Abstract
The biological effects of estrogens are mediated by the estrogen receptors ERα and ERβ. These receptors regulate gene expression through binding to DNA enhancer elements and subsequently recruiting factors such as coactivators that modulate their transcriptional activity. Here we show that ARNT (aryl hydrocarbon receptor nuclear translocator), the obligatory heterodimerization partner for the aryl hydrocarbon receptor and hypoxia inducible factor 1α, functions as a potent coactivator of ERα- and ERβ- dependent transcription. The coactivating effect of ARNT depends on physical interaction with the ERs and involves the C-terminal domain of ARNT and not the structurally conserved basic helix–loop–helix and PAS (Per-ARNT-Sim) motifs. Moreover, we show that ARNT/ER interaction requires the E2-activated ligand binding domain of ERα or ERβ. These observations, together with the previous role of ARNT as an obligatory partner protein for conditionally regulated basic helix–loop–helix–PAS proteins like the aryl hydrocarbon receptor or hypoxia inducible factor 1α, expand the cellular functions of ARNT to include regulation of ERα and ERβ transcriptional activity. ARNT was furthermore recruited to a natural ER target gene promoter in a estrogen-dependent manner, supporting a physiological role for ARNT as an ER coactivator.
Keywords: cross-talk, chromatin immunoprecipitation
Estrogens regulate important physiological processes, such as development and function of the male and female reproductive system and maintenance of bone mass in women, and represent a protective factor against cardiovascular disease (1). The physiological effects of estrogens are mediated by estrogen receptors α (ERα) and β (ERβ), which belong to the nuclear receptor superfamily (1, 2). Nuclear receptors are ligand-dependent transcription factors characterized by a conserved structural arrangement composed of a centrally located DNA binding domain (DBD) containing two highly conserved Zn finger motifs. This domain is flanked in the N terminus by a variable A/B region, which contains an activation function (AF-1). The ligand binding domain (LBD), located C-terminally of the DBD, contains a second AF (AF-2) and is also responsible for ligand binding, receptor dimerization, and cofactor interaction (3).
The ERs are, in the absence of ligand, present in the nucleus in a nonactivated form. The latent ERs interact with corepressors such as N-Cor, SHP, or SMRT, which inhibit constitutive transcriptional activity (1). Binding of agonists induces release of repressor proteins and allows interaction with coactivators of the p160 class like SRC-1 or transcription intermediary factor 2 (TIF-2). These proteins have been shown to increase access to chromatin through acetylation of histones, mediate contact with general transcription factors, and enhance receptor AF-1/AF-2 synergy (4, 5).
Interestingly, p160 coactivators share considerable sequence homology to basic helix–loop–helix (bHLH)–PAS (Per-ARNT-Sim) transcription factors. This family also includes factors such as the aryl hydrocarbon receptor (AhR), which mediates the biological effects of environmental pollutants like 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD); the hypoxia inducible factor HIF-1α, the regulator of the cellular response to low oxygen tension; the circadian regulatory protein bMAL; and Clock, which is involved in regulation of diurnal–nocturnal gene expression. The general bHLH–PAS dimerization partner ARNT (AhR nuclear translocator) functions as a necessary partner for the AhR and HIF-1α (reviewed in ref. 6). Interaction between ARNT and, e.g., the ligand-activated AhR generates a DNA binding complex that regulates expression of dioxin-inducible genes, such as the cytochrome P450 1A1 (6). Recent studies have identified three isoforms of ARNT. ARNT and ARNT-2 are highly homologous proteins, whereas bMAL (also known as MOP-3/ARNT-3) shares considerable sequence homology to ARNT and ARNT-2 in the bHLH and PAS domain but is less well conserved in the C-terminal domain. Interestingly, ARNT and ARNT-2 are functionally exchangeable, whereas bMAL interacts only with selected members of the bHLH–PAS family of transcription factors, such as the circadian regulator Clock (7, 8).
bHLH–PAS proteins share a conserved structural arrangement with the bHLH domain located in the N-terminal part of the protein where the basic region is required for specific DNA binding and the HLH domain is a key dimerization interface of bHLH–PAS proteins (6). The PAS domain is located immediately after the bHLH domain. This domain spans ≈250 aa and harbors two conserved hydrophobic repeats termed A and B. The PAS A domain mediates dimerization specificity between bHLH–PAS proteins (9), and the PAS B domain serves as a secondary dimerization interface and represents an interaction site for the hsp90 molecular chaperone complex for selected members of the bHLH–PAS family (6).
Given the structural similarity among p160 coactivators and bHLH–PAS transcription factors, we decided to test whether bHLH–PAS transcription factors could functionally interact with and influence ERα and ERβ activity. ARNT was selected because this protein appears to be constitutively active and not conditionally regulated like AhR or HIF-1α. Furthermore, ARNT displays a broad spectrum of partner proteins and interacts with numerous bHLH–PAS factors (6).
In this study, we show that ARNT functions as a potent coactivator of both ERα- and ERβ-dependent transcription.
Materials and Methods
Recombinant Plasmids. Human pCMV-ARNT (10), deletion constructs of ARNT (10), human pSG5-ERα, human pSG5-ERβ, human ERα-ΔA/B (lacking the 182 most N-terminal amino acids), human ERβ-ΔA/B (93 most N-terminal amino acids deleted), Gal4 ERα-LBD (amino acids 247–599 of mouse ERα linked to the Gal4 DBD), and Gal4 ERβ-LBD (amino acids 173–485 of mouse ERβ; ref. 11), and the 3xERE-TATA-Luc reporter construct (12) have been described.
Mouse pCMV4-ARNT-2 (13) and pCDNA-bMAL (14) were generous gifts from Bill Wilson (Karolinska Institutet) and Y. Fujii-Kuriyama (Tohuko University, Sendai, Japan), respectively.
Protein Expression and Immunoprecipitation. Coimmunoprecipitation of ERα and ERβ attached proteins was performed as described. In short, COS-7 cells were grown in 10-cm plates, and expression vectors for human ARNT and ERα or ERβ were introduced by transient tranfection with Lipofectamine (Invitrogen) according to the manufacturer's suggestions. After transfection, cells were grown for 48 h in phenol red-free medium supplemented with dextran-coated, charcoal-treated serum before treatment with 10 nM E2, 1 μM tamoxifen, or vehicle as described for 1 h. At this point, whole-cell extracts were prepared and coimmunoprecipitation experiments of ERα- or ERβ-associated proteins were performed as described with antibodies directed against ERα or ERβ (Abcam, Cambridge, U.K.) or preimmune serum as a negative control as indicated in the figure legends. After coimmunoprecipitation, the beads were extensively washed and precipitated material was fractionated through 7.5% SDS/PAGE. Proteins were subsequently transferred to nitrocellulose membrane, and the presence of ARNT was investigated by using a polyclonal ARNT antibody (H-172, Santa Cruz Biotechnology) and an ECL (Amersham Biosciences) visualization system according to the manufacturer's instructions.
Transient Transfection Assays. HeLa cells were transfected as described (15). Typically, cells were seeded in 6-well plates 24 h before transfection. Cells were transfected by using Lipofectamine in dextran-coated, charcoal-treated serum and phenol red-free medium with 0.5 μg of appropriate reporter plasmid, as described in the figure legends, together with 100 ng of pCMV5-β-galactosidase (β-gal) as internal transfection control in the presence of 1–5 ng of pSG5-ERα and pSG5-ERβ and 100–500 ng of pCMV4-ARNT expression plasmids according to the manufacturer's recommendations. After transfection, the medium was exchanged with phenol red-free medium supplemented with 5% dextran-coated, charcoal-treated FBS, and the cells were allowed to grow for an additional 48 h. At this point, cells were harvested and luciferase and β-gal activities were determined. Data are presented as percent activity ± SD of at least three independent experiments performed in duplicate, where reporter activity obtained at 10 nM E2 was arbitrarily set to 100% for ERα or ERβ, respectively, and the effect of ARNT was calculated as the increase in percent activity for each receptor subtype or as mean of fold induction ± SD, where activity of reporter plasmid alone without hormone treatment was arbitrarily set to one.
Chromatin Immunoprecipitation (ChIP) Assay. ChIP assays were performed as described (16) with minor modifications. Briefly, T47D cells were grown to 80–90% confluency in phenol red-free DMEM/F12 supplemented with 5% dextran-coated, charcoal-treated FBS for 3 days. Cells were cross-linked with 1% formaldehyde for 10 min at room temperature, then quenched with 125 mM glycine. The cells were harvested and resuspended in 50 mM Tris, pH 8.0/1% Triton X-100/0.1% deoxycholate/1 mM EDTA/0.5 mM EGTA/140 mM NaCl for 10 min at 4°C. Samples were sonicated (10 × 10 s) and soluble chromatin was immunoprecipitated as described (17). The washed resin was resuspended in 120 μl of elution buffer (Tris-EDTA/1% SDS) and reverse-cross-linked overnight at 66°C. DNA fragments were isolated and purified (QIAquick Spin kit, Qiagen, Valencia, CA), and the PCR-amplified (human pS2 promoter region -353 to -54) fragments were separated on 2% agarose gels.
Results
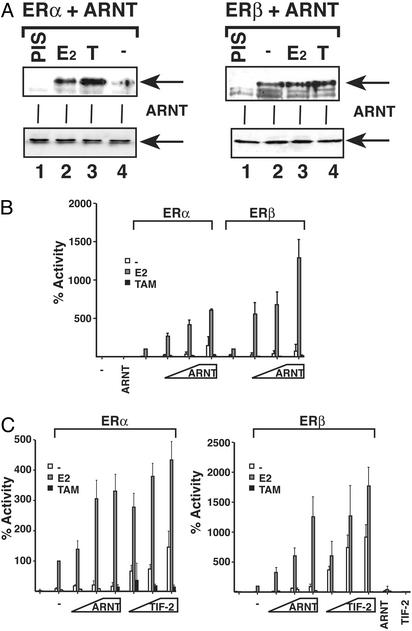
ARNT Interacts with and Enhances the Transcriptional Activity of ERα and ERβ Coactivators like TIF-2 are structurally related to members of the bHLH–PAS family. Given the similarities between p160 coactivators and bHLH–PAS transcription factors, we decided to investigate whether ARNT has the ability to interact with ERα and ERβ. COS-7 cells were cotransfected with expression plasmids for ARNT and ERα (Fig. 1A Left) or ERβ (Fig. 1 A Right). After transfection, the cells were grown for 48 h before treatment with 10 nM E2 or 1 μM tamoxifen for 1 h. After this treatment the cells were harvested and whole-cell extracts were prepared. The extracts were incubated with antibodies against ERα or ERβ, precipitated by using protein G-Sepharose, and resolved by SDS/PAGE. Western blotting was then performed with a polyclonal ARNT antibody (18). As shown in Fig. 1 A, both ERα and ERβ were able to efficiently precipitate ARNT, both in the presence of 10 nM E2 and 1 μM tamoxifen and, to a lesser extent, in the absence of hormone.
Fig. 1.
ARNT interacts with ERα and ERβ and enhances E2-dependent transcription. (A) COS-7 cells were seeded in 10-cm plates and transfected with 5 μg of pCMV4-ARNT and 5 μg of pSG5-ERα or -ERβ as shown and treated with 10 nM E2 or 1 μM tamoxifen (T) or vehicle. After transfection, whole-cell extracts were prepared and immunoprecipitation experiments were performed with ERα or ERβ antibodies as described in Materials and Methods. The precipitated material was transferred to nitrocellulose, and the presence of ARNT was monitored by Western blot experiments. (Upper) Precipitated material under different treatments. (Lower) Ten percent input material. (B) HeLa cells were transfected with 10 ng of expression vectors for ERα or ERβ together with 3xERE-TATA-luciferase reporter (500 ng) construct and pCMV5-β-gal (50 ng) reporter gene construct. In selected reactions, increasing amounts of ARNT (100–500 ng) were also introduced as indicated. After transfection, cells were treated with 10 nM E2 or 1 μM tamoxifen and incubated for 48 h before luciferase measurement. The activity of ERα or ERβ in the presence of E2 was arbitrarily set to 100%, and the effects of ARNT were compared with this result. Mean of three transfections performed in duplicate is presented. (C) HeLa cells were transfected as above with expression vectors for ERα (Left), ERβ (Right), and ARNT or TIF-2 as indicated together with a 3xERE-luciferase reporter construct. After transfection, cells were treated with 10 nM E2 or 1 μM tamoxifen, and luciferase activity was measured and plotted.
These results prompted us to test whether the interaction between ARNT and the ERs had any effect on ERα/β transcriptional activity. For this purpose, HeLa cells were cotransfected with a 3xERE-TATA luciferase reporter gene construct together with expression vectors for ERα or ERβ and increasing amounts of ARNT plasmid. After transfection, the cells were incubated with 10 nM E2, 1 μM tamoxifen, or vehicle. To compare the effect of ARNT on the individual activity of ERα and ERβ, the transcriptional response of E2-treated ERα or ERβ, respectively, was arbitrarily set to 100%, and the effect of ARNT was calculated as the increase in percent activity for each receptor subtype. Addition of ARNT alone had no effect on the activity of the estrogen response element (ERE) reporter. However, in cells cotransfected with ERα or ERβ, ARNT induced a robust increase in reporter gene activity. In cells cotransfected with ARNT and ERα and treated with E2, the presence of ARNT induced a 6-fold increase in luciferase activity compared with what was observed with ERα alone (Fig. 1B, gray bars). The effect of ARNT on ERβ was more dramatic with a 13-fold increase in E2-induced transcriptional activity (Fig. 1B, gray bars). These results indicate ER subtype-specific differences in the ability of ARNT to enhance ER-dependent transcriptional activity. In the absence of ligand ARNT induced a moderate increase in luciferase activity of both ER subtypes. In the presence of tamoxifen neither ERα nor ERβ was able to induce reporter gene expression, and ARNT did not enhance ERα or ERβ transcriptional activity under these conditions (Fig. 1B, black bars).
These experiments suggest that ARNT can function as a coactivator of ERα- and ERβ-dependent transcription. We decided to compare ARNT with the p160 coactivator TIF-2, which modulates the response of numerous transcription factors, including nuclear hormone receptors. For this purpose, HeLa cells were transfected with expression vectors for ERα (Fig. 1C Left) or ERβ (Fig. 1C Right) and increasing concentrations of ARNT or TIF-2 expression vectors, and we compared the activity of the ERE-regulated luciferase reporter gene. Interestingly, both in the case of ERα and ERβ, the effect of ARNT in the presence of E2 is fully comparable with TIF-2 (Fig. 1C). A considerable increase in the transcriptional response by both ERα and ERβ in the presence of TIF-2 was also observed in the absence of ligand, whereas the effects of ARNT on both ERα and ERβ in the absence of ligand were considerably weaker, suggesting notable functional differences between ARNT and TIF-2. Interestingly, although ARNT interacts with both ERα and ERβ in the absence of ligand and in the presence of tamoxifen, a strong enhancing effect of ARNT on the transcriptional response was evident only in the presence of E2.
Taken together, these experiments demonstrate that ARNT functions as a potent coactivator of ER-dependent transcription, which probably is mediated via a direct interaction between ARNT and the ERs.
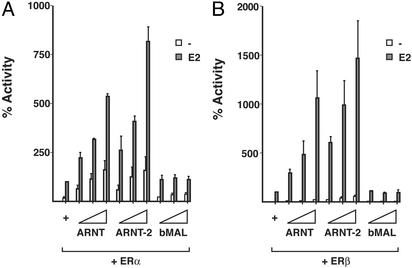
Differential Regulation of ER Activity by Different ARNT Family Members. The results presented above show that the bHLH–PAS factor ARNT strongly enhances ERα- and ERβ-dependent transcriptional activity. In mammalian cells two additional ARNT isoforms have been identified, ARNT-2 (13) and ARNT-3/bMAL (14). At the structural level, ARNT and ARNT-2 are very closely related, and homology with bMAL is relatively well conserved up to the PAS domain, whereas the C-terminal region differs considerably between bMAL and ARNT/ARNT-2. We tested the effect of the other ARNT family members on the transcriptional activity of ERα and ERβ. HeLa cells were cotransfected with expression vectors for ERα or ERβ together with the ERE-regulated luciferase reporter and increasing amounts of ARNT, ARNT-2, or bMAL. ARNT, as before, induced a significant increase in ERα and ERβ activity, and interestingly, ARNT-2 was even more efficient in enhancing ERα and ERβ activity. bMAL, however, failed to increase the transcriptional activity of either ERα or ERβ (Fig. 2 A and B, respectively). These results suggest that structural differences between ARNT family members dictate their ability to enhance ERα or ERβ transcriptional activity.
Fig. 2.
Differential enhancement of ER activity by different ARNT family members. HeLa cells were transfected as described in Fig. 1 with expression vectors for ERα (A) or ERβ (B), an ERE-regulated luciferase reporter gene construct, β-gal internal control, and increasing concentrations of ARNT, ARNT-2, or bMAL. After transfection, cells were treated with 10 nM E2 for 48 h. After this incubation, cells were harvested and luciferase andβ-gal activity was determined.
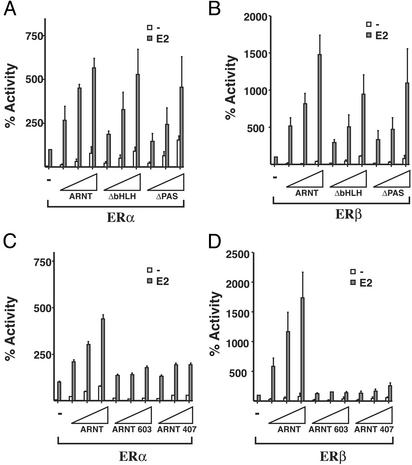
The C-Terminal Domain of ARNT Is Required for Coactivation of ERα and ERβ The observation that bMAL fails to enhance ERα or ERβ transcriptional activity suggests that functional differences among ARNT isoforms exist in their ability to function as coactivators for ERα or ERβ. Several studies have shown that the bHLH and PAS domains of ARNT are required to support the transcriptional activity of HIF-1α or AhR. We decided to investigate which domains of ARNT are required to enhance ERα or ERβ activity. For this purpose we used mutant forms of ARNT where various functional domains of ARNT are deleted. ARNT mutants lacking the bHLH or the PAS domains are unable to interact with AhR, thus obliterating TCDD-dependent transcription (19, 20). In contrast, deletion of the C-terminal transactivating region of ARNT does not interfere with its ability to dimerize with AhR, and the resulting complex is still transcriptionally active in response to TCDD (19, 21). We set out to investigate the role of the bHLH and PAS domains in ER/ARNT interaction. Furthermore, two deletion constructs, ARNT 603 (lacking 171 most C-terminal amino acids) or ARNT 407 (where 367 of the most C-terminal amino acids containing the entire transactivation domain is removed), were tested. HeLa cells were cotransfected with the ARNT mutants ΔbHLH or ΔPAS together with expression vectors for ERα or ERβ and the ERE-luciferase reporter. Interestingly, deletion of the bHLH or PAS domains of ARNT had very minor, if any, effect on the coactivating ability of ARNT on ERα or ERβ (Fig. 3 A and B, respectively). In contrast, deletion of the C-terminal domain severely compromised ARNT-dependent coactivation of both ERα and ERβ (Fig. 3 C and D, respectively). These observations demonstrate that ARNT requires the C-terminal domain for coactivation of ERα- or ERβ-dependent transcription. This domain, in contrast, is dispensable for activity of the AhR/ARNT complex, which instead relies on both the bHLH and PAS domains.
Fig. 3.
The C-terminal domain of ARNT is required to enhance ER activity. HeLa cells were transiently transfected with ERα (A and C)or ERβ (B and D) expression vectors, ERE-regulated reporter gene construct, β-gal internal control plasmid, and equal amounts of expression vectors for full-length ARNT or different deletion constructs as shown. Cells were treated and harvested as described in Fig. 1, and luciferase activity was determined and plotted.
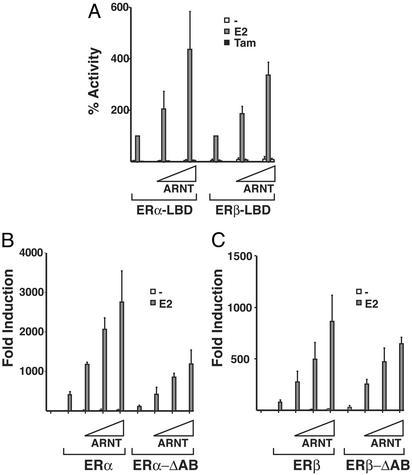
ARNT Interacts with the LBD of ERα and ERβ We decided to identify the domains of ERα and ERβ that are involved in the interaction with ARNT. Coactivators such as TIF-2 have been shown to interact both with the LBD and the N-terminal AF-1 transcriptional AF of nuclear receptors. Binding of agonists induces a rearrangement of the helical structure of the LBD, creating a hydrophobic cleft, which allows interaction with LxxLL motifs, known as NR boxes, present in p160 coactivators. ARNT is related to the p160 class of coactivators (PAS domain family), but contains no obvious consensus LxxLL motifs, indicating that the interaction of ARNT with the ERs may differ from that of classical coactivators. To investigate the role of the LBD of the ERs, in interaction with ARNT without interference from the N-terminal transactivation function, fusion constructs of the LBDs of ERα and ERβ linked to the DBD of the yeast factor Gal4 were used. These chimeric constructs were cotransfected into HeLa cells together with ARNT and a Gal4-regulated luciferase reporter plasmid. As shown in Fig. 4A, ARNT was able to increase the transcriptional response of both Gal4-ERα LBD and Gal4-ERβ LBD, demonstrating that the ERα/β LBD is sufficient to mediate interaction with ARNT. However, in contrast to our observations with the full-length proteins where the effects of ARNT were considerably stronger on ERβ, Gal4-ERα LBD and Gal4-ERβ LBD displayed a similar increase in transactivation in the presence of ARNT, suggesting differential involvement of N-terminal domains of the ERs. We verified these observations with mutant forms of ERα and ERβ with deletions of the N-terminal A/B domains, ERα-ΔA/B or ERβ-ΔA/B, respectively. These receptor forms retain their ERE-binding ability and activate transcription in a ligand-dependent manner through the AF-2 region (12).
Fig. 4.
ARNT interacts with the C-terminal domain of the ERs. (A) HeLa cells were transfected with expression vectors coding for Gal4 fusion proteins of ERα or ERβ together with a Gal4-regulated luciferase reporter construct. After transfection, cells were treated with ligands as in Fig. 1, and the transcriptional response was measured. Luciferase activity from cells transfected with E2-activated Gal-ERα LBD or Gal-ERβ LBD in the absence of ARNT was arbitrarily set to 100%, and the effect of ARNT was compared with this value. HeLa cells were transfected with expression vectors for ERα (B) or ERβ (C) and corresponding deletion constructs where the N-terminal A/B domain was removed, ERE-regulated luciferase reporter gene construct, β-gal internal control, and increasing concentrations of ARNT. The cells were treated with E2 for 48 h and harvested, and luciferase activity was measured and corrected against the internal β-gal transfection control. The reporter activity in nontreated cells in the absence of ARNT and ERα (B)or ERβ (C) was arbitrarily set to one for comparison.
Deletion of the AF-1 domain of ERα has a strong negative effect on ERα transcriptional activity; however, a similar deletion in ERβ causes only a minor effect, suggesting clear functional differences between the AF-1 domains of ERα and ERβ (12).
Expression vectors encoding ERα-ΔA/B or ERβ-ΔA/B were introduced into HeLa cells together with ARNT and the ERE reporter. ARNT amplified the transcriptional responses of ERα-ΔA/B and ERβ-ΔA/B to a similar extent (Fig. 4 B and C, respectively). These experiments suggest that the LBDs of ERα and ERβ are involved in the interaction with ARNT and that ARNT augments AF-2-dependent transcriptional activity. In the absence of the respective AF-1, ARNT coactivated ERα and ERβ to a similar extent, whereas the effect of ARNT was significantly more potent on ERβ in the context of the full-length receptors. These observations suggest that ARNT is able to mediate cooperativity between the ER AF-1 and AF-2, but that this occurs more efficiently in ERβ than ERα (see Fig. 1C for comparison).
ARNT Is Recruited to an Endogenous E2-Regulated Promoter. The results presented above suggest that ARNT can function as an ERα or ERβ coactivator. To further elucidate the role of ARNT as a coactivator for E2-dependent transcription, we performed ChIP experiments using the pS2 promoter. pS2 is a well characterized E2 responsive gene expressed in breast cancer cell lines. For this experiment we used T47D, a breast cancer cell line, because these cells have been shown to express endogenous ERα and ARNT. T47D cells are also able to sustain both E2 and dioxin signaling pathways, confirming functionality of the expressed ERα and ARNT proteins.
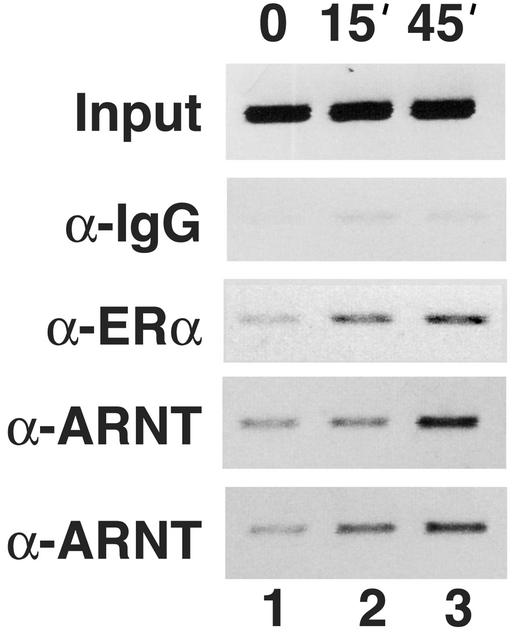
T47D cells were treated with 10 nM E2 for 0, 15, or 45 min. After treatment, the cells were harvested, and proteins were cross-linked to chromatin and immunoprecipitated with antibodies against ERα or ARNT (commercial antibody or ref. 18). The ChIP analysis was performed with primers specific for the pS2 promoter. ERα displayed a clear time-dependent recruitment to the pS2 promoter (Fig. 5). Interestingly, ChIP using two different ARNT antibodies also revealed a distinct time-dependent recruitment of ARNT to the pS2 promoter (Fig. 5). This experiment shows that ARNT is recruited to an endogenous ER-regulated promoter in an E2-dependent fashion, demonstrating a functional interaction between ERα and ARNT occurring in an in vivo setting.
Fig. 5.
Time-dependent recruitment of ARNT to the estrogen-responsive pS2 promoter. T47D cells, cultured in the absence of estrogen, were treated without (time 0) and with 10 nM E2 for 15 and 45 min. Soluble chromatin was prepared and immunoprecipitated by using antibodies raised against rabbit IgG (α-IgG) as negative control, ERα (α-ERα), and two different ARNT antibodies (α-ARNT1). Immunoprecipitated DNA was PCR-amplified with primers that span the -353 to -54 region of the pS2 promoter.
Taken together, these findings establish ARNT as a potent ER coactivator. The result from the ChIP assay strongly supports a physiological role of ARNT in ER-dependent transcription.
Discussion
The transition of the ERs to a transcriptionally active form is a multistep process where binding of an agonist results in a structural rearrangement of the receptor, leading to release of repressors and allowing interaction with coactivators like TIF-2 (22). Given the structural similarities between ARNT and p160 coactivators we investigated whether ARNT proteins might be involved in ER signaling. In this study, we observed a physical interaction between ARNT and both ER subtypes. In functional assays, ARNT was found to potently enhance ER-dependent transcription, with a noteworthy preference for the ERβ subtype. Moreover, ARNT was efficiently recruited together with ERα to the pS2 promoter in an E2-dependent fashion. ARNT-2 (13) and bMAL (14) are two mammalian factors closely related to ARNT. Interestingly, whereas ARNT-2, similar to ARNT, efficiently enhanced ERα and ERβ transcriptional activity, bMAL was unable to act as a coactivator for either ERα or ERβ. Sequence alignment of the three ARNT subtypes shows extensive similarity in the bHLH and PAS domains. However, the region located C-terminally of the PAS domain is well conserved only between ARNT and ARNT-2. This finding suggests that the C-terminal part of ARNT and ARNT-2 may be involved in the coactivating effect of ERα and ERβ. These results prompted us to delineate the domains of ARNT conferring the enhancing effect on ER activity. For this purpose, different deletion constructs of ARNT were used. The C-terminal transactivating region of ARNT is required for coactivation of ERα and ERβ, whereas removal of the bHLH or the PAS domains had little effect on ARNT's ability to enhance ER-dependent transcription. These observations demonstrate that the interaction between the ERs and ARNT represents a functionally distinct mechanism compared with ARNT's role as a dimerization partner for the AhR or HIF-1α. Deletion of the bHLH or PAS domain of ARNT severely compromises AhR and HIF-1α activity because of failure of the mutant forms of ARNT to dimerize with the AhR (20) and HIF-1α (23). Deletion of the C-terminal transactivation function of ARNT, on the other hand, does not restrict ARNT/AhR interaction and has no effect on AhR-mediated transcription (19, 24). In terms of TCDD signaling, the transactivation function of ARNT is thus dispensable for AhR-dependent transcription, and its role in this respect has remained unclear (19, 24). In contrast, our results show that the C-terminal domain of ARNT is clearly required for coactivation of ERα and ERβ. The mechanism whereby ARNT enhances ER-dependent transcription may involve cooperativity between the transactivation functions of ERs and ARNT. This type of crosstalk has been described between STAT5 and the glucocorticoid receptor (GR), where GR has been demonstrated to enhance prolactin-induced STAT5-dependent transcription of the β-casein promoter. Interaction between GR and STAT5 directs the strong N-terminal transactivation function of GR to work in concert with STAT5 and increase transcription (25). The precise region of ARNT involved in interaction with the ERs remains to be determined; however, it is interesting to note that the critical domain required to sustain ERα or ERβ transcriptional activity is distinct from those used in dimerization of ARNT with the AhR or HIF-1α. The mechanism of ARNT/ER interaction also appears to differ from that of “classical” coactivators such as TIF-2. The interaction between the ERs and TIF-2 depends on so-called NR boxes, present in TIF-2, which are composed of LxxLL motifs. These motifs mediate contact with the hydrophobic cleft presented in the agonist bound conformation of the ER LBD. The C-terminal domain of ARNT, however, contains no obvious LxxLL sequences, and its interaction with the ERs may occur independently of the hydrophobic pocket, suggesting a novel coactivation mechanism of the ERs.
Interaction between p160 coactivators and the nuclear receptor depends on the LBD. The absence of consensus LxxLL motifs in the C-terminal domain of ARNT prompted us to investigate the role of the ER LBD in ARNT interaction. We observed that ARNT was able to enhance the transcriptional activity of the LBD of both ERα and ERβ when fused to the Gal4 DBD. It is interesting to note that the Gal4–ER–LBD fusion constructs did not display any ligand-independent activity in the presence of ARNT, suggesting that the E2-independent increase in the ER's transcriptional activity by ARNT requires the ER's N terminus. This model is corroborated by our results using the A/B domain deletion mutants, where the effect of ARNT also was strictly ligand-dependent.
Comparison of the transcriptional magnitude of the ERs in the presence of ARNT revealed differences between the ER subtypes, where ARNT enhanced the transcriptional activity of ERβ more than twice as potently as that of ERα. However, when coactivation by ARNT was studied in context of the isolated LBDs of ERα and ERβ, there were no significant differences in the effect of ARNT on the two ER subtypes, indicating that ARNT may be more efficient in supporting synergy between AF-1 and AF-2 of ERβ, compared with ERα. This is an interesting observation in view of several reports describing low or absent ERβ AF-1 activity (12, 26). Apparently, ERβ AF-1 differs from ERα in the specificity of interaction with accessory factors, where classic coactivators of the p160 class apparently fail to sustain activity of ERβ AF-1 in most systems tested. The fact that ARNT is equally efficient in enhancing the activity of the respective ERα and ERβ AF-2s suggests that the ability of ARNT to more potently enhance ERβ transcription is not caused by the inherent differences in the LBDs of the ERs, but rather depends on differences in the respective A/B domains. Confirming this notion is the observation that ARNT displayed similar potency in enhancing the activity of ERα and ERβ N-terminal deletions. The effects of ARNT on ER signaling were also compared with a classic nuclear receptor coactivator, TIF-2. Interestingly, the effects of ARNT on ERα and ERβ transcriptional activity are fully comparable with those elicited by TIF-2, indicating that ARNT is indeed a bona fide coactivator of ER-dependent transcription. Moreover, our results showing E2-induced recruitment of endogenous ARNT present in T47D cells to the promoter of the natural ER target gene pS2 demonstrate a functional interaction between ARNT and ERα taking place in a physiological setting.
The functional implications of ARNT/ER interaction need to be further elucidated. Our results may add an additional level to the TCDD-mediated interference of E2 signaling, a phenomenon often referred to as endocrine disruption. TCDD inhibits estrogen-dependent biological responses, such as increase in uterine wet weight and peroxidase activity, and also decreases cellular estrogen and progesterone receptor levels (27). In addition, TCDD inhibits transcription of estrogen-regulated genes such as cathepsin D, c-fos, pS2, and Hsp27 in the human breast cancer cell line MCF-7 (28, 29, 30, 31). These effects involve DNA binding interference by the AhR/ARNT complex on ERE-regulated promoters. Our results presented here add an additional possibility, namely that cellular availability of ARNT may also be an important factor in regulating ERα and ERβ transcriptional activity.
In conclusion, our results expand the previously known cellular functions of the bHLH–PAS factor ARNT to include a potent coactivating function for ERα and especially ERβ.
Acknowledgments
This work was supported by grants to I.P. from the Swedish Cancer Foundation, the Åke Wiberg Foundation, and the Magnus Bergvall Foundation, and grants to A.H. and S.B. from the Foundation for Strategic Environmental Research and the Swedish Council for Environment Agricultural Sciences and Spatial Planning, and grants to K.P. from the Tore Nilsson Fund for Medical Research and the Lars Hierta Foundation. I.P. is supported by a fellowship from the Swedish Medical Research Council. K.P. is a postdoctoral fellow with the Swedish Society for Medical Research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ER, estrogen receptor; TIF, transcription intermediary factor; DBD, DNA binding domain; LBD, ligand binding domain; TCDD, 2,3,7,8-tetrachloro-dibenzo-p-dioxin; bHLH, basic helix–loop–helix; PAS, Per-ARNT-Sim; AF, activation function; AhR, aryl hydrocarbon receptor; ARNT, AhR nuclear translocator; HIF, hypoxia inducible factor; β-gal, β-galactosidase; ChIP, chromatin immunoprecipitation; ERE, estrogen response element.
References
- 1.Nilsson, S., Makela, S., Treuter, E., Tujague, M., Thomsen, J., Andersson, G., Enmark, E., Pettersson, K., Warner, M. & Gustafsson, J. A. (2001) Physiol. Rev. 81, 1535–1565. [DOI] [PubMed] [Google Scholar]
- 2.Pettersson, K. & Gustafsson, J. (2001) Annu. Rev. Physiol. 63, 165–192. [DOI] [PubMed] [Google Scholar]
- 3.Aranda, A. & Pascual, A. (2001) Physiol. Rev. 81, 1269–1304. [DOI] [PubMed] [Google Scholar]
- 4.Urnov, F. D. & Wolffe, A. P. (2001) Oncogene 20, 2991–3006. [DOI] [PubMed] [Google Scholar]
- 5.Narlikar, G. J., Fan, H. Y. & Kingston, R. E. (2002) Cell 108, 475–487. [DOI] [PubMed] [Google Scholar]
- 6.Gu, Y.-Z., Hogenesch, J. B. & Bradfield, C. A. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 519–561. [DOI] [PubMed] [Google Scholar]
- 7.Gekakis, N., Staknis, D., Nguyen, H. B., Davis, F. C., Wilsbacher, L. D., King, D. P., Takahashi, J. S. & Weitz, C. J. (1998) Science 280, 1564–1569. [DOI] [PubMed] [Google Scholar]
- 8.Darlington, T. K., Wager-Smith, K., Ceriani, M. F., Staknis, D., Gekakis, N., Steeves, T. D. L., Weitz, C. J., Takahashi, J. S. & Kay, S. A. (1998) Science 280, 1599–1603. [DOI] [PubMed] [Google Scholar]
- 9.Pongratz, I., Antonsson, C., Whitelaw, M. L. & Poellinger, L. (1998) Mol. Cell. Biol. 18, 4079–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitelaw, M. L., Gustafsson, J. A. & Poellinger, L. (1994) Mol. Cell. Biol. 14, 8343–8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pettersson, K., Delaunay, F. & Gustafsson, J. A. (2000) Oncogene 19, 4970–4978. [DOI] [PubMed] [Google Scholar]
- 12.Delaunay, F., Pettersson, K., Tujague, M. & Gustafsson, J. A. (2000) Mol. Pharmacol. 58, 584–590. [DOI] [PubMed] [Google Scholar]
- 13.Hirose, K., Morita, M., Ema, M., Mimura, J., Hamada, H., Fujii, H., Saijo, Y., Gotoh, O., Sogawa, K. & Fujii-Kuriyama, Y. (1996) Mol. Cell. Biol. 16, 1706–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda, M. & Nomura, M. (1997) Biochem. Biophys. Res. Commun. 233, 258–264. [DOI] [PubMed] [Google Scholar]
- 15.Berg, P. & Pongratz, I. (2001) J. Biol. Chem. 276, 43231–43238. [DOI] [PubMed] [Google Scholar]
- 16.Burakov, D., Crofts, L. A., Chang, C. P. & Freedman, L. P. (2002) J. Biol. Chem. 277, 14359–14362. [DOI] [PubMed] [Google Scholar]
- 17.Shang, Y., Hu, X., DiRenzo, J., Lazar, M. A. & Brown, M. (2000) Cell 103, 843–852. [DOI] [PubMed] [Google Scholar]
- 18.Mason, G. G. F., Witte, A. M., Whitelaw, M. L., Antonsson, C., McGuire, J., Wilhelmsson, A., Poellinger, L. & Gustafsson, J. A. (1994) J. Biol. Chem. 269, 4438–4449. [PubMed] [Google Scholar]
- 19.Reisz-Porszasz, S., Probst, M. R., Fukunaga, B. N. & Hankinson, O. (1994) Mol. Cell. Biol. 14, 6075–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitelaw, M., Pongratz, I., Wilhelmsson, A., Gustafsson, J. A. & Poellinger, L. (1993) Mol. Cell. Biol. 13, 2504–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, H., Dong, L. & Whitlock, J. P. J. (1994) J. Biol. Chem. 269, 28098–28105. [PubMed] [Google Scholar]
- 22.Klinge, C. M. (2000) Steroids 65, 227–251. [DOI] [PubMed] [Google Scholar]
- 23.Wood, S. M., Gleadle, J. M., Pugh, C. W., Hankinson, O. & Ratcliffe, P. J. (1996) J. Biol. Chem. 271, 15117–15123. [DOI] [PubMed] [Google Scholar]
- 24.Ko, H. S. P., Okino, S. T., Ma, Q. & Whitlock, J. P. (1996) Mol. Cell. Biol. 16, 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cella, N., Groner, B. & Hynes, N. E. (1998) Mol. Cell. Biol. 18, 1783–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowley, S. M. & Parker, M. G. (1999) J. Steroid Biochem. Mol. Biol. 69, 165–175. [DOI] [PubMed] [Google Scholar]
- 27.Ahlborg, U. G., Lipworth, L., Titus-Ernstoff, L., Hsieh, C. C., Hanberg, A., Baron, J., Trichopoulos, D. & Adami, H. O. (1995) Crit. Rev. Toxicol. 25, 463–531. [DOI] [PubMed] [Google Scholar]
- 28.Porter, W., Wang, F., Duan, R., Qin, C., Castro-Rivera, E., Kim, K. & Safe, S. (2001) J. Mol. Endocrinol. 26, 31–42. [DOI] [PubMed] [Google Scholar]
- 29.Krishnan, V., Porter, W., Santostefano, M., Wang, X. H. & Safe, S. (1995) Mol. Cell. Biol. 15, 6710–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gillesby, B. E., Stanostefano, M., Porter, W., Safe, S., Wu, Z. F. & Zacharewski, T. R. (1997) Biochemistry 36, 6080–6089. [DOI] [PubMed] [Google Scholar]
- 31.Duan, R., Porter, W., Samudio, I., Vyhlidal, C., Kladde, M. & Safe, S. (1999) Mol. Endocrinol. 13, 1511–1521. [DOI] [PubMed] [Google Scholar]