Abstract
A microscopy-based screen of a large collection of maize Mutator (Mu) transposon lines identified the supernumerary aleurone layers 1-1 (sal1-1) mutant line carrying up to seven layers of aleurone cells in defective kernel endosperm compared with only a single layer in wild-type grains. Normal, well filled endosperm that is homozygous for the sal1-1 mutant allele contains two to three layers of aleurone cells. Cloning of the sal1 gene was accomplished by using Mu tagging, and the identity of the cloned gene was confirmed by isolating an independent sal1-2 allele by reverse genetics. Homozygous sal1-2 endosperm has two to three layers of aleurone cells in normal, well filled grains. In situ hybridization experiments reveal that the sal1 gene is ubiquitously expressed in vegetative as well as zygotic grain tissues, with no difference being detected between aleurone cells and starchy endosperm cells. Northern blot analysis failed to detect the sal1-2 transcript in leaves of homozygous plants, suggesting that the allele is a true sal1 knockout allele. The sal1 gene encodes a homologue of the human Chmp1 gene, a member of the conserved family of the class E vacuolar protein sorting genes implicated in membrane vesicle trafficking. In mammals, CHMP1 functions in the pathway targeting plasma membrane receptors and ligands to lysosomes for proteolytic degradation. Possible roles for the function of the sal1 gene in aleurone signaling, including a defect in endosome trafficking, are discussed.
The cereal endosperm is a major source for food, feed, and raw materials, thus forming an important basis for our civilization. In addition, endosperm is an attractive system for developmental biology studies because of its simple anatomy, the mature grain consisting of three cell types, the aleurone layer, the starchy endosperm, and the transfer layer (1–4). The endosperm develops from the fertilized central cell of the megagametophyte or embryo sac. Shortly after fertilization, repeated rounds of mitosis (but no cell-wall formation or cytokinesis) takes place, which leads to the formation of the endosperm coenocyte. Cellularization of the coenocyte is initiated by the formation of nuclear-based radial microtubular systems from the nuclear envelope of all nuclei followed by cell-wall deposition in the interzones between individual radial microtubular systems (5). Mitotic divisions of the nuclei within each cell-wall compartment or alveoli lead to one peripheral layer of cells and a new layer of alveoli. Repeated rounds of the same cycle of events lead to cell files that eventually completely fill the central cell 4 days after pollination. Based on the presence of a full complement of cytoskeletal arrays in the first endosperm cell layer, aleurone cell-fate specification is believed to occur after the first periclinal cell division (5). The fully developed endosperm consists of a central mass of starchy endosperm cells, a one-cell-thick layer of aleurone cells surrounding the starchy endosperm, and a basal layer of transfer cells (2).
Naturally occurring variants of corn, displaying a wide variety of grain colors produced in the aleurone layer, have provided insight into the developmental mechanisms underlying aleurone cell-fate specification. In the mutants crinkly4 (cr4) (6) and defective kernel (dek1) (7), mature kernels lack aleurone cells partially or completely, respectively. In these mutants, starchy endosperm cells are found in the periphery of the endosperm, suggesting a defect in the mechanisms specifying aleurone cell fate. Both of these genes have been cloned; Cr4 encodes a tumor necrosis factor receptor-like receptor kinase (6), and Dek1 a member of the calpain gene super family (7). For CR4, the structural similarity to tumor necrosis factor receptor led to a model in which the receptor-like kinase is activated by a positional signal in the form of a CR4 ligand that specifies aleurone cell identity in the peripheral cell layers of the endosperm (8). In addition to a targeting signal for membranes, the DEK1 protein contains 21 membrane-spanning domains, a loop region predicted to be on the extracellular side of the plasma membrane, and a calpain, or cysteine proteinase domain, on the cytosolic side. Revertant sector analysis with the dek1 gene strongly supports the role of positional signaling in aleurone cell formation, with the presence of the positional signal being required for continued aleurone cell-fate specification. In this analysis, loss of Dek1 function led to loss of aleurone cell identity and respecification to starchy endosperm identity (9). The exact role of dek1 in aleurone signaling remains unknown.
In addition to mutants lacking aleurone cells, studies of mutants with increased number of aleurone cell layers is expected to add valuable insight into the mechanisms underlying aleurone cell-fate specification. Interestingly, barley wild-type endosperm contains three layers of aleurone cells (10, 11), whereas most cereal endosperms have only a single layer of cells. One exception to this rule is rice, in which the number of aleurone cell layers ranges from one to three in different positions of the endosperm (12). No information is currently available on the mechanism of aleurone cell-fate specification from these species.
In maize, several cases of variants with more than one aleurone cell layer have been reported, including the Coroico maize population (13). In this case, multiple aleurone was transmitted as a partial dominant character over the normal single aleurone condition. Also, in a second case, the multiple-aleurone trait appeared to be inherited as a semidominant character, although firm conclusions could not be reached (14).
To identify monogenic multiple-aleurone-layer mutants in maize, we conducted a microscopy screen using a subset of lines from a Mutator (Mu) mutagenized maize population (7, 15). One of the multiple-aleurone lines identified in this screen, supernumerary aleurone 1 (sal1)-1 carries up to seven layers of aleurone cells in the endosperm of defective kernels, compared with one layer in wild-type grains. Homozygous mutant seeds for a second allele, sal1-2, carry endosperm with two to three layers of aleurone cells and have normal grains. We cloned the sal1 gene by Mu tagging and found that it encodes a homologue of the human charged multivesicular body protein 1/chromatin-modifying protein 1 (Chmp1) gene, a member of a conserved family of the class E vacuolar protein sorting genes implicated in membrane vesicle trafficking.
Materials and Methods
Mutant Screening and Microscopy. Visual inspection of the 41,610 ears in Pioneer Hi-Bred International's Trait Utility System for Corn (TUSC) collection (15) revealed 12,600 ears that contained a mixture of plump (wild-type) and nonplump grains. Mature nonplump grains from all segregating ears, representing potential endosperm mutant grains, were fixed overnight at room temperature in 2% glutaraldehyde/0.3 M sodium-phosphate buffer (pH 6.5). Hand sections were stained with Toluidine blue and mounted on glass slides by using 15% Mowiol/35% glycerol/0.07 M Tris (pH 7.5). Selected sections were also stained with Sudan red 7B for neutral lipids (16).
Allelism Test, Cosegregation Analysis, and Cloning of sal1. Cosegregation analysis was performed in a population of plants segregating for the sal1-1 allele. Plants carrying ears with only wild-type grains were classified as wild type, and plants with ears that segregated for the multiple-aleurone-layer trait were classified as sal1-1/+. Homozygous sal1-1 embryos do not germinate. DNA for the cosegregation analysis was isolated from freeze-dried leaf material according to a cetyltrimethylammonium bromide protocol (17), digested with restriction enzymes, and separated on 1.0% agarose gels. DNA fragments were transferred to Hybond N+ membranes in 10× standard saline citrate (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7). Genomic DNA gel blots were probed with 32P-labeled Mu-element-specific probes (18) kindly provided by V. Chandler (Tucson, AZ). A subgenomic library of the polymorphic DNA fragment from sal1-1/+ plants was prepared in pUC18. A single clone containing the cosegregating 1.6-kb fragment was identified by using the Mu1 probe. The DNA flanking the Mu1 insertion was identified by sequencing the insert. The sal1-2 allele was isolated by reverse genetics on the TUSC population by using the sal1-1-specific primer 5′-ACGAGCCTTCACAAATTGCCCTCTCAG-3′ in combination with the MuTIR primer 5′-AGAGAAGCCAACGCCAWCGCCTCYATTTCGTC-3′.
Northern Blot Analysis. Total RNA was isolated from maize leaves and immature kernels by using the Trizol reagent according to manufacturer directions (GIBCO/BRL). RNA electrophoresis, membrane transfer, and hybridization were performed according to standard protocols (19). The 32P-labeled probes were prepared by random primer labeling with a RediPrime kit (Amersham Pharmacia).
In Situ Hybridization Analysis. Digoxigenin-11-UTP-labeled RNA probes were used to localize corresponding mRNA in cells according to the protocol described by Jackson (20). RNA probes were made by using digoxigenin-11-UTP-labeled NTP mix and SP6 and T7 RNA polymerases (Roche Molecular Biochemicals). A 764-bp fragment of sal1 cDNA starting from the 5′ UTR was subcloned into pSPORT I vector (Invitrogen). The clone that contains the insert was linearized by NotI (sense) and SalI (antisense) and transcribed into RNA in vitro. Unincorporated ribonucleotides were removed by using Qiagen (Valencia, CA) RNeasy purification kit, and probes were subjected to carbonate hydrolysis to reduce probe length to ≈150 nt. Microscopy evaluation was carried out in a dark field by using a Nikon Eclipse E800.
Results
A Light-Microscopy Screen of the Pioneer TUSC Population Revealed Many Lines with Multiple Aleurone Layers. Among the collection of 41,610 F2 maize ears derived from crosses between inbred lines and high Mu copy-number lines, 12,600 lines segregated for incomplete grain filling, the mutant grains ranging from an empty pericarp phenotype to grains that were slightly less filled than normally filled wild-type grains. We infer that the high number of lines segregating for grain filling is caused by mutations in genes affecting endosperm growth, and that homozygous endosperm carried on heterozygous (phenotypically wild type) plants represents a sensitive screen for such mutations. A high frequency of these mutants represents dek mutants that do not germinate and must be propagated as heterozygous plants. The number of independent mutations in the TUSC population affecting grain filling is difficult to assess based on the number of lines segregating for incomplete grain filling, because the number of mutations existing in the parental Mu lines used to derive the TUSC population is unknown.
To identify lines with multiple aleurone layers, transverse sections from the midgrain level of the 12,600 lines described above were examined with light microscopy. During the initial round of screening, several hundred lines with multiple layers of aleurone cells were discovered. Due to the high Mu load in the TUSC population, we routinely crossed selected lines carrying ears that segregated for a multiple-aleurone phenotype to standard maize inbred lines (B73 or A188) for several generations. In line with previously reported investigations, in most of these mutants the penetrance of the multiple-aleurone trait was variable, and firm conclusions about the mode of inheritance of the trait were difficult to draw.
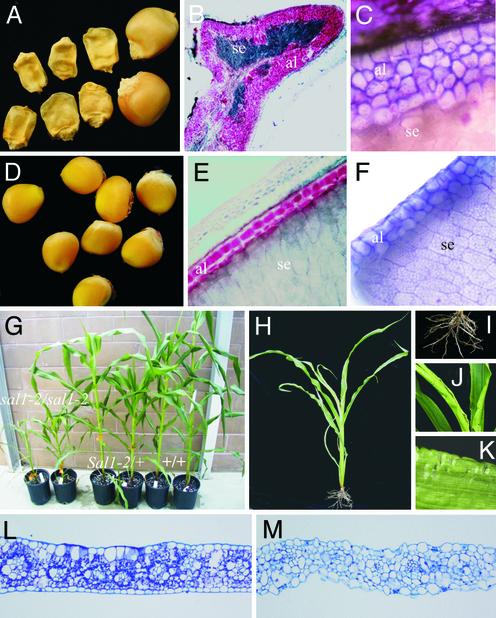
The sal1-1 Mutant Has up to Seven Layers of Aleurone Cells in Defective Kernels. In the microscopy investigation of the F2 TUSC lines, a block of lines sharing a high-copy Mu parent plant segregated for defective kernels that consistently contained multiple layers of aleurone cells. One representative line, sal1-1, was selected for further studies. The defective kernel seeds from this line contained up to seven layers of aleurone cells, the highest number of aleurone layers found in this screen (Fig. 1 A–C). As shown by the stain with Sudan red, a lipid stain (Fig. 1B), and by cell morphology (Fig. 1C), the multiple aleurone layers of sal1-1 endosperms have true aleurone cell identity. The multiple-aleurone-layer phenotype is transmitted as a Mendelian trait, with multiple aleurone layers being the recessive condition. Homozygous defective kernels have highly defective embryos that do not germinate. The mutation was therefore propagated by growing heterozygous plants for the sal1 trait. Over the generations that have been observed, the multiple-aleurone phenotype has remained stable.
Fig. 1.
The phenotype of sal1 endosperm and vegetative plant tissues. (A) Homozygous sal1-1 defective kernels. Two wild-type siblings are shown to the right. (B) Section of the homozygous sal1-1-defective kernel shown in A. The cells of the supernumerary aleurone layer (al) are stained red with the lipid stain Sudan red. The highly reduced starchy endosperm (se) is shown in dark blue. (C) Micrograph showing part of a transverse section of the multilayered aleurone of the homozygous sal1-1-defective kernel shown in A.(D) Homozygous doubleal1 (sal1-1′) grains. (E) Transverse section of homozygous sal1-1′ endosperm of the grains shown in D. The two-cell-thick aleurone layer is stained red with Sudan red. (F) Micrograph showing part of the two-cell-thick aleurone of the homozygous sal1-1′ kernel shown in D. (G) Family of plants segregating for the sal1-2 allele. (H) homozygous sal1-2 plant. (I) Roots of homozygous sal1-2 plant. (J and K) Morphology of homozygous sal1-2 leaves. (L) Section of wild-type leaf. (M) Section of sal1-2 leaf.
In addition to sal1-1, many more lines with two to three layers of aleurone cells were detected in segments of the population that were related to sal1 through a common pedigree sharing the same Mu donor stock. We selected the doubleal1 mutant line as a representative of these mutants. In contrast to sal1-1, this line carried two layers of aleurone cells in well filled but slightly smaller grains than wild type. After four rounds of backcrosses of one of these lines to A188, the aleurone phenotype showed reduced penetrance, with the number of aleurone layers varying from one to two layers. The doubleal1 line was shown to be allelic to sal1 in an allelism test in which two heterozygous plants were crossed. For reasons to be described below, this mutant allele was named sal1′-1.
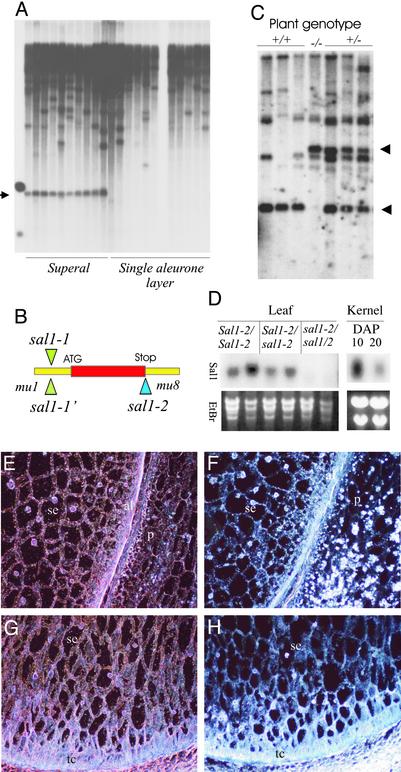
Cloning of the sal1 Gene. To identify the sal1 gene, Southern blot analysis with probes specific for different classes of Mu transposons was used to search for Mu bands cosegregating with the sal1-1 phenotype. By using the restriction enzyme BglII, a 1.6-kb Mu1 band was identified that cosegregated with the multiple-aleurone-layer phenotype in a population of 47 selfed individuals after four backcrosses between sal1-1/+ and A188 (Fig. 2A). This 1.6-kb band was cloned and sequenced. Based on the Mu1 flanking sequence contained in this band, an EST clone was identified from our EST collections. Further sequence analysis of the sal1-1 mutant revealed that the Mu1 transposon was inserted in the 5′-UTR region of the putative sal1 gene (Fig. 2B). Sequence analysis of the sal1-1′ allele revealed that Mu1 was inserted in the exact same position as in sal1-1, strongly suggesting that the sal1-1 Mu-insertion allele existed already in the Mu-active parental line used to derive both the sal1-1 and the doubleal1 mutant lines.
Fig. 2.
Cloning and expression analysis of sal1.(A) DNA blot from heterozygous sal1/+ (superal) and wild-type (single aleurone layer) plants digested with BglII and probed with a Mu1-specific probe. The blot identifies a 1.6-kb BglII band cosegregating with the sal1 phenotype (arrow). (B) Relative position of Mu insertion in the two sal1 alleles. (C) DNA blot from F2 population segregating for the sal1-2 allele probed with the sal1 DNA. (D) Northern blot analysis of sal1 expression in leaves from sal1-2/sal1-2, sal1-2/+, and +/+ plants. (Right) Northern blot analysis from wild-type kernels harvested at 10 and 20 DAP. (E) In situ hybridization analysis with sal1 antisense probe for wild-type 10 DAP peripheral endosperm. p, pericarp; al, aleurone; se, starchy endosperm. (F) Same as E with sense (control) probe. (G) In situ hybridization analysis with sal1 antisense probe on wild-type 10 DAP basal endosperm. tc, transfer cells. (H) Same as G with sense (control) probe.
To confirm that the sal1 gene is responsible for the multiple-aleurone phenotype, we searched the TUSC population for additional sal1 alleles using reverse genetics. A second allele, sal1-2 was isolated, in which a Mu8 element was inserted in the region next to the stop code of the gene (Fig. 2B). To investigate whether the Mu8 insertion in the sal1 gene results in a multiple-aleurone-layer phenotype, we determined the genotype of 20 F2 kernels from a segregating ear using PCR. Of these, the five sal1-2/sal1-2 kernels all showed a multiple-aleurone phenotype, whereas eight sal1-2/+ and seven nonmutant grains all had single layers of aleurone cells.
Genomic DNA then was isolated from segregating sal1-2 F2 plants and hybridized with sal1 full-length cDNA (Fig. 2C). The arrow indicates the segregating sal1 bands. The size difference corresponds to the size of the Mu8 transposon. The phenotype of homozygous sal1-2 endosperm is similar to that of sal1-1′ (data not shown), confirming the identity of the sal1 gene. Homozygous sal1-2 plants grow to a size of ≈30% of wild type (Fig. 1 G and H), have a reduced root mass (Fig. 1I), and set small ears after self-pollination with ≈10–40 seeds (data not shown). All seeds from homozygous sal1-2 plants show the multiple-aleurone-layer phenotype, again supporting the conclusion that the Mu insertion in the sal1 gene is responsible for the multiple-aleurone-layer phenotype in both mutants. Similar to homozygous plants for the two other known genes affecting aleurone cell development, cr4 and dek1 (21), homozygous sal1-2 plants have crinkly leaves (Fig. 1 J–K). In contrast to the regular-shaped epidermis cells of wild-type siblings (Fig. 1L), transverse sections of homozygous sal1-2/sal1-2 leaves show enlarged, irregularly shaped epidermal cells (Fig. 1M). The epidermal cell phenotype of sal2 plants is very similar to that of plants that are homozygous for weak cr4 alleles (21), suggesting that the sal1 and cr4 genes function in the same or interacting pathways.
sal1 Is Ubiquitously Expressed in Most if Not All Plant Tissues. To evaluate sal1 expression, we probed RNA blots with a full-length cDNA (Fig. 2D). A single transcript of ≈1.3 kb was detected in leaves and kernels of wild-type plants. sal1 was not expressed in homozygous sal1-2 plants, consistent with the plant phenotype. Expression of the sal1 gene in heterozygous plants is similar to wild type. Expression of sal1 at 10 days after pollination (DAP) is much stronger than at 20 DAP kernels. An indication that sal1 is expressed in most plant tissues is also obtained from the distribution of sal1 ESTs in our EST collection covering most if not all maize tissues. In total, 88 ESTs were identified from several tissue-specific cDNA libraries, including kernel, tassel, seedling, root, silk, immature ear, and callus. The highest number of ESTs was found in kernel libraries followed by seedling, tassel, and root. By using in situ hybridization, the sal1 transcript was found to be uniformly distributed in all grain tissues including pericarp, aleurone cells, starchy endosperm, transfer cells, and pedicel (Fig. 2 E–H).
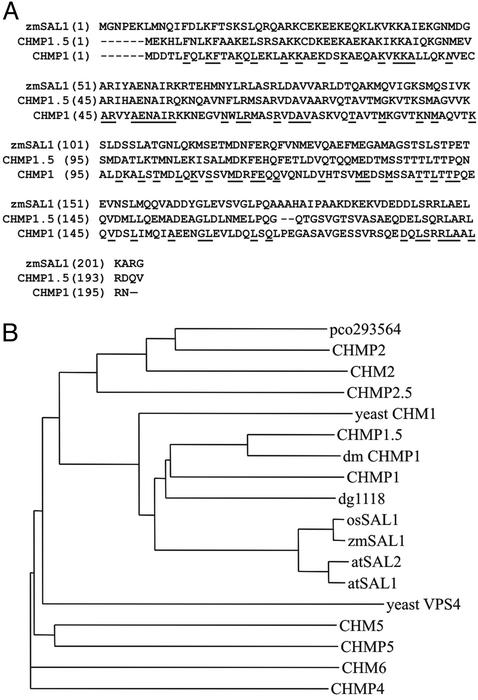
The sal1 Gene Encodes a 204-aa Protein That Is Strongly Implicated in Multivesicular-Body Formation in Animal Systems. SAL1 encodes a highly conserved protein found in yeast, Dictyostelium, plant, Drosophila, and human. SAL1 is 43% identical and 61% similar to human CHMP1, which localizes to early endosomes and physically interacts with SKD1/VPS4, a highly conserved protein directly linked to multivesicular body sorting in yeast (22). The similarity is equally distributed over the whole protein (Fig. 3A). Based on sequence analysis of the sal1 cDNAs in our EST collection, maize expresses only one sal1 gene. In contrast, Arabidopsis has two sal1 homologs, both encoding proteins that are >89% identical to the predicted maize SAL1 protein. Based on available sequence information, the rice genome contains only one sal1 homologue, encoding a protein that is 96% identical to the maize SAL1 protein. Phylogenetic analysis showed that SAL1 belongs to the CHMP1 gene group (Fig. 3B). The proteins in this group are ≈200 aa residues long with a conserved, distinctive charge distribution, with the N- and C-terminal halves of the protein having a predicted average isoelectric point of ≈10 and 4, respectively. A search for structural motifs identified a predicted bipartite nuclear localization signal that is also present in the human CHMP1 protein, which targets a distinctive CHMP1 isoform to the nuclear matrix (23).
Fig. 3.
SAL1 sequence and evolution. (A) Alignment of the SAL1 protein with its human homologue, CHMP1. (B) Dendrogram showing the evolutionary relationship within the vacuolar E sorting protein family. GenBank accession numbers for proteins are atSAL1 (Arabidopsis developmental protein, At1g73030); atSAL2 (Arabidopsis developmental protein, At1g17730); osSAL1 (rice, AA072640); dg1118 (Dictyostelium developmental protein, AAC67541); dmCHMP1 (Drosophila, AAL28346); CHMP1 (human, AAG01448); CHMP1.5 (human, AAG01449); CHMP2 (human, AAC00005); CHMP2.5 (human, AAD34079); CHMP4 (human, AAF29098); CHMP5 (human, AAD27743); yeast CHM1 (yeast, YKR035w); CHM2 (yeast, YKL002w); CHM5 (yeast, YDR486c); and CHM6 (yeast, YMR077c).
Discussion
This study is part of our effort to identify components of the signal transduction pathway(s) regulating aleurone cell formation in maize grains. Previous investigations have led to the cloning of two genes from mutants lacking aleurone cells, cr4 (6) and dek1 (7), encoding a putative tumor necrosis factor receptor-like receptor kinase and a member of the calpain gene super family, respectively. Here we describe sal1, a mutant with supernumerary aleurone layers, and the identification of the gene responsible for this phenotype.
Earlier investigations of maize lines with multiple layers of aleurone cells describe segregation ratios deviating significantly from a Mendelian mode of inheritance. Similar to those experiments, the pattern of inheritance for the multiple-aleurone trait was also difficult to follow in most of the TUSC lines identified in our mutant screen because of an apparent low degree of penetrance. Further investigations are needed to understand the underlying mechanisms for this phenomenon. Identification of the gene(s) giving rise to the Coroico multiple aleurone lines (13, 14) and extra cell layers 1 (xcL1) (32) will hopefully shed light on factors influencing the number of aleurone cell layers in maize.
This report describes three sal1 TUSC lines. Of these, the sal1-1 (Fig. 1 A–C) and sal1-1′ (Fig. 1 D–E) lines carry identical Mu-insertional alleles but have dramatically different aleurone phenotypes with up to seven and three layers of aleurone cells, respectively (Fig. 1). Based on the TUSC pedigree information, the sal1-1 and doubleal1 lines were related through a common Mu-active line, which was confirmed by using reverse genetics, demonstrating that approximately half the plants from individual crosses were heterozygous for the sal1-1 allele. We interpret this result to mean that the insertion allele sal1-1 preexisted in a plant that gave rise to the sal1-1 and doubleal1 pollen donor plants and that these plants were heterozygous for sal1-1 (i.e., sal1-1/+). The mechanisms causing the difference in the phenotype between sal1-1 and sal1-1′ are currently unknown, but one possibility is that a second, independent enhancer mutation occurred in the sal1-1/+ Mu-active plant used as a pollen donor for the sal1-1 lines. Alternatively, the doubleal1 phenotype may be caused by a suppression of the Mu-insertion allele, a phenomenon that has been reported for several other Mu-insertion alleles (24, 25, 26).
The sal1-2 allele was identified by using the sequence information from the cloning of the sal1-1 allele in a reverse-genetics experiment on DNA from the TUSC population (Fig. 1 G–K). Homozygous sal1-2 plants are shorter than wild-type plants, have crinkly leaves and underdeveloped roots, and carry normal grains with two to three layers of aleurone cells similar to sal1-1′. The phenotype of sal1-2 homozygous plants has been stable over two generations of selfing. These plants lack the sal1 transcript, thus most likely representing a true knockout of the sal1 gene. From these experiments we conclude that the cloned sal1 gene is involved in the determination of the number of aleurone cell layers in maize endosperm, and that loss of function of this gene leads to supernumerary aleurone layers. Interestingly, the phenotype of the sal1-2 homozygous plants implicate the sal1 gene also in leaf development, a characteristic shared with the two previously identified aleurone-signaling genes Cr4 and Dek1. In our interpretation, this finding strongly suggests that sal1 functions in the same or interacting pathways as Dek1 and Cr4, and that this pathway(s) regulates not only aleurone cell development but also other aspects of plant development. A role for the sal1 gene in several tissues and cell types is also supported by the ubiquitous expression of sal1 in all grain cell types (Fig. 2 E–H) as well as in leaves (Fig. 2D).
The cloning of the sal1 gene identifies the SAL1 protein as a homologue of the human CHMP1 protein (22), which is present in early endosomes and physically interacts with the multivesicular-body-sorting protein SKD1/VPS4 (27). A multivesicular body is a vesicle-filled endosome that targets proteins to the interior of lysosomes. Functional evidence for a role of CHMP1 in multivesicular-body formation includes abnormal distribution of endosomal markers and dilation of endosomal compartments after the overexpression of CHMP1 in yeast (22). Furthermore, a Saccharomyces cerevisiae CHM1 (homologue of CHMP1) deletion mutant has abnormal, multilamellar prevacuolar compartments accompanied by a defect in carboxypeptidases S and Y sorting, classifying CHM1 as a member of the class E vacuolar protein sorting genes (22).
We previously proposed a model for aleurone cell-fate specification in which the tumor necrosis factor receptor-like protein kinase receptor CR4 is activated in the surface cell layer of the endosperm by an unidentified ligand concentrated in the periphery of the endosperm (8). In homozygous loss-of-function sal1 endosperm, aleurone cell-fate specification occurs in deeper cell layers of the endosperm than in wild type. One possible mechanism for the increased numbers of aleurone cells is that sal1 endosperm has an increased density of CR4 receptors on the surface of the peripheral endosperm cell layers that normally become starchy endosperm cells. Based on the identification of SAL1 as a homologue of CHMP1, increased CR4 density could come from an inability to target internalized membranes to lytic vacuoles, the plant equivalent of lysosomes. In animals, many receptors including G protein-coupled receptors and EGFR are endocytosed after concentration in clathrin-coated pits (reviewed in ref. 28). Recent evidence also suggests that this process can differentially regulate specific classes of receptors, as shown for example with H-Ras and K-Ras signaling (29). Although much less-detailed knowledge is available for plant endocytosis, rapidly accumulating evidence points to similarities with animal endocytosis, including the identification of Arabidopsis thaliana homologues of nearly all genes known to be involved in endocytosis in animals (30). Among other functions, calpains are implicated in endocytosis and exocytosis and more specifically in formation of coated vesicles and vesicle fusion to endosomes (31). A second possible mechanism underlying the sal1 phenotype is the mistargeting of proteins as seen in the case of carboxypeptidases in class E vacuolar mutants (22). The activity of mistargeted proteases and glycosidases to the extracellular space could potentially alter the surface proteins or cell-wall polysaccharides, thereby affecting cell–cell interactions through mechanisms that might be independent of Cr4 (6) and Dek1 (7). A third possibility is that some unexpected effect from loss of function of sal1 results in degradation or abnormal persistence of a completely uncharacterized regulator of aleurone cell fate. For example, SAL1 is predicted to have a bipartite nuclear localization signal, and we cannot exclude that it might function in transcriptional regulation in the nucleus to affect expression of cell-fate regulators. Further research would help to determine the exact subcellular localization of the SAL1 protein as well as the spatial distribution of CR4 and DEK1 in wild-type, sal1, cr4, and dek1 plant tissues.
Acknowledgments
Jane Spauldine, Berit Morken, Peter Sekkelsten, and Karin O. Stenberg Olsen are thankfully acknowledged for the mutant screening, as are Karla Kurth for molecular analysis and Lizabeth C. Meeley for field support. Guri Johal and Dilbag Multani are also acknowledged for advise and discussion of Mu-tagging and cloning methods. The Biotechnology Program of the Bioprocessing and Production Division of the Research Council of Norway is acknowledged for support throughout the early phase of this project.
Abbreviations: DAP, days after pollination; Dek, defective kernel; Chmp1, charged multivesicular body protein 1/chromatin-modifying protein 1; Cr4, crinkly4; Mu, mutator; sal1, supernumerary aleurone 1; TUSC, Trait Utility System for Corn; XcL1, extra cell layers 1.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY243475).
References
- 1.Lopes, M. A. & Larkins, B. A. (1993) Plant Cell 5, 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olsen, O.-A. (2001) Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 233–267. [DOI] [PubMed] [Google Scholar]
- 3.Berger, F. (1999) Curr. Opin. Plant Biol. 2, 28–32. [DOI] [PubMed] [Google Scholar]
- 4.Becraft, P. W., Brown, R. C., Lemmon, B. E., Olsen, O.-A. & Opsahl-Ferstad, H.-G. (2000) Developmental Biology of Endosperm Development (Kluwer, Dordrecht, The Netherlands).
- 5.Brown, R. C., Lemon, B. E. & Olsen, O.-A. (1994) Plant Cell 6, 1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becraft, P. W., Stinard, P. S. & McCarty, D. (1996) Science 273, 1406–1409. [DOI] [PubMed] [Google Scholar]
- 7.Lid, S. E., Gruis, D., Jung, R., Lorentzen, J. A., Ananiev, E., Chamberlin, M., Niu, X., Meeley, R., Nichols, S. E. & Olsen, O.-A. (2002) Proc. Natl. Acad. Sci. USA 99, 5460–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsen, O.-A., Lemmon, B. E. & Brown, R. C. (1998) Trends Plant Sci. 3, 168–169. [Google Scholar]
- 9.Becraft, P. W. & Asuncion-Crabb, Y. (2000) Development (Cambridge, U.K.) 127, 4039–4048. [DOI] [PubMed] [Google Scholar]
- 10.Bosnes, M., Harris, E., Ailgeltiger, L. & Olsen, O.-A. (1987) Theor. Appl. Genet. 74, 177–187. [DOI] [PubMed] [Google Scholar]
- 11.Bosnes, M., Weideman, F. & Olsen, O.-A. (1992) Plant J. 2, 661–674. [Google Scholar]
- 12.Hoshikawa, K. (1993) in Science of the Rice Plant I: Morphology, eds. Matsuo, T. & Hoshikawa, K. (Nobunkyo, Tokyo), pp. 339–376.
- 13.Wolf, M. J., Cutler, H. C., Zūber, M. S. & Khoo, U. (1972) Crop Sci. 12, 440–442. [Google Scholar]
- 14.Nelson, O. & Chang, M. T. (1974) Crop Sci. 14, 374–376. [Google Scholar]
- 15.Bensen, R. J., Johal, G. S., Crane, V. C., Tossberg, J. T., Schnable, P. S., Meeley, R. B. & Briggs, S. P. (1995) Plant Cell 7, 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brundrett, M. C., Kendrick, B. & Peterson, C. A. (1991) Biotech. Histochem. 66, 111–116. [DOI] [PubMed] [Google Scholar]
- 17.Saghai Maroof, M. A., Biyashev, R. M., Yang, G. P., Zhang, Q. & Allard, R. W. (1994) Proc. Natl. Acad. Sci. USA 91, 5466–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chomet, P. S. (1994) in The Maize Handbook, eds. Freeling, M. & Walbot, V. (Springer, New York), pp. 243–249.
- 19.Sambrook, J., Frisch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 20.Jackson, D. P. (1992) in Molecular Plant Pathology: A Practical Approach., eds. Bowles, D. J., Gurr, S. J. & McPherson, M. (Oxford Univ. Press, Oxford), pp. 163–174.
- 21.Jin, P., Guo, T. & Becraft, P. W. (2000) Genesis 27, 104–116. [DOI] [PubMed] [Google Scholar]
- 22.Howard, T. L., Stauffer, D. R., Degnin, C. R. & Hollenberg, S. M. (2001) J. Cell Sci. 114, 2395–2404. [DOI] [PubMed] [Google Scholar]
- 23.Stauffer, D. R., Howard, T. L., Nyun, T. & Hollenberg, S. M. (2001) J. Cell Sci. 114, 2383–2393. [DOI] [PubMed] [Google Scholar]
- 24.Girard, L. & Freeling, M. (2000) Genetics 154, 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martienssen, R., Barkan, A., Taylor, W. C. & Freeling, M. (1990) Genes Dev. 4, 331–343. [DOI] [PubMed] [Google Scholar]
- 26.Martienssen, R. A., Barkan, A., Freeling, M. & Taylor, W. C. (1989) EMBO J. 8, 1633–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bishop, N. & Woodman, P. (2000) Mol. Biol. Cell 11, 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Claing, S. A., Laporte, M. G. & Caron, R. J. L. (2002) Prog. Neurobiol. 66, 61–79. [DOI] [PubMed] [Google Scholar]
- 29.Roy, S., Wyse, B. & Hancock, J. F. (2002) Mol. Cell. Biol. 22, 5128–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holstein, S. E. H. (2002) Traffic 3, 614–620. [DOI] [PubMed] [Google Scholar]
- 31.Sato, K., Saito, Y. & Kawashima, S. (1995) Eur. J. Biochem. 230, 25–31. [DOI] [PubMed] [Google Scholar]
- 32.Kessler, S., Seiki, S. & Sinha, N. (2002) Development (Cambridge, U.K.) 129, 1859–1869. [DOI] [PubMed] [Google Scholar]