Abstract
Cytokine-provided survival signals are known to suppress apoptosis through inhibition of mitochondrial pathways that involve Bcl-2 family members. Here we show that in hematopoietic cells, cytokines also regulate death receptor-mediated pathways. We demonstrate that hematopoietic cytokines such as IL-3 and erythropoietin in normal cells, as well as BCR-ABL oncoprotein in transformed cells, inhibit transcription of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). Using small interfering RNAs, we show that the inhibition of TRAIL function is sufficient to partially rescue cytokine-deprived cells from apoptosis. Finally, we demonstrate that cytokine and BCR-ABL suppression of TRAIL transcription is mediated through phosphorylation and inhibition of the forkhead FOXO3a transcription factor. BCR-ABL-induced inhibition of TRAIL transcription in hematopoietic cells may provide a novel mechanism for tumorigenicity in chronic myeloid leukemia.
The development of hematopoietic cells requires a delicate balance between proliferation and apoptosis that is regulated in part by hematopoietic cytokines (1). Apoptosis occurs as a default mechanism in cells that fail to receive extracellular survival signals and plays a critical role in controlling the number of cells by eliminating unneeded or damaged cells. In the mammalian system, apoptosis is regulated by two known pathways, the extrinsic death receptor and intrinsic mitochondrial pathways, either alone or through cross talk (1). Stimulation of either of these pathways results in activation of caspases (2). The mitochondrial pathway involves a set of proapoptotic (Bid, Bad, Hrk, and Bim) and antiapoptotic (Bcl-2, Bcl-xL, and Mcl-1) members of the Bcl-2 family that together control mitochondrial membrane permeability (1). The extrinsic apoptotic pathway is activated by stimulation of death receptors, including those for tumor necrosis factor (TNF), CD95/Fas ligand (FasL) or TNF-related apoptosis-inducing ligand (Apo-2L/TRAIL), through binding of their ligands, resulting in activation of a caspase cascade and subsequent cell death (3). This pathway plays an important role in the immune response involved in the elimination of transformed cells (3).
Hematopoietic progenitor cells deprived of essential cytokines undergo mitogenic arrest followed by apoptosis (4). In contrast, oncogenically transformed leukemic cells continuously proliferate and evade apoptosis even in the absence of cytokines (5). A well characterized example of one such protein causing leukemic transformation is the constitutively active protein tyrosine kinase BCR-ABL (6). Expression of the chimeric 210-kDa BCR-ABL (P210) in hematopoietic stem cells results in chronic myeloid leukemia, a disease characterized by abnormal cell cycling during the initial chronic phase and resistance to apoptosis during the blast crisis (7, 8). These features are induced by sustained activation of BCR-ABL protein tyrosine kinase signaling pathways in hematopoietic cells (7, 8). Many of the same signaling pathways are also activated by cytokines such as IL-3, granulocytic–macrophage colony-stimulating factor (GM-CSF), or erythropoietin (Epo) in hematopoietic cells (9, 10).
In cytokine-stimulated or oncogenically transformed cells, such as cells transformed by BCR-ABL, survival signals are generated partly through functional or transcriptional regulation of Bcl-2 family members (11–13).
Hematopoietic cytokines have not been found to regulate FasL, a member of the death-inducing TNF family (14), and it is unclear whether cytokines promote survival by regulation of the extrinsic apoptotic pathway in addition to functional or transcriptional modulation of Bcl-2 proteins. Here, we demonstrate that hematopoietic cytokines in normal cells, and BCR-ABL oncoprotein in transformed cells, promote survival through inhibition of the expression of the TNF family member TRAIL, an activator of the extrinsic apoptotic pathways. We further demonstrate that cytokine or BCR-ABL regulation of the extrinsic apoptotic pathway is mediated, at least partly, through phosphorylation and inhibition of a member of FOXO family of transcription factors.
Materials and Methods
Cells. BaF3 cells were maintained in RPMI supplemented with 10% FCS and 10% WEHI cell conditioned media (source of IL-3). BaF3 cells were retrovirally transduced with murine stem-cell virus–internal ribosomal entry site-GFP (MIG)-P210 (fluorescence-activated cell sorting of GFP-positive cells) or with MSCV-P210-pac (puromycin-resistant cells were selected; experiments in Fig. 5) to generate BaF3P210 (10, 15). UT7 cells were electroporated with the ecotropic retroviral receptor, and puromycin-resistant UT7/Eco cells were selected (16) and transduced with MIG-P210 (15) to generate UT7-P210 cells. UT7-P210 were maintained in α-MEM (GIBCO/BRL) containing 10% FCS and 2 ng/ml GM-CSF. AS-E2 cells (obtained from Chugai Pharmaceutical, Tokyo; ref. 17) were maintained in Iscove's modified Dulbecco's media (GIBCO/BRL) supplemented with 20% FCS and 2 units/ml Epo. Expression of tyrosine-phosphorylated P210 was verified in BaF3P210 and UT7-P210 cells.
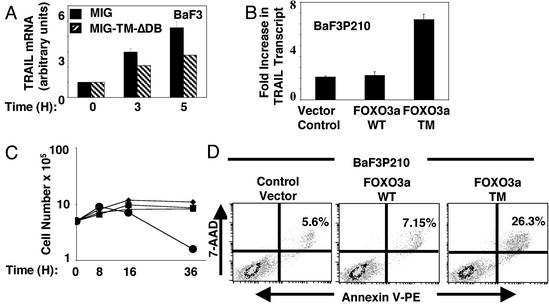
Fig. 5.
IL3 and BCR-ABL inhibition of TRAIL expression is mediated by FOXO3a. (A) BaF3 cells were retrovirally transduced with either an empty vector (MIG) or a vector containing a dominant inhibitory form of FOXO3a TM (MIG-TM-δDB). Cells were maintained in BaF3 media for 24 h, washed four times in RPMI, and cultured in RPMI with 10% FCS in the absence of IL-3 for the indicated times, and the fold increase in TRAIL transcript was measured. Results represent the mean and SEM of three independent experiments. (B) BaF3P210 cells were retrovirally transduced with either MIG vector alone or a vector containing WT (FOXO3a WT) or constitutively active FOXOa3 (FOXO3a TM), and GFP-positive cells were sorted with fluorescence-activated cell sorting and subjected to IL-3 withdrawal for 8 h, after which TRAIL transcript was measured by real-time PCR as indicated in Fig. 1B. Results are shown as fold increase of TRAIL transcript in BaF3P210 cells cultured in the absence as compared with the presence of IL-3. One representative of two independent experiments performed in triplicate is shown. (C) Growth curve of BaF3P210 cells from B cultured for the indicated times in RPMI containing 10% FCS in the absence of IL-3 [vector control (▪), FOXO3a WT (▴), or FOXO3a TM (•)] or BaF3P210 cells transduced with vector alone cultured in RPMI containing 10% FCS with IL-3 (♦). (D) BaF3 P210 cells from B cultured in RPMI containing 10% FCS in the absence of IL-3 for 8 h and used in the annexin V-binding assay. Percentage of cells positive for both annexin V and 7-amino actinomycin D is shown.
Plasmid Construction. The MIG-FOXO3a WT and MIG-FOXO3a triple mutant vectors were constructed by blunt-end ligation of corresponding cDNAs into the HpaI site of the MIG vector. Reporter genes were generated by inserting annealed oligonucleotides containing either the WT forkhead consensus-binding site within the human TRAIL promoter (FBST) or its mutants (Fig. 3) into the KpnI/PstI site of pGL-3-TATA-Luc (kindly provided by X. Hua, University of Pennsylvania, Philadelphia).
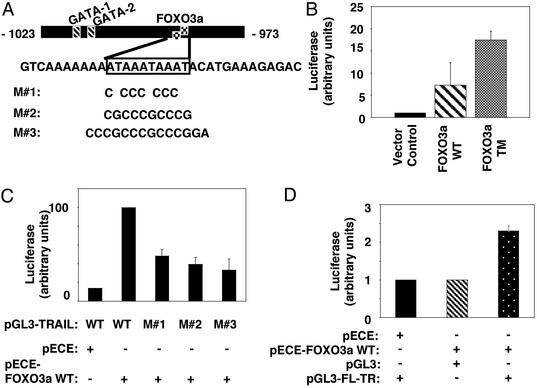
Fig. 3.
FOXO3a activates TRAIL transcription. (A) WT pGL3-TRAIL luciferase construct contains the sequence–1023 to –973 relative to the translation initiation site of the human TRAIL promoter that encompasses FBST. pGL3-TRAIL with partial (M#1) or complete (M#2) mutated FBST, or with the two FBST flanking oligonucleotides mutated in addition to the complete mutation of FBST (M#3) were used in C.(B) 293T cells were transfected with 50 ng of the pRL-TK vector (Promega) along with 2 μg of the empty vector pECE, pECE-FOXO3a WT, or pECE-FOXO3a TM, and 2 μg of WT pGL3-TRAIL. Twenty-four hours after transfection, cells were serum starved in 0.5% FCS for 16–18 h, harvested, and lysed in 250 μl of PLB lysis buffer (Promega). Luciferase (firefly and Renilla) activities were determined by using the dual luciferase assay system. Firefly luciferase activity was normalized to the corresponding Renilla luciferase activity, and results represent mean and SEM of luciferase activity of at least three independent experiments performed in triplicate. (C) Experiments performed as in B; luciferase activity obtained from cotransfection of pECE-FOXO3a WT or pECE empty vector with WT or mutant pGL3-TRAIL reporter constructs in 293T cells is shown. Results represent mean and SEM of luciferase activity of at least three independent experiments. (D) Experiments performed as in B; full-length human TRAIL promoter (–1152 to +1) (27) driving the luciferase gene (pGL-3-FL-TR) was cotransfected with either an empty (pECE) vector or with pECE-FOXO3a WT in 293T cells. An empty pGL3 luciferase vector cotransfected with pECE-FOXO3aWTwas used as a control. Results represent mean and SEM of luciferase activity from at least three independent experiments.
Real-Time Quantitative RT-PCR. Total RNA was isolated by using the RNeasy Mini Kit (Qiagen, Chatsworth, CA); 3 μg for detection of mouse TRAIL or 1 μg for mouse Bim and human TRAIL were included in a 50-μl reaction containing 25 μl of 2× TaqMan universal PCR master mix, 1.25 μl of 40× Multscribe Reverse Transcriptase/RNase inhibitor mix (PE Applied Biosystems), forward and reverse primers (900 nM), fluorogenic TaqMan probe (200 nM), and RNase/Dnase free water. The thermal cycling conditions of 30 min at 48°C, an initial denaturation step for 10 min at 95°C, and 40 cycles at 95°C for 15 s and 60°C for 1 min were used. Thermal cycling was performed on an ABI Prism 7700 Sequence Detector (PE Applied Biosystems). Primers and Probes are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
RNA Interference. Twenty-one nucleotide complementary RNAs with Phos-symmetrical two-nucleotide overhangs to the following regions of mouse TRAIL (bases 278–300 or 328–350) and to 801–823 of Lamin A/C were obtained from Dharmacon (Lafayette, CO). On average, 200 million BaF3 cells were electroporated with TRAIL or with control siRNA duplexes. Eight million BaF3 cells in 0.8 ml of RPMI were aliquoted per cuvette and electroporated with 160 nM annealed oligonucleotides. Cells were coelectroporated with FITC-conjugated randomly generated annealed oligonucleotides 5′-ACUGAACCCGUUUCCACCUTT-3′ and 5′-FITC-AGGUGGAAACGGGUUCAGUTT-3′.
Electrophoretic Mobility-Shift Assay. Radiolabeled probes were prepared by filling in a 5′ overhang in a Klenow reaction in the presence of α32P-labeled nucleotides. Oligonucleotides 5′-CTGTCAAAAAAATAAATAA ATACATGA A AGAGA-3′, WT, or 5′-CTGTCTTAATAGTTTAATTATACATGAAAGAGA-3′, mutant TRAIL probe were annealed to 5′-TCTCTTTCA-3′; the WT insulin-responsive sequence probe was described (18). Ten micrograms of nuclear extract and 2× DNA–protein-binding reaction buffer [25 mM Tris, pH 8.0/200 mM NaCl/2.5 mM MgCl2/15% glycerol/1mMDDT/100 μg/ml BSA)/1% Nonidet P-40/4 μg of poly dI-dC/poly dI-dC (Pharmacia)] were used in reactions, run on a 6% polyacrylamide gel in Tris-glycine buffer (25 mM Tris/190 mM glycine/1 mM EDTA) containing 5% glycerol, at 250 V for 120 min. Signals were detected on a Biomax MR (Kodak) film at–80°C.
Results
Cytokines and BCR-ABL Inhibit Transcription of TRAIL. TRAIL transcription, as measured by quantitative real-time PCR, was up-regulated as a result of withdrawal of IL-3, GM-CSF, or Epo in cytokine-dependent hematopoietic cell lines such as IL-3-dependent pro-B BaF3, GM-CSF-dependent primitive hematopoietic UT7, and Epo-dependent erythroid UT7/Epo, AS-E2, and HCD57 cells (Table 1). Cytokine-dependent hematopoietic cell lines that overexpress BCR-ABL can survive and proliferate in the absence of cytokines (ref. 9 and reviewed in ref. 19). Overexpression of BCR-ABL oncoprotein significantly suppressed the increase of TRAIL transcription that was observed in response to cytokine deprivation in both human UT7 and murine BaF3 cells. In particular, in IL-3-deprived BaF3 cells, the induction of TRAIL transcript and protein expression, which is suppressed by the expression of BCR-ABL, correlates with cell death (data not shown). Because BCR-ABL and IL-3 activate many of the same signaling pathways in BaF3 cells (reviewed in ref. 19), this observation suggests a common mechanism for the suppression of TRAIL transcript by BCR-ABL and IL-3. In contrast to TRAIL transcript, transcription of FasL is not detectable (Table 1). These findings are in agreement with the previous demonstration that FasL is not involved in apoptosis of cytokine-deprived hematopoietic cells (14).
Table 1. Real-time PCR analysis of apoptosis-inducing transcripts in cytokine-dependent hematopoietic cell lines.
| Cell lines | TRAIL | Bim | FasL | Time, h |
|---|---|---|---|---|
| Murine | ||||
| BaF3 | ||||
| + IL-3 | 1 | 1 | NDT | |
| - IL-3 | 6.2 ± 0.3 | 2.1 ± 0.73 | NDT | 8 |
| - IL-3/BCR-ABL | 2.2 ± 0.7 | 1.1 ± 0.15 | NDT | 8 |
| HCD57 | ||||
| + Epo | 1 | 1 | ND | |
| - Epo | 3.6 ± 0.02 | 4.8 ± 1.4 | ND | 19 |
| + Epo | 1 | ND | ND | |
| - Epo | 4.71 ± 1.37 | ND | ND | 33 |
| Human | ||||
| UT7 | ||||
| + GM-CSF | 1 | NDT | ||
| - GM-CSF | 2.23 ± 0.35 | NDT | 24 | |
| - GM-CSF/BCR-ABL | 1.05 ± 0.23 | NDT | 24 | |
| UT7/Epo | ||||
| + Epo | 1 | NDT | ||
| - Epo | 1.6 ± 0.21 | NDT | 18 | |
| AS-E2 | ||||
| + Epo | 1 | ND | ||
| - Epo | 2.35 ± 0.78 | ND | 24 | |
| + Epo | 1 | ND | ||
| - Epo | 4.6 ± 0.42 | ND | 50 |
A time course of induction of TRAIL, FasL, and Bim transcripts in response to cytokine deprivation was performed for each cell line, and the value shown corresponds to a particular time point during the starvation. The time points were chosen according to the known time of apoptotic induction and progression on cytokine withdrawal in each cell line. The fold induction in TRAIL transcript on cytokine deprivation is shown as the mean fold induction ± standard deviation. An internal GAPDH control was included throughout the experiments, and the relative ΔCt method was used to calculate the fold change in transcript in comparison to control unstarved cells, whose fold change value is set to 1. Results shown are average from at least two independent experiments performed in triplicate. NDT, not detectable transcript; ND, not done.
We, like others (20), found that Bim, a proapoptotic member of the Bcl-2 family of proteins, was moderately up-regulated in response to IL-3 withdrawal of IL-3-dependent pro-B BaF3 cells (Table 1) and significantly up-regulated in response to Epo withdrawal of Epo-dependent erythroleukemic HCD57 cells (Table 1). BCR-ABL expression in BaF3 cells reversed the transcriptional up-regulation of Bim observed in response to IL-3 withdrawal (Table 1). These findings indicate that, in addition to the intrinsic mitochondrial pathway, the extrinsic death receptor pathway, specifically TRAIL, is also transcriptionally regulated by cytokines and BCR-ABL oncoprotein in both murine and human cells.
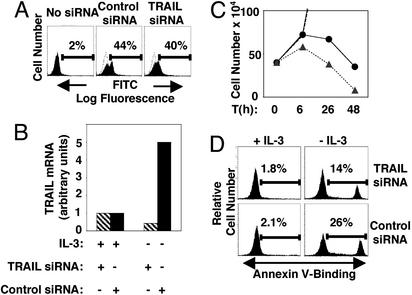
Inhibition by siRNA of TRAIL Production in IL-3-Deprived BaF3 Cells Enhances Survival. Using siRNA (21) to decrease levels of TRAIL that are normally induced in IL-3-starved BaF3 cells, we asked whether induction of TRAIL expression contributes to apoptosis of IL-3-deprived BaF3 cells (Fig. 1). BaF3 cells were electroporated either with siRNA duplices targeting TRAIL transcript or control duplices, along with FITC-labeled oligonucleotide duplices. FITC-positive electroporated cells were sorted with fluorescence-activated cell sorting (Fig. 1 A) and subjected to IL-3 withdrawal for 6 h. As shown in Fig. 1B, the expected 5-fold induction of TRAIL transcript observed in the cells electroporated with control-targeted siRNA was absent in cells with TRAIL-specific targeted siRNA (Fig. 1B). The annexin V-binding assay (Fig. 1D) showed that after6hof IL-3 deprivation, BaF3 cells containing TRAIL-specific siRNAs entered apoptosis at a significantly lower rate than cells with control siRNAs under the same conditions (Fig. 1D). In parallel, suppression of TRAIL transcription that is normally induced as a result of IL-3 withdrawal resulted in a slower and delayed apoptotic response of the BaF3 cells (Fig. 1C). These data implicate TRAIL as one of the factors inducing apoptosis in BaF3 cells in the absence of IL-3. Our data also suggest the existence of additional apoptotic signals, because suppression of TRAIL up-regulation delayed but did not prevent cell death in IL-3-deprived BaF3 cells.
Fig. 1.
Inhibitory targeting of TRAIL transcript by siRNA enhances survival of IL-3-deprived BaF3 cells. (A) BaF3 cells were electroporated with either control siRNA duplices or duplices of siRNAs targeting specifically the TRAIL transcript along with the FITC-conjugated duplex of randomly generated oligonucleotides. Overlapping histograms with dotted lines represent nonelectroporated BaF3 cells. (B) FITC-positive BaF3 cells from A were sorted with fluorescence-activated cell sorting and cultured for 48 h, then seeded at 4 × 105 cells per well with or without IL-3 for 6 h, and the TRAIL transcript was measured by real-time PCR. (C) Growth curve of trypan blue excluded live cells prepared in B: BaF3 cells in normal media with siRNA targeting TRAIL (▪), or with control siRNA (♦) (points showing number of live cells cultured in normal media after 26- and 48-h fall outside of the graph), and BaF3 cells cultured in the absence of IL-3 electroporated with siRNA targeting TRAIL (•, solid line) or with control siRNA (▴, dashed line). Time (T) is in hours (h). (D) After 6 h, IL-3-starved cells from B were used in an annexin V-binding assay; 7-amino actinomycin D negative cells are gated. One representative of two independent experiments is shown.
FOXO3a Activates TRAIL Transcription by Binding to TRAIL Promoter Sequences. Our previous work (15) suggested that the 14–3-3-binding region (22) of the BCR-ABL oncoprotein that is also required for optimum BCR-ABL activation of PI3-kinase/AKT (23) is essential for BCR-ABL support of survival and maturation of red cell progenitors in the absence of cytokines. 14-3-3 adaptor proteins (24) bind serine/threonine-phosphorylated targets of AKT and AKT-related kinases including the proapoptotic FOXO3a transcription factor (18), which, when unphosphorylated and active, enhances transcription of proapoptotic genes such as FasL and Bim (18, 20). Using the PATSCAN (www-unix.mcs.anl.gov/compbio/PatScan/HTML/patscan.html) and TFSEARCH (www.cbrc.jp/research/db/TFSEARCH.html) programs, we found that the human TRAIL promoter contains two overlapping FOXO3a consensus-binding sites [(FBST)–995 to –986 relative to the translation initiation site] and thus may be a putative target of forkhead FOXO3a transcription factor (Fig. 3A) that by binding to TRAIL promoter mediates activation of TRAIL transcription.
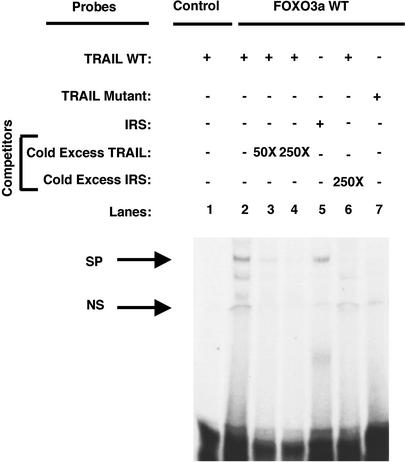
WT (Fig. 2, lane 2) and a constitutively active (data not shown) FOXO3a bound a radiolabeled TRAIL promoter probe (–1006 to –974 relative to the translation initiation site) encompassing FBST resulting in a shift in its mobility in a nondenaturing gel. The formation of the FOXO3a/TRAIL promoter sequence complex was inhibited by an excess of unlabeled TRAIL oligonucleotide (Fig. 2, lanes 3 and 4) or by an excess of insulin-responsive sequence oligonucleotide (Fig. 2, lane 6), known to bind FOXO3a (18) (Fig. 2, lane 5). Importantly, WT FOXO3a did not bind to a probe containing a mutant FBST (Fig. 2, lane 7), indicating the interaction between FOXO3a and TRAIL promoter sequence is specific.
Fig. 2.
FOXO3a binds to TRAIL promoter sequences. Gel shift: 293 T cells were transfected with pECE-FOXO3a WT (FOXO3a WT), and nuclear extracts were prepared. Radiolabeled probes (100,000 cpm) of either TRAIL WT (lanes 1–4 and 6) or TRAIL mutant (lane 7) probe, in which the putative forkhead-binding site was mutated, or of the insulin-responsive sequence (IRS) (used as a positive control, lane 5; ref. 18) were incubated at room temperature with 0 (lane 1) or 10 μg of nuclear extract (lanes 2–7) in binding reactions in the presence of 0 (lane 2), 50 (lane 3), or 250 (lanes 4 and 6) molar excess of either unlabeled TRAIL WT probe (lanes 3 and 4) or unlabeled IRS probe (lane 6). The upper arrow marks a specific (SP) DNA–protein shift. A nonspecific protein/DNA complex is also found (NS).
To examine the potential of FOXO3a to mediate the activation of TRAIL transcription, reporter constructs with a short sequence (–1023 to –973 relative to the translation initiation site of the human TRAIL promoter) containing the two canonical forkhead consensus-binding sites, or mutants in these sites, were used in a reporter gene expression assay (Fig. 3A). In the absence of serum, WT FOXO3a or the constitutively active FOXO3a (FOXO3a TM) induced high levels of luciferase reporter gene expression (Fig. 3B). Partial or complete mutation of FBST itself (Fig. 3C, M#1 and M#2), or its flanking residues (Fig. 3C, M#3), resulted in 30–40% of maximal luciferase reporter gene expression. Absence of complete abolition of luciferase activity by these mutants (Fig. 3C) suggests that nonconsensus forkhead-binding sequences are located outside of the mutated sequences but within the region–1023 to –973 of the human TRAIL promoter. These results are consistent with regulatory functions of residues outside of the core forkhead consensus-binding sites in high-affinity binding of forkhead transcription factors to their targets (25). Importantly, FBST falls within the region of the human TRAIL promoter with a major function in regulating basal TRAIL activity (26). In addition, FOXO3a transactivated the full-length ≈1.2-kb TRAIL promoter (pGL3-FL-TR) (27) in a reporter gene expression assay (Fig. 3D). Taken together, these data indicate that FOXO3a activates TRAIL expression by binding to its promoter.
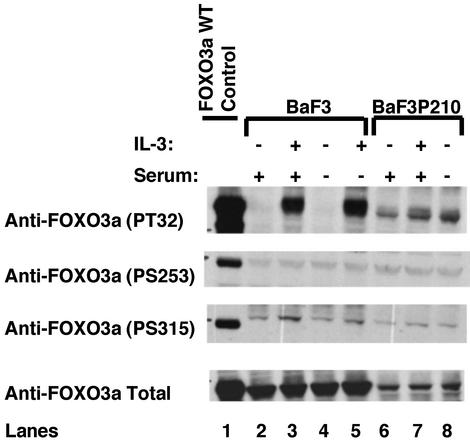
Suppression of TRAIL by IL-3 or BCR-ABL Is Mediated at Least Partly Through Inhibition of FOXO3a Transcription Factor. To address whether BCR-ABL and cytokines suppress TRAIL expression through inhibition of FOXO3a transcription factor, we first investigated whether cytokine stimulation and/or BCR-ABL expression results in FOXO3a phosphorylation. As shown in Fig. 4, FOXO3a was phosphorylated in response to IL-3 stimulation of starved BaF3 cells on T32 and S315 but not on S253. Proportionately more of the same FOXO3a residues were constitutively phosphorylated in BaF3P210 cells even in the absence of IL-3 and/or serum [Fig. 4, lanes 6 and 8; note that total FOXO3a protein levels (Fig. 4 Bottom) are lower in BaF3P210 cells, and thus a proportionately higher fraction of FOXO3a is phosphorylated]. Furthermore, IL-3 stimulation of IL-3-starved BaF3P210 cells did not induce any major changes in FOXO3a phosphorylation (Fig. 4, lane 7). In these experiments, 293T cells overexpressing FOXO3a were included as a positive control (lane 1). The FOXO3a (T32, S253, and S315) residues are phosphorylated with various degrees of specificity by AKT (18) and AKT-related serine threonine kinases of the SGK family in a PI3-kinase dependent manner (28). Both AKT and SGK kinases are known to phosphorylate T32, but AKT favors S253, and SGK has a marked preference for S315 (28). Our results suggest that SGK and not AKT may be the preferred kinase in phosphorylating FOXO3a in response to IL-3 stimulation or BCR-ABL expression in BaF3 cells. In agreement with previous reports, we found that FOXO3a was relocalized to the cytoplasm, thus inhibited from activating its target, in response to IL-3 stimulation of starved BaF3 cells and in IL-3-deprived BaF3P210 cells (Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 4.
FOXO3a is phosphorylated and inhibited in BCR-ABL-expressing BaF3 cells. BaF3 (lanes 2–5) and BaF3P210 (lanes 6–8) cells starved from either IL-3 alone (in RPMI with 10% FCS; lanes 2, 3, 6, and 7) or IL-3 and serum (in RPMI with 1% BSA; lanes 4, 5, and 8) for 8 h were stimulated with 50 ng/ml IL-3 for 5 min (lanes 3, 5, and 7). Cell lysates were prepared as described (15), fractionated on a gradient SDS/PAGE, and transferred to nitrocellulose, and blots were probed with antiphospho-Thr-32, [1:750; anti-phospho-Ser-253, 1:500 (Upstate Biotechnology)] or antiphospho-Ser-315 (1:750; kindly provided by M. Greenberg) or with antitotal FOXO3a antibody. Bound antibodies were detected by the enhanced chemiluminescence system (DuPont–NEN). A control lysate of 293 T cells overexpressing FOXO3a WT was used as a positive control.
Next, we used an inhibitory mutant of FOXO3a (FOXO3a TM-δDB) (29) in BaF3 cells that is a deletion mutant of FOXO3a TM and, like FOXOa3 TM, is constitutively nuclear. FOXO3a TM-δDB also lacks the FOXO3a DNA-binding domain and acts in a dominant-negative fashion possibly by binding and depleting FOXO3a transcriptional cofactors (29). Ectopic expression of the FOXO3a TM-δDB (MIG-TM-δDB) but not the control vector (MIG) in BaF3 cells resulted in a moderate but significant decrease in TRAIL transcription that is normally induced in response to IL-3 withdrawal (Fig. 5A), indicating that induction of TRAIL expression in response to IL-3 withdrawal occurs at least partly through activation of FOXO3a. The moderate effect may be due to a limited dominant-negative function of FOXO3a TM-δDB. Overexpression of FOXO3a TM induced massive and rapid cell death in BaF3 cells even in the presence of IL-3 (data not shown), and a stable line could not be established. Our observations that (i) TRAIL expression was suppressed in BaF3P210 cells, (ii) FOXO3a was phosphorylated and inhibited in Baf3P210 cells, and (iii) FOXO3a activates TRAIL transcription through binding to TRAIL promoter suggest that inhibition of FOXO3a in BCR-ABL-transformed cells mediates suppression of TRAIL expression. Ectopic expression of a constitutively active mutant of FOXO3a (FOXO3a TM) but not WT FOXO3a in BaF3P210 cells indeed induced TRAIL transcription (Fig. 5B). In addition, expression of FOXO3a TM, but not WT FOXO3a, in BaF3P210 cells resulted in a rapid decline of the growth of these cells (Fig. 5C) and significant induction of apoptosis as detected by the annexin V-binding assay (Fig. 5D). These results demonstrate that inhibition of forkhead transcription factor in response to hematopoietic cytokines or BCR-ABL oncoprotein blocks TRAIL expression at the level of transcription, and that this results in suppression of apoptosis.
Discussion
TRAIL Induces Apoptosis of Cytokine-Deprived Hematopoietic Cells and Is Inhibited by BCR-ABL. TRAIL is a type II membrane protein and a member of the TNF family of ligands (30–32). TRAIL binds to its cognate death receptors 4 and 5 (DR4 and DR5) in humans and DR5 in mice leading to the activation of caspases and cell death. The physiological role of TRAIL in normal nonimmune hematopoietic development remains unknown (33). As a promising anticancer agent, recombinant soluble TRAIL induces apoptosis in a broad range of transformed cells, including in BCR-ABL positive chronic myeloid leukemia cells but rarely in nontransformed cells with little or no toxicity (reviewed in refs. 30 and 34–36). The regulation of TRAIL production is critical for determining the rate of apoptosis vs. survival of hematopoietic cells, because blocking TRAIL synthesis by siRNA significantly delayed apoptosis induced by cytokine deprivation. Understanding the physiological role of TRAIL in hematopoiesis will require further investigation. These findings have broad implications, because they suggest that cytokines, in addition to their direct cellular targets, regulate selectively survival of neighboring cells that express TRAIL receptors. By modulation of the extrinsic death pathway, TRAIL may thus affect the balance of apoptosis and cell division that controls hematopoietic cell production.
Cytokine and BCR-ABL-Mediated Inhibition of TRAIL Transcription Occurs via Phosphorylation and Inhibition of FOXO3a Transcription Factor. The forkhead family is a large class of eukaryotic winged helix transcription factors playing important roles in embryonic and adult development, including cellular proliferation, differentiation, apoptosis, cell cycle regulation, and DNA repair (37). In mammals, three members of the FOXO subfamily of the forkhead family have been identified that are homologs of the PI3-kinase/AKT target DAF-16 in Caenorhabditis elegans (38). FOXO transcription factors have also been identified at the sites of chromosomal rearrangements in certain human tumors and leukemias (37). Forkhead transcription factors share a 100-aa sequence, called the forkhead domain (forkhead box or FOX), that mediates their interaction with DNA (37). We found that TRAIL promoter contains two overlapping sequences, highly similar to the forkhead transcription factor consensus-binding sequence (RTAAAY), which bind FOXO3a, resulting in activation of TRAIL transcription (Figs. 2 and 3). We further demonstrate that inhibition of TRAIL transcription by cytokine stimulation or BCR-ABL expression is mediated by phosphorylation and inhibition of FOXO3a transcription factor (Figs. 4 and 5). Thus, by regulating both TRAIL and FasL (18), FOXO3a appears to be a key transcriptional regulator of several components of extrinsic death receptor pathways. However, FOXO3a activation of TRAIL (as demonstrated here) but not FasL (ref. 14 and Table 1) results in cell death in cytokine-deprived hematopoietic cells, indicating that FOXO3a induction of the extrinsic death pathways in response to cytokine withdrawal is mediated by TRAIL.
We have demonstrated that hematopoietic cytokines, in addition to modulating several intrinsic apoptotic pathways, suppress at least one extrinsic apoptotic pathway through inhibition of TRAIL synthesis. Equally importantly, we identified FOXO3a transcription factor as a mediator of transcriptional regulation of TRAIL expression downstream of cytokine receptors and the cytoplasmic BCR-ABL protein tyrosine kinase. In addition, our results provide a rationale for the use of TRAIL in the treatment of chronic myeloid leukemia patients (35). We propose that cytokine regulation of TRAIL expression may be a critical step in the control of hematopoietic cell development.
Supplementary Material
Acknowledgments
We thank Drs. Michael Greenberg and Anne Brunet (Harvard Medical School) for FOXO3a cDNAs and antibodies, especially the antiphospho FOXO3a-(S315) antibody, before publication. We thank Drs. Alex Almasan (Cleveland Clinic Foundation, Cleveland) for ApoP1152-Luc; and Olivier Kocher and Laura Benjamin (Beth Israel Hospital, Boston) for mouse and human GAPDH primer and probe sequences, respectively. We also thank Drs. Steven Johnston and Robert Latek (Whitehead Institute Biocomputing) for help with database analyses of forkhead targets and the TRAIL promoter; Brent Stockwell (Whitehead Institute) and Dominique Dumesnil (Institut Gustave Roussy, Villejuif, France) for advice on siRNA protocols; James Evans (Whitehead Institute) for advice on fluorescence microscopy; Noredin Benhaga (Beth Israel Hospital) for technical help; and Michael Doire (Massachusetts Institute of Technology Cancer Center) for cell sorting. We are grateful to Drs. George Daley, Joseph Marszalek, Moonkyoung Um, Chang-Zheng Chen, and Guang Wong (Whitehead Institute) and Shirley Stiver (Beth Israel Hospital) for helpful discussions and for critically reading the manuscript. This research was supported by Award EEC 9843342 through the Biotechnology Process Engineering Center (Massachusetts Institute of Technology) from the Engineering Research Centers Program of the National Science Foundation; National Institutes of Health (NIH) Grant HL 32262 (to H.F.L.); National Cancer Institute Clinician Scientist Career Award NIH5K08 (CA77675 to S.G.); NIH Howard Temin Award (CA78396); NIH Shannon Award (CA87675); and the Beth Israel Hospital Pathology Foundation fund (to R.K.). Z.J. was supported by NIH Training Grant T32GMO7196.
Abbreviations: TNF, tumor necrosis factor; TRAIL, TNF-related apoptosis-inducing ligand, FBST, forkhead consensus-binding site within the human TRAIL promoter; Epo, erythropoietin; FasL, CD95/Fas ligand; MIG, murine stem-cell virus–internal ribosomal entry site–GFP; siRNA, small interfering RNA; TM, triple mutant.
References
- 1.Cory, S. & Adams, J. M. (2002) Nat. Rev. Cancer 2, 647–656. [DOI] [PubMed] [Google Scholar]
- 2.Strasser, A., O'Connor, L. & Dixit, V. M. (2000) Annu. Rev. Biochem. 69, 217–245. [DOI] [PubMed] [Google Scholar]
- 3.Wallach, D., Varfolomeev, E. E., Malinin, N. L., Goltsev, Y. V., Kovalenko, A. V. & Boldin, M. P. (1999) Annu. Rev. Immunol. 17, 331–367. [DOI] [PubMed] [Google Scholar]
- 4.Pawson, T. & Hunter, T. (1994) Curr. Opin. Genet. Dev. 4, 1–4. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan, D. & Weinberg, R. A. (2000) Cell 100, 57–70. [DOI] [PubMed] [Google Scholar]
- 6.Sawyers, C. L. (1997) Baillieres Clin. Haematol. 10, 223–231. [DOI] [PubMed] [Google Scholar]
- 7.Sawyers, C. L. (1999) N. Engl. J. Med. 340, 1330–1340. [DOI] [PubMed] [Google Scholar]
- 8.Druker, B. J., Sawyers, C. L., Capdeville, R., Ford, J. M., Baccarani, M. & Goldman, J. M. (2001) in Hematology (Am. Soc. Hematol. Educ. Prog., Washington, DC), pp. 87–112. [DOI] [PubMed]
- 9.Daley, G. Q. & Baltimore, D. (1988) Proc. Natl. Acad. Sci. USA 85, 9312–9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghaffari, S., Wu, H., Gerlach, M., Han, Y., Lodish, H. F. & Daley, G. Q. (1999) Proc. Natl. Acad. Sci. USA 96, 13186–13190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cirinna, M., Trotta, R., Salomoni, P., Kossev, P., Wasik, M., Perrotti, D. & Calabretta, B. (2000) Blood 96, 3915–3921. [PubMed] [Google Scholar]
- 12.Neshat, M. S., Raitano, A. B., Wang, H. G., Reed, J. C. & Sawyers, C. L. (2000) Mol. Cell. Biol. 20, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gesbert, F. & Griffin, J. D. (2000) Blood 96, 2269–2276. [PubMed] [Google Scholar]
- 14.Dijkers, P. F., Birkenkamp, K. U., Lam, E. W., Thomas, N. S., Lammers, J. W., Koenderman, L. & Coffer, P. J. (2002) J. Cell Biol. 156, 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghaffari, S., Kitidis, C., Fleming, M. D., Neubauer, H., Pfeffer, K. & Lodish, H. F. (2001) Blood 98, 2948–2957. [DOI] [PubMed] [Google Scholar]
- 16.Komatsu, N., Nakauchi, H., Miwa, A., Ishihara, T., Eguchi, M., Moroi, M., Okada, M., Sato, Y., Wada, H., Yawata, Y., et al. (1991) Cancer Res. 51, 341–348. [PubMed] [Google Scholar]
- 17.Miyazaki, Y., Kuriyama, K., Higuchi, M., Tsushima, H., Sohda, H., Imai, N., Saito, M., Kondo, T. & Tomonaga, M. (1997) Leukemia 11, 1941–1949. [DOI] [PubMed] [Google Scholar]
- 18.Brunet, A., Bonni, A., Zigmond, M. J., Lin, M. Z., Juo, P., Hu, L. S., Anderson, M. J., Arden, K. C., Blenis, J. & Greenberg, M. E. (1999) Cell 96, 857–868. [DOI] [PubMed] [Google Scholar]
- 19.Ghaffari, S., Daley, G. Q. & Lodish, H. F. (1999) Leukemia 13, 1200–1206. [DOI] [PubMed] [Google Scholar]
- 20.Dijkers, P. F., Medema, R. H., Lammers, J. W., Koenderman, L. & Coffer, P. J. (2000) Curr. Biol. 10, 1201–1204. [DOI] [PubMed] [Google Scholar]
- 21.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 22.Reuther, G. W., Fu, H., Cripe, L. D., Collier, R. J. & Pendergast, A. M. (1994) Science 266, 129–133. [DOI] [PubMed] [Google Scholar]
- 23.Skorski, T., Bellacosa, A., Nieborowska-Skorska, M., Majewski, M., Martinez, R., Choi, J. K., Trotta, R., Wlodarski, P., Perrotti, D., Chan, T. O., et al. (1997) EMBO J. 16, 6151–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu, H., Xia, K., Pallas, D. C., Cui, C., Conroy, K., Narsimhan, R. P., Mamon, H., Collier, R. J. & Roberts, T. M. (1994) Science 266, 126–129. [DOI] [PubMed] [Google Scholar]
- 25.Pierrou, S., Hellqvist, M., Samuelsson, L., Enerback, S. & Carlsson, P. (1994) EMBO J. 13, 5002–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, Q., Ji, Y., Wang, X. & Evers, B. M. (2000) Biochem. Biophys. Res. Commun. 276, 466–471. [DOI] [PubMed] [Google Scholar]
- 27.Gong, B. & Almasan, A. (2000) Cancer Res. 60, 5754–5760. [PubMed] [Google Scholar]
- 28.Brunet, A., Park, J., Tran, H., Hu, L. S., Hemmings, B. A. & Greenberg, M. E. (2001) Mol. Cell. Biol. 21, 952–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran, H., Brunet, A., Grenier, J. M., Datta, S. R., Fornace, A. J., Jr., DiStefano, P. S., Chiang, L. W. & Greenberg, M. E. (2002) Science 296, 530–534. [DOI] [PubMed] [Google Scholar]
- 30.Abe, K., Kurakin, A., Mohseni-Maybodi, M., Kay, B. & Khosravi-Far, R. (2000) Ann. N.Y. Acad. Sci. 926, 52–63. [DOI] [PubMed] [Google Scholar]
- 31.Ashkenazi, A. & Dixit, V. M. (1999) Curr. Opin. Cell Biol. 11, 255–260. [DOI] [PubMed] [Google Scholar]
- 32.Wiley, S. R., Schooley, K., Smolak, P. J., Din, W. S., Huang, C. P., Nicholl, J. K., Sutherland, G. R., Smith, T. D., Rauch, C., Smith, C. A., et al. (1995) Immunity 3, 673–682. [DOI] [PubMed] [Google Scholar]
- 33.Cretney, E., Takeda, K., Yagita, H., Glaccum, M., Peschon, J. J. & Smyth, M. J. (2002) J. Immunol. 168, 1356–1361. [DOI] [PubMed] [Google Scholar]
- 34.Nimmanapalli, R., Porosnicu, M., Nguyen, D., Worthington, E., O'Bryan, E., Perkins, C. & Bhalla, K. (2001) Clin. Cancer Res. 7, 350–357. [PubMed] [Google Scholar]
- 35.Uno, K., Inukai, T., Kayagaki, N., Goi, K., Sato, H., Nemoto, A., Takahashi, K., Kagami, K., Yamaguchi, N., Yagita, H., et al. (2003) Blood 101, 3658–3667. [DOI] [PubMed] [Google Scholar]
- 36.Walczak, H., Miller, R. E., Ariail, K., Gliniak, B., Griffith, T. S., Kubin, M., Chin, W., Jones, J., Woodward, A., Le, T., et al. (1999) Nat. Med. 5, 157–163. [DOI] [PubMed] [Google Scholar]
- 37.Kops, G. J. & Burgering, B. M. (2000) J. Anat. 197, 571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, K., Dorman, J. B., Rodan, A. & Kenyon, C. (1997) Science 278, 1319–1322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.