. [DOI] [PubMed] [Google Scholar]
- 14.Liu, X., Ito, K., Lee, R. J. & Uyeda, T. Q. P. (2000) Biochem. Biophys. Res. Commun. 271, 75–81. [DOI] [PubMed] [Google Scholar]
- 15.Ruppel, K. M., Uyeda, T. Q. P. & Spudich, J. A. (1994) J. Biol. Chem. 269, 18773–18780. [PubMed] [Google Scholar]
- 16.Egelhoff, T. T., Brown, S. S. & Spudich, J. A. (1991) J. Cell Biol. 112, 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sussman, S. (1987) Methods Cell Biol. 28, 9–29. [DOI] [PubMed] [Google Scholar]
- 18.Parent, C. A., Blacklock, B. J., Froehlich, W. M., Murphy, D. B. & Devreotes, P. N. (1998) Cell 95, 81–91. [DOI] [PubMed] [Google Scholar]
- 19.Levi, S., Polyakov, M. V. & Egelhoff, T. T. (2002) Cell Motil. Cytoskeleton 53, 177–188. [DOI] [PubMed] [Google Scholar]
- 20.LeBlanc-Straceski, J. M., Fukui, Y., Sohn, R. L., Spudich, J. A. & Leinwand, L. (1994) Cell Motil. Cytoskeleton 27, 313–326. [DOI] [PubMed] [Google Scholar]
- 21.Berger, B., Wilson, D. B., Wolf, E., Tonchev, T., Milla, M. & Kim, P. S. (1995) Proc. Natl. Acad. Sci. USA 92, 8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shoffner, J. D. & De Lozanne, A. (1996) Biochem. Biophys. Res. Commun. 218, 860–864. [DOI] [PubMed] [Google Scholar]
- 23.Ganguly, C., Atkinson, M. A. L., Sathyamoorthy, V., Bowers, B. & Korn, E. D. (1990) J. Biol. Chem. 265, 9993–9998. [PubMed] [Google Scholar]
- 24.Burns, C. G., Larochelle, D. A., Erickson, H., Reedy, M. & De Lozanne, A. (1995) Proc. Natl. Acad. Sci. USA 92, 8244–8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burns, C. G., Reedy, M., Heuser, J. & De Lozanne, A. (1995) J. Cell Biol. 130, 605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]