Abstract
Changes in homeotic gene expression patterns or in the functions of the encoded proteins are thought to play a prominent role in the evolution of new morphologies. The floral homeotic APETALA3 (AP3) and PISTILLATA (PI) genes encode MADS domain-containing transcription factors required to specify petal and stamen identities in Arabidopsis. We have previously shown that perianth expression of AP3 and PI homologs varies in different groups of angiosperms with diverse floral structures, suggesting that changes in expression may contribute to changing morphology. We have investigated the possibility that changes in the functions of the encoded gene products may also have played a role in the evolution of different floral morphologies. AP3 and PI are members of paralogous gene lineages and share extensive similarity along the length of the protein products. Genes within these lineages encode products with characteristic C-terminal motifs that we show are critical for functional specificity. In particular, the C terminus of AP3 is sufficient to confer AP3 functionality on the heterologous PI protein. Furthermore, we have shown that the evolution of the divergent AP3 C-terminal domain in the core eudicots is correlated with the acquisition of a role in specifying perianth structures. These results suggest that divergence in these sequence motifs has contributed to the evolution of distinct functions for these floral homeotic gene products.
Diversification in the function of homeotic gene products has been implicated in the evolution of new morphologies in a variety of animal taxa. For instance, insects have only three pairs of legs, as compared with the many-limbed crustaceans and onychophorans. This distinction appears to be due in large measure to differences in the function of the homeotic Hox gene product UBX (1, 2). In contrast, the identification of similar changes in homeotic protein function, and the roles that such differences play in the generation of novel body plans, has largely been lacking in plants.
Here we examine the function of different conserved motifs that are present in members of the APETALA3 (AP3) and PISTILLATA (PI) gene lineages in angiosperms. The Arabidopsis thaliana AP3 and PI genes encode MIKC-type MADS domain-containing transcription factors and are required to specify petal and stamen identities in the flower (3, 4). Mutations in the two genes exhibit similar phenotypes in which the second-whorl petals are transformed into sepals and the third-whorl stamens are transformed into carpels (4–6). The AP3 and PI proteins contain an N-terminal DNA-binding MADS domain of 58 aa, an I domain, a K region that is predicted to form a coiled-coil domain, and a divergent C terminus (Fig. 1B). The AP3 and PI proteins must heterodimerize to bind to DNA in vitro (7); furthermore, expression of both proteins is necessary to confer an ectopic phenotype (6, 8). Orthologs of AP3 and PI have been identified and characterized in Antirrhinum majus [snapdragon (9, 10)] as well as in Petunia (11) and other core eudicot species and appear to function in a manner similar to that of their Arabidopsis counterparts (12).
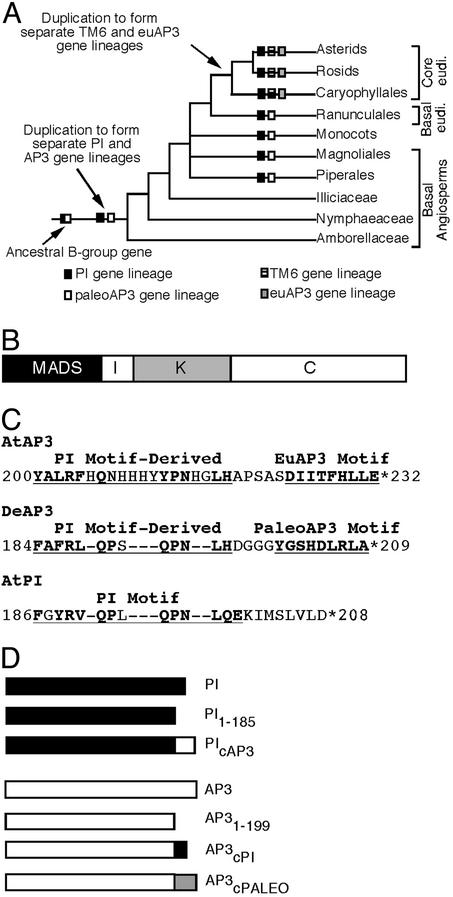
Fig. 1.
Evolution of PI and AP3 lineages. (A) Simplified phylogeny of the angiosperms (after ref. 16), showing key events in PI and AP3 gene lineage evolution (22). (B) Domains of the plant MADS domain proteins. The N-terminal MADS domain is required for DNA binding and is followed by the I (intervening) region, the K domain, and the C-terminal divergent region. (C) Amino acid sequences of the C-terminal motifs of AtAP3, DeAP3, and AtPI (22). (D) PI and AP3 constructs used in this study.
Angiosperm flowers differ in a variety of floral features, including phyllotaxy, organ number, and type of sterile organs (13). Fossil evidence indicates that early angiosperms did not possess a well-developed perianth (sterile organs) and may have been apetalous (13, 14). It has been hypothesized that petals have been independently derived either from bracts or from stamens multiple times during angiosperm evolution (15). Recent resolution of angiosperm phylogeny with molecular data has helped to define where and when these acquisitions may have taken place (16, 17). Both Arabidopsis and Antirrhinum are members of a large clade of angiosperms, the core eudicots (Fig. 1 A), characterized in part by the fixation of set numbers of floral organs, whorled floral phyllotaxy, and the independent acquisition of distinct petals (18, 19). In contrast, noncore eudicot species are characterized by more plasticity in their floral architecture. For instance, species in the basal eudicot ranunculid clade, thought to have diverged from other eudicots ≈140 million years ago (20), have flowers with a variable number of organs often organized in a spiral phyllotaxy (21).
The AP3 and PI lineage genes are thought to represent paralogous gene lineages that arose from a duplication event before the origin of the angiosperms (Fig. 1 A; refs. 12, 22, and 23). In addition, the AP3 lineage underwent another major duplication event at the base of the core eudicots, giving rise to two AP3 sublineages found in all groups of core eudicots: the euAP3 and the TM6 gene lineages (Fig. 1 A; refs. 22 and 24). Each of these lineages, although they share significant sequence similarity, is characterized by diagnostic residues found throughout the coding sequence but especially by short lineage-specific motifs found in the C terminus of the predicted proteins (Fig. 1C; ref. 22). The PI lineage genes encode a short hydrophobic region at the C terminus termed the PI motif. Subsequent to the duplication leading to the PI and AP3 gene lineages, the PI genes retained this motif while the AP3 lineage genes diverged to give rise to a related sequence, the PI-derived motif. In basal angiosperms, the paleoAP3 gene lineage has an additional motif C-terminal to the PI-derived motif, the paleoAP3 motif. Within the core eudicots, genes of the TM6 lineage have largely retained the ancestral paleoAP3 sequence. In contrast, the euAP3 lineage genes have lost this sequence and instead acquired a completely new motif, the euAP3 motif. The well characterized Arabidopsis AP3 and orthologous Antirrhinum DEFICIENS (DEF) genes are members of the euAP3 lineage. Most core eudicot species examined have both euAP3 and TM6 lineage representatives (22); Arabidopsis is unusual, however, because it appears to have lost the TM6 gene and retained only the euAP3 lineage gene (unpublished observations), although other rosids have TM6 representatives (for example, see ref. 25).
It has been demonstrated that the MADS domain does not contribute significantly to the functional specificity of floral homeotic proteins. Transgenic Arabidopsis plants containing chimeric constructs in which the MADS domain from one floral homeotic gene product is fused to the IKC regions of another display ectopic phenotypes characteristic of the IKC component of the construct (26, 27). Similarly, chimeric protein and mutagenesis studies have shown that the I and K boxes of both AP3 and PI are necessary both for dimerization specificity in vitro and for protein function in vivo (27–30). Although there are some suggestions that the C-terminal region of at least the AP3 protein (26, 28) may be important for function, it has been unclear what role this region may play.
To test whether these C-terminal lineage-specific motifs are critical for function, a series of truncation and chimeric constructs have been generated and expressed in Arabidopsis (Fig. 1D). Our analyses suggest that these motifs are necessary for the full function of AP3 and PI, and that the AP3 and PI C termini have distinct roles. Furthermore, we show that the C terminus of AP3 is sufficient to confer AP3 function, because a chimeric gene composed of PI coding sequences fused in frame to the 3′ end of AP3 rescues an ap3 mutant phenotype. We have also shown that these C-terminal domains have acquired unique roles in the paleoAP3 and euAP3 lineages. Together, these results suggest that the evolution of different C-terminal motifs was a key component in the functional diversification of these gene lineages.
Materials and Methods
Construction of Transgenic Plants. Plasmid constructs were made using standard protocols. PCR products were generated that contained the coding sequence derived from cDNA of AP3 or PI up to the start of the PI-derived or PI motif, respectively, with a MluI site in frame at the 3′ end. Other PCR products containing the C termini of AtAP3, AtPI, or DeAP3 (22) were generated with a 5′ MluI site in frame. PCR products were cloned into the pCR2.1 plasmid (Invitrogen Life Technologies) and sequenced. Restriction digests were used to create the chimeric constructs in pCR2.1 and then to clone constructs into pSPI (29) or pIa19 (30). The constructs were then cloned into the binary transformation vector pPZP221 (31) and used to transform Landsberg erecta by the floral dip method (32). For each transgenic construct, three independent single-insert lines, as determined by both Southern blotting and progeny testing, were analyzed in the homozygous condition.
Strain Construction and Genotyping. Homozygous transgenic lines were used as the pollen donor for crosses to homozygous mutant strains [pi-1, pi-2, pi-3, ap3-1, ap3-3; (3, 4)]. The resulting progeny were allowed to self-fertilize, and plants containing the transgene and homozygous for the appropriate mutant allele were identified in the next generation. Mutant alleles were identified by dCAPS genotyping (33) with at least one intronic primer to amplify only the endogenous gene. All pi alleles were amplified by using the same reverse primer: PIINT-2, 5′-CCAATTTCATGATATCTAGCTCAG-3′. The following forward primers were used to identify pi alleles (the underlined bases are introduced mismatches): PI-1, 5′-TACCAGAAGTTATCTGGCAAGAAATCATCATG-3′; PI-2E, 5′-GTGCTATGTTGGACCAATACCAGAAGATAT-3′; and PI-3S, 5′-GTGATGCAAAAGTTGCCCTCATAATCTACG-3′. The pi-1 fragment is cleaved by BspHI to reduce a 237-bp fragment to 212 bp. The wild-type amplicon is cleaved by EcoRV to produce a fragment of 226 bp as distinguished from the pi-2 254-bp fragment. The pi-3 fragment is cleaved by SnaB1 to reduce a 333-bp band to 304 bp. The ap3 alleles were genotyped by using the following primer pairs: AP3-3C, 5′-GATCAAGAGGATAGAGAACCAGACAAATCGA-3′; AP3INT-2, 5′-CGCATCAAGAATTTAACCAACCAGCG-3′; AP3-1N, 5′-CTTGGGAATCAGATCGAGCCACCAAGCATA-3′; and AP3INT, 5′-GTTCTGCAATTGTGTAAACCACAAGAG-3′. The fragment amplified from ap3-3 is cleaved by ClaI to produce a 380-bp fragment from a 408-bp fragment. The ap3-1 fragment is cleaved by NdeI to generate a 285-bp fragment from a 313-bp fragment.
Analyses of Transgenic Plants. After genotyping, homozygous mutant plants containing the transgene were analyzed. At least 100 flowers taken from at least 10 plants were examined for each genotype, with each independent transgenic line represented by at least 25 flowers from at least three plants. Flowers between flowers 5 and 20 on the primary inflorescence were scored.
Protein Extraction and Western Blotting. These procedures are described in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
Results
Experimental Design. The PI and AP3 lineage gene products are characterized in part by distinct C-terminal motifs (Fig. 1C). To determine whether these motifs are important for the function of these floral homeotic proteins, a series of deletion and domain-swap constructs were made (Fig. 1D) and introduced into transgenic Arabidopsis plants. In the first set of experiments, the full-length AP3 promoter (5D3) was used to drive expression of the transgenes in the wild-type expression domain of AP3, the developing petal and stamen primordia (34). The phenotypes of these transgenic lines were assayed in pi or ap3 mutant backgrounds to determine whether they were able to compensate for the lack of wild-type PI or AP3 protein product. Mutant alleles of differing strengths were used to assess the functions of the engineered constructs. pi-1 is a strong allele containing a stop codon that results in a truncated protein product of 79 aa, pi-2 is a moderate allele, and pi-3 is a weak allele (4). ap3-3 is likely to be a null allele as the nonsense mutation associated with this allele causes the AP3 protein to truncate at amino acid 18. ap3-1 behaves as a moderate to weak allele (5, 6). In the second set of experiments, the cauliflower mosaic virus 35S promoter (35S; ref. 35) was used to drive nearly ubiquitous expression of the transgenes. The ectopic phenotypes conferred by these constructs were assayed in a wild-type background.
As controls, transgenic plants expressing wild-type PI or AP3 were generated and their ability to cause ectopic phenotypes or to rescue the pi and ap3 mutants was assessed (see Tables 1 and 2 and Fig. 2 B, E, and H). PI expression driven by the 35S promoter caused a partial transformation of the first whorl sepals to petaloid organs (Fig. 2B). This transformation was due to the simultaneous expression of both AP3 and PI in the first whorl by an autoregulatory mechanism (6, 8). The 5D3::PI construct was able to largely rescue the pi-1 mutant (Table 1; Fig. 2E) and nearly completely rescue the weak pi-3 phenotype (data not shown). The rescue of organ identity defects by 5D3::PI was weaker in the third whorl, where only a few fertile stamens per flower were present (1.2 ± 1.5 in the pi-1 background) and many carpel-like organs remained, than in the second whorl, where most organs had petaloid characteristics.
Table 1. Organ numbers of transgenic plants containing PI constructs.
| Whorl 1
|
Whorl 2
|
Whorl 3
|
Whorl 4
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | No. of plants | No. of flowers | S | S/P | P | S | S/P | P | St | O | C | St | O | C |
| Landsberg erecta | 21 | 114 | 4 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4 (0) | 5.8 (0.4) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (0) |
| pi-1 | 10 | 100 | 4 (0) | 0 (0) | 0 (0) | 4 (0) | 0 (0) | 0 (0) | 0 (0) | 1.6 (0.7) | 4.9 (1.0) | 0 (0) | 0 (0) | 2 (0) |
| 5D3::PI; pi-1 | 22 | 140 | 0.4 (1.2) | 3.6 (1.2) | 0 (0) | 0.2 (0.9) | 1.6 (1.9) | 2.5 (1.9) | 1.2 (1.5) | 3.4 (1.2) | 1.6 (1.2) | 0 (0) | 0 (0) | 2 (0) |
| 5D3::PI1-185; pi-1 | 17 | 157 | 4 (0) | 0 (0) | 0 (0) | 3.3 (1.5) | 0.7 (1.5) | 0 (0) | 0.1 (0.7) | 0.9 (0.6) | 4.7 (1.3) | 0 (0) | 0 (0) | 2 (0) |
| 5D3::PIcAP3; pi-1 | 12 | 110 | 4 (0) | 0 (0) | 0 (0) | 4 (0) | 0 (0) | 0 (0) | 0 (0) | 0.7 (0.8) | 5.1 (1.1) | 0 (0) | 0 (0) | 2 (0) |
| 5D3::PIcAP3; ap3-3 | 18 | 102 | 4 (0) | 0 (0) | 0 (0) | 0 (0) | 0.6 (0.9) | 3.4 (0.9) | 4.3 (1.1) | 2.9 (0.6) | 0.3 (0.7) | 0 (0) | 0 (0) | 2 (0) |
S, sepal; P, petal; S/P, sepal/petal mosaic; St, stamen and stamen-like (stamen-shaped, but infertile and often immature-looking anther); O, filaments and mosaic organs; C, carpeloid organs. The numbers indicate the average number of organs of that type found in that genotype. Standard deviations are in parentheses.
Table 2. Organ numbers of transgenic plants containing AP3 constructs.
| Whorl 1
|
Whorl 2
|
Whorl 3
|
Whorl 4
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | No. of plants | No. of flowers | S | S/P | P | S | S/P | P | St | O | C | St | O | C |
| Landsberg erecta | 21 | 114 | 4 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4 (0) | 5.8 (0.4) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (0) |
| ap3-3 | 11 | 103 | 4 (0) | 0 (0) | 0 (0) | 4 (0) | 0 (0) | 0 (0) | 0 (0) | 1.3 (0.7) | 4.4 (0.9) | 0 (0) | 0 (0) | 2 (0) |
| 5D3::AP3; ap3-3 | 21 | 123 | 4 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 4 (0) | 5.7 (0.5) | 0.1 (0.3) | 0 (0) | 0 (0) | 0 (0) | 2 (0) |
| 5D3::AP31-199; ap3-3 | 13 | 119 | 4 (0) | 0 (0) | 0 (0) | 4 (0) | 0 (0) | 0 (0) | 0 (0) | 0.6 (0.6) | 5.0 (1.2) | 0 (0) | 0 (0) | 2 (0) |
| 5D3::AP3cPI; pi-1 | 17 | 115 | 4 (0) | 0 (0) | 0 (0) | 4 (0) | 0 (0) | 0 (0) | 0 (0) | 1.7 (0.8) | 4.2 (1.0) | 0 (0) | 0 (0) | 2 (0) |
| 5D3::AP3cPI; ap3-3 | 14 | 129 | 4 (0) | 0 (0) | 0 (0) | 4 (0) | 0 (0) | 0 (0) | 0.2 (0.5) | 2.1 (0.9) | 3.6 (1.1) | 0 (0) | 0 (0) | 2 (0) |
| 5D3::AP3cPALEO; ap3-3 | 18 | 172 | 4 (0) | 0 (0) | 0 (0) | 4 (0) | 0 (0) | 0 (0) | 1.2 (1.3) | 3.5 (1.1) | 1.1 (1.2) | 0 (0) | 0 (0) | 2 (0) |
Notation as in Table 1.
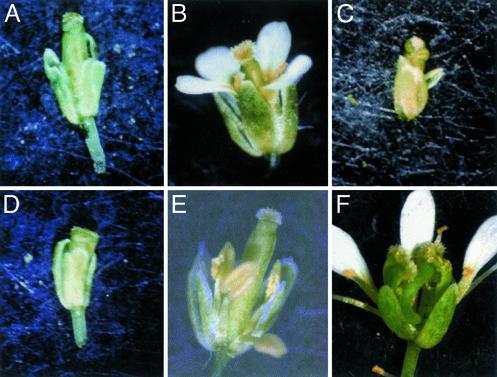
Fig. 2.
The C-terminal motifs are necessary for PI and AP3 function. Light micrographs of mature Arabidopsis flowers are shown. In some micrographs, one or more first-whorl organs have been removed to reveal the inner whorls of the flower. (A) Landsberg erecta. (B) 35S::PI. (C) 35S::PI1–185. (D) pi-1. (E) 5D3::PI; pi-1. (F) 5D3::PI1–185; pi-1. (G) ap3-3. (H) 5D3::AP3; ap3-3. (I) 5D3::AP31–199; ap3-3.
The single-insert 35S::AP3 lines used in this study displayed a weak ectopic expression phenotype. In some studies, 35S::AP3 transgenic plants have been reported to display a conversion of fourth-whorl carpels into stamens (6, 8, 36). We did observe occasional bent carpels and/or siliques, suggesting some mild perturbation of fourth-whorl development (data not shown); however, we were unable to recapitulate the strong ectopic phenotype seen in other studies. Presumably the levels of ectopic AP3 expression in the fourth whorl produced by our single-insert lines were insufficient to cause such a strong transformation.
The 5D3::AP3 transgene was able to completely rescue strong ap3-3 mutants, such that 5D3::AP3; ap3-3/ap3-3 plants were indistinguishable from wild type (Table 2; Fig. 2H; ref. 36). This result was expected because the 5D3 promoter can recapitulate all aspects of the AP3 expression pattern when driving a reporter gene such as GUS (34).
The C-Terminal Domains of PI and AP3 Are Necessary for Protein Function. To assess the function of the C-terminal motifs, truncated versions of Arabidopsis PI (PI1–185) and AP3 (AP31–199) were generated. The proteins encoded by these constructs terminate just before the first amino acid of the PI motif or PI-derived motif and therefore are missing both the PI motif and the eight amino acids beyond the PI motif found in Arabidopsis PI or both motifs characteristic of euAP3 proteins.
PI1–185 driven by the 35S promoter is unable to generate an ectopic gain-of-function phenotype in a wild-type background as compared with that produced by full-length PI (Fig. 2C). This observation suggests that the C-terminal region of the PI protein that includes the PI motif is necessary for the sepal-to-petal transformation. The region of PI necessary for heterodimerization with AP3 has been identified and includes the MADS domain, the I domain, and part of the K box (26, 28). Because these regions of the protein are intact in PI1–185, this implies that another function of the PI protein is impaired.
As suggested by the fact that 35S::PI1–185 confers no ectopic phenotype, this truncated protein product is unable to replace endogenous PI in the plant. There is little or no rescue of the strong pi-1 allele by 5D3::PI1–185 (Table 1; Fig. 2F), although PI protein expression can be detected (Fig. 4A, which is published as supporting information on the PNAS web site). For example, 5D3::PI; pi-1 plants contain on average 2.5 ± 1.9 petals in their second whorl, whereas plants of the genotype 5D3::PI1–185; pi-1 completely lack petals. Rescue is also minimal in the third whorl. There may be some slight rescue provided by the 5D3::PI1–185 construct in the weak pi-3 background (as seen by a reduction in the amount of transformation in the third whorl of this genotype; data not shown), but this rescue is much reduced from that seen with control constructs. This observation suggests that the PI motif is necessary for PI to function in the specification of organ identity.
When the 35S promoter is used to drive AP31–199 in Arabidopsis plants, there is no ectopic phenotype in a wild-type background (data not shown). When crossed into the null ap3-3 mutant background, the 5D3::AP31–199 construct is unable to rescue the mutant phenotype (Table 2; Fig. 2I), although protein can be detected on a Western blot (Fig. 4B, which is published as supporting information on the PNAS web site). Both the second whorl, which remains sepaloid, and the third whorl, which does not make stamens, fail to be rescued. This evidence implies that the C-terminal region is required for full function of the AP3 protein.
The AP3 and PI C Termini Are Not Functionally Equivalent and Contribute to Functional Specificity. To determine whether the C termini of AP3 and PI have diverged in function since the separation of the AP3 and PI gene lineages, a set of mutual motif-swap constructs were made between Arabidopsis AP3 and PI to generate AP3cPI and PIcAP3 (Fig. 1D). The 35S::PIcAP3 construct produces no ectopic phenotype in the first whorl of the flower (data not shown), implying that the PI-derived motif is not able to support the conversion of sepals into petals seen when PI is expressed in the first whorl. As would be expected on the basis of this lack of an ectopic phenotype, the 5D3::PIcAP3 construct, like the 5D3::PI1–185 construct, is unable to rescue the strong pi-1 mutant (Table 1; Fig. 3A), although protein accumulates (Fig. 4A). These results indicate that the C terminus of AP3 is not functionally equivalent to that of PI. In turn, this implies that the PI-derived motif has diverged to acquire unique functions, or that the euAP3 motif interferes with the function of the PI gene product, or both. However, when the 5D3::PIcAP3 construct was expressed in an ap3-3 mutant background, both petals and stamens were restored, completely in the case of the petals and almost fully in the case of the stamens (Table 1; Fig. 3B). This result indicates that the AP3 C terminus containing the PI-derived motif and euAP3 motifs is sufficient to confer AP3 functional specificity.
Fig. 3.
The C-terminal motifs have unique functions. Light micrographs of Arabidopsis flowers are shown. In some micrographs, one or more first-whorl organs have been removed to reveal the inner whorls of the flower. (A) 5D3::PIcAP3; pi-1. (B) 5D3::PIcAP3; ap3-3. (C) 5D3::AP3cPI; pi-1. (D) 5D3::AP3cPI; ap3-3. (E) 5D3::AP3cPALEO; ap3-3. (F) 35S::AP3cPALEO.
Similar tests were done with the AP3cPI chimeric protein. When the 5D3 promoter was used to drive this construct, little rescue of the null ap3-3 allele was observed (Table 2; Fig. 3D). However, some amelioration of the phenotype was seen in the third whorl, as these organs displayed less carpeloid character in this background than in ap3-3 plants (Table 2). This observation implies that the C terminus of PI may be able to partially replace that of the Arabidopsis AP3 protein. The 5D3::AP3cPI construct was unable to rescue the pi-1 mutant phenotype (Table 2; Fig. 3C). This result suggests that sequences upstream of the PI C-terminal domain are required to confer PI function. Together, these results demonstrate that the C termini of PI and AP3 cannot fully replace each other, and they imply that the divergence of these sequences over evolutionary time reflects the acquisition of unique functions. Furthermore, it appears that AP3 specific function is conferred by the AP3 C-terminal domain.
The euAP3 Lineage May Have Acquired New Functions. To assess whether the PI-derived and euAP3 motifs found in euAP3 gene lineage in core eudicots have diverged in function from the PI-derived motif and paleoAP3 motifs found in both the TM6 and paleoAP3 gene lineages, a version of Arabidopsis AP3, AP3cPALEO, was constructed. In this chimeric construct, the Arabidopsis AP3 C terminus was replaced by the C terminus of the Dicentra eximia (fringed bleeding heart) paleoAP3 gene, which contains PI-derived and paleoAP3 motifs (Fig. 1 C and D).
When AP3cPALEO was expressed under the control of the 5D3 promoter in Arabidopsis, we observed differential rescue of the second and third whorls of the ap3-3 mutant (Table 2; Fig. 3E). These plants displayed a reduction in the amount of carpel-like tissue in the third whorl of 5D3::AP3cPALEO; ap3-3 plants. Although no fertile stamens were produced, stamen-like organs (1.2 ± 1.3) as well as mosaic organs were observed (3.5 ± 1.1; Table 2; Fig. 3E). This result is in contrast to the results seen with 5D3::AP31–199; ap3-3 plants, which make far fewer mosaic organs. Although in 5D3::AP3cPALEO; ap3-3 plants the third-whorl mutant phenotype was ameliorated, the second whorl remained sepaloid. This observation implies that some function of the paleoAP3 motif can partially rescue stamen development but not petal development. 5D3::AP3cPALEO was completely unable to replace the endogenous PI protein when expressed in a pi-1 mutant background (data not shown), underscoring the fact that the paralogous PI and AP3 gene lineages have diverged in function.
Despite its inability to fully rescue the ap3 mutant phenotype when driven by the 5D3 promoter, the AP3cPALEO construct under the control of the 35S promoter was able to generate an ectopic expression phenotype in the fourth whorl in a wild-type background. This phenotype was both more frequent and more extreme than the ectopic expression phenotype caused by full-length AP3. In the fourth whorl, these plants contained extra carpels and/or malformed carpels (Fig. 3F) and some stamenoid tissue, and they displayed reduced female fertility (data not shown). This stands in contrast to the ectopic phenotypes of 35S::AP31–199 and 35S::AP3cPI, which do not affect fourth-whorl development. This result suggests that the paleoAP3 motif is functional in this context and is able to promote stamen identity in the fourth whorl.
Discussion
The C-Terminal Domains Are Required for Function and Are Critical for Functional Specificity. The PI and AP3 lineages arose by gene duplication and remain highly similar to each other throughout the length of the proteins. However, characteristic residues are found in each lineage (22). Particularly striking are the short motifs found at the C terminus of these proteins that are lineage specific and show high levels of sequence conservation across a wide range of angiosperms. Analyses of both ectopic expression phenotypes and rescue experiments indicate that the lineage-specific motifs found in Arabidopsis PI and AP3 are important for function of these proteins (Table 3). Truncation of PI or AP3 proteins, which removes their characteristic C-terminal motifs, reduces or eliminates function of these proteins. Additionally, the C termini of the two proteins cannot replace each other, implying that they have diverged and acquired unique functions.
Table 3. Summary of the phenotypes of chimeric PI and AP3 transgenic lines.
| Rescue
|
|||
|---|---|---|---|
| Construct | Ectopic | Whorl 2 | Whorl 3 |
| AP3 | - | +++ | +++ |
| AP31-199 | - | - | - |
| AP3cPI | - | - | +/- |
| AP3cPALEO | + | - | + |
| PI | ++ | ++ | +/- |
| PI1-185 | - | - | - |
| PIcAP3; pi-1 | - | - | - |
| PIcAP3; ap3-3 | - | ++ | +++ |
The degree of ectopic phenotype or rescue is indicated. -, The inability to generate an ectopic phenotype or complement the mutant phenotype; +/-, marginal ability to generate an ectopic phenotype; +, some ectopic phenotype or complementation; ++, moderate ectopic phenotype or rescue; +++, strong ectopic phenotype or full complementation.
Intriguingly, the replacement of the PI C terminus with that of AP3 is sufficient to confer AP3 activity on the chimeric protein, although the converse is not true. Previous work involving ectopic expression studies have shown the I and K regions of PI and AP3 are required to confer an ectopic expression phenotype, but do not explicitly demonstrate that such sequences are sufficient for conferring specificity (26, 27). Furthermore, in agreement with our analyses, these earlier studies show that a C terminus is necessary for function because constructs lacking the C terminus are nonfunctional (27). One model to explain these results is as follows. In Arabidopsis, PI and AP3 are thought to be components of multiprotein transcriptional complexes including other MADS domain transcription factors (37, 38). Stability of these complexes would depend on the C termini of PI and AP3, perhaps by recruitment one or more cofactors. In this model, the AP3 C terminus, but not the PI C terminus, would be essential for stability of petal- and stamen-specific complexes, whereas only the presence of a C terminus on PI is required. PI would be necessary to recruit the AP3 C terminus to the complex by dimerizing with AP3 through sequences in the I and K domains. The ability of 5D3::PIcAP3 to rescue ap3-3 but not the converse suggests that PI (and PIcAP3) can homodimerize (thereby providing the AP3 C terminus) whereas AP3 (and AP3cPI) cannot. In fact, the PI orthologous gene product from lily can homodimerize in vitro (39). DEF and GLOBOSA (GLO), the AP3 and PI orthologous gene products in Antirrinhum, have been shown to form a ternary complex with SQUAMOSA (SQUA) (40). This complex has higher DNA-binding affinity in vitro than either the DEF/GLO heterodimer or the SQUA homodimer. The formation and stability of these ternary complexes requires the C termini of DEF and GLO (40), supporting the hypothesis that the C-terminal motifs found in these proteins are essential for formation and/or maintenance of higher-order transcriptional complexes. Based on work done with MADS box proteins in animals (41, 42) and in rice (43), it is likely that other types of transcription factors will be identified in these complexes in addition to MADS domain proteins. The C termini of AP3 and PI could function in recruitment of such factors. As the MADS box proteins identified to date as present in these larger complexes have roles in specification of multiple organ types within the flower, non-MADS cofactors could determine target selectivity, explaining the role of the AP3 C terminus in functional specificity.
A Model for the Evolution of Organ Identity Functions. The gene duplication event that led to the PI and AP3 gene lineages is thought to have occurred before the radiation of the angiosperms. The functional divergence between these two gene lineages may have contributed to evolution of angiosperm-specific structures in the flower. There has been a second major duplication within the AP3 gene lineage at the base of the core eudicots. Most of the well-studied AP3 lineage genes fall within the euAP3 lineage of core eudicots, including both Arabidopsis AP3 and its ortholog from Antirrinhum, DEF (22). The paleoAP3 gene lineage, found in monocots and other basal angiosperms, has been functionally studied only in grasses, including maize (44) and rice (45, 46). In maize, it appears that the paleoAP3 gene is involved in determining the identity of the lodicules and the stamens (44). The variability of expression of these genes seen in other groups of noncore eudicots, such as the ranunculids (47), suggests differences in the mechanisms of sterile organ identity establishment. This observation suggests that the roles in lodicule identity in grasses and petal identity in the core eudicots arose independently, which agrees with morphological data (48). The gene duplication that resulted in the euAP3 gene lineage correlates with the proposed derivation of petals in the core eudicots, and it suggests that novel sequences present in this lineage may have been associated with petal evolution in this group of angiosperms.
To test the functional specificity of the lineage-specific motifs from both groups of AP3 genes present in core eudicots, we expressed a chimeric protein in which the PI-derived motif and paleoAP3 motif from a basal eudicot paleoAP3 gene replaced the PI-derived motif and euAP3 motif of Arabidopsis AP3. The 5D3::AP3cPALEO construct is unable to fully rescue ap3-3 mutants (Tables 2 and 3; Fig. 3E). This result suggests that the C-terminal motifs of these two gene lineages are not interchangeable and further suggests that the euAP3 lineage, which resulted from the duplication and divergence of an ancestral paleoAP3 lineage gene, has acquired new functions. The ability of the 5D3:AP3cPALEO chimeric construct to replace endogenous AP3 was stronger in the third whorl of the flower, where more stamen-like and mosaic organs were formed than with truncated AP3 (Table 2). The second whorl was not rescued at all by this chimeric protein, demonstrating that the PI-derived motif and paleoAP3 motif from Dicentra eximia does not support petal development in Arabidopsis. Additionally, the 35S::AP3cPALEO transgene caused strong disruption of the fourth whorl. These data suggest that the paleoAP3 C terminus retains a stamen-promoting activity but is unable to promote petal formation. Although perianth evolution remains poorly understood, the correlation of a possible independent derivation of petaloid organs at the base of the core eudicots with a gene duplication resulting in two AP3 lineages suggests that the acquisition of the euAP3 motif in addition to the PI-derived motif may have played a role in the independent origin of petals in this clade.
Our data support a model in which the ancestral PI and AP3 genes were involved in specification of male reproductive development in the nonflowering ancestor of the angiosperms, similar to the role that homologs of these genes are thought to play in extant gymnosperms (49, 50, 51, 52). Within the angiosperms, this role in specification of male reproductive structures (stamens) has been retained, but different clades of plant species have deployed these genes independently [i.e., grasses and core eudicots (53)] to specify sterile organs within the flower and, in at least some instances, have evolved new protein motifs to support these new functions.
It has long been thought that gene duplications can play an important role in the generation of new morphologies by providing material for the evolution of new gene functions (54). Our previous studies have shown diversification of expression patterns within the AP3 and PI gene lineages (47), and now we have shown a diversification of protein function encoded by this family of floral homeotic genes. These changes in expression patterns and protein functions appear to correlate with the advent of novel floral morphologies within the angiosperms, suggesting that changes in both MADS box homeotic gene regulation and function have been an important force in the evolution of floral form.
Supplementary Material
Acknowledgments
We thank G. deMartino, E. Kramer, and A. Litt for critical reading of the manuscript, M. Zik for PCR primers, and members of the Irish laboratory for advice. This work was supported by National Science Foundation Grant IBN-0110731 (to V.F.I.).
References
- 1.Galant, R. & Carroll, S. B. (2002) Nature 415, 910–913. [DOI] [PubMed] [Google Scholar]
- 2.Ronshaugen, M., McGinnis, N. & McGinnis, W. (2002) Nature 415, 914–917. [DOI] [PubMed] [Google Scholar]
- 3.Jack, T., Brockman, L. L. & Meyerowitz, E. M. (1992) Cell 68, 683–697. [DOI] [PubMed] [Google Scholar]
- 4.Goto, K. & Meyerowitz, E. M. (1994) Genes Dev. 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- 5.Bowman, J. L., Smyth, D. R. & Meyerowitz, E. M. (1989) Plant Cell 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jack, T., Fox, G. L. & Meyerowitz, E. M. (1994) Cell 76, 703–716. [DOI] [PubMed] [Google Scholar]
- 7.Riechmann, J. L., Wang, M. & Meyerowitz, E. M. (1996) Nucleic Acids Res. 24, 3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krizek, B. A. & Meyerowitz, E. M. (1996) Development (Cambridge, U.K.) 122, 11–22. [DOI] [PubMed] [Google Scholar]
- 9.Sommer, H., Beltran, J.-P., Huijser, P., Pape, H., Lonnig, W.-E., Saedler, H. & Schwarz-Sommer, Z. (1990) EMBO J. 9, 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trobner, W., Ramirez, L., Motte, P., Hue, I., Huijser, P., Lonnig, W. E., Saedler, H., Sommer, H. & Schwarz-Sommer, Z. (1992) EMBO J. 11, 4693–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Krol, A. R., Brunelle, A., Tsuchimoto, S. & Chua, N. H. (1993) Genes Dev. 7, 1214–1228. [DOI] [PubMed] [Google Scholar]
- 12.Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J. T., Muenster, T., Winter, K.-W. & Saedler, H. (2000) Plant Mol. Biol. 42, 115–149. [PubMed] [Google Scholar]
- 13.Endress, P. K. (1994) Plant Syst. Evol. 192, 79–97. [Google Scholar]
- 14.Friis, E. M., Pedersen, K. R. & Crane, P. R. (2001) Nature 410, 357–360. [DOI] [PubMed] [Google Scholar]
- 15.Takhtajan, A. (1991) Evolutionary Trends in Flowering Plants (Columbia Univ. Press, New York).
- 16.Soltis, P. S., Soltis, D. E. & Chase, M. W. (1999) Nature 402, 402–404. [DOI] [PubMed] [Google Scholar]
- 17.Qiu, Y.-L., Lee, J., Bernasconi-Quadroni, F., Soltis, D. E., Soltis, P. S., Zanis, M., Zimmer, E. A., Chen, Z., Savolainen, V. & Chase, M. W. (1999) Nature 402, 404–407. [DOI] [PubMed] [Google Scholar]
- 18.Magallon, S., Crane, P. R. & Herendeen, P. S. (1999) Ann. Mo. Bot. Gard. 86, 297–372. [Google Scholar]
- 19.Drinnan, A. N., Crane, P. R. & Hoot, S. B. (1994) in Early Evolution of Flowers, eds. Endress, P. K. & Friis, E. M. (Springer, New York), pp. 93–122.
- 20.Wikstrom, N., Savolainen, V. & Chase, M. W. (2001) Proc. R. Soc. London Ser. B 268, 2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosuge, K. (1994) Plant Syst. Evol. 8, 185–191. [Google Scholar]
- 22.Kramer, E. M., Dorit, R. L. & Irish, V. F. (1998) Genetics 149, 765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purugganan, M. D. (1997) J. Mol. Evol. 45, 392–396. [DOI] [PubMed] [Google Scholar]
- 24.Kramer, E. M. & Irish, V. F. (2000) Int. J. Plant Sci. 161, S29–S40. [Google Scholar]
- 25.Sheppard, L. A., Brunner, A. M., Krutovskii, K. V., Rottmann, W. H., Skinner, J. S., Vollmer, S. S. & Strauss, S. H. (2000) Plant Physiol. 124, 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krizek, B. A. & Meyerowitz, E. M. (1996) Proc. Natl. Acad. Sci. USA 93, 4063–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krizek, B. A., Riechmann, J. L. & Meyerowitz, E. M. (1999) Sex. Plant Reprod. 12, 14–26. [Google Scholar]
- 28.Yang, Y., Fanning, L. & Jack, T. (2003) Plant J. 33, 47–59. [DOI] [PubMed] [Google Scholar]
- 29.McGonigle, B., Bouhidel, K. & Irish, V. F. (1996) Genes Dev. 10, 1812–1821. [DOI] [PubMed] [Google Scholar]
- 30.Irish, V. F. & Yamamoto, Y. T. (1995) Plant Cell 7, 1635–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hajdukiewicz, P., Svab, Z. & Maliga, P. (1994) Plant Mol. Biol. 25, 989–994. [DOI] [PubMed] [Google Scholar]
- 32.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- 33.Neff, M. M., Neff, J. D., Chory, J. & Pepper, A. E. (1998) Plant J. 14, 387–392. [DOI] [PubMed] [Google Scholar]
- 34.Hill, T. A., Day, C. D., Zondlo, S. C., Thackeray, A. G. & Irish, V. F. (1998) Development (Cambridge, U.K.) 125, 1711–1721. [DOI] [PubMed] [Google Scholar]
- 35.Benfey, P. N. & Chua, N.-H. (1990) Science 250, 959–966. [DOI] [PubMed] [Google Scholar]
- 36.Jenik, P. D. & Irish, V. F. (2001) Development (Cambridge, U.K.) 128, 13–23. [DOI] [PubMed] [Google Scholar]
- 37.Pelaz, S., Gustafson-Brown, C., Kohalmi, S. E., Crosby, W. L. & Yanofsky, M. F. (2001) Plant J. 26, 385–394. [DOI] [PubMed] [Google Scholar]
- 38.Honma, T. & Goto, K. (2001) Nature 409, 469–471. [DOI] [PubMed] [Google Scholar]
- 39.Winter, K. U., Weiser, C., Kaufmann, K., Bohne, A., Kirchner, C., Kanno, A., Saedler, H. & Theissen, G. (2002) Mol. Biol. Evol. 19, 587–596. [DOI] [PubMed] [Google Scholar]
- 40.Egea-Cortines, M., Saedler, H. & Sommer, H. (1999) EMBO J. 18, 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Treisman, R. (1994) Curr. Opin. Genet. Dev. 4, 96–101. [DOI] [PubMed] [Google Scholar]
- 42.Shore, P. & Sharrocks, A. D. (1995) Eur. J. Biochem. 229, 1–13. [DOI] [PubMed] [Google Scholar]
- 43.Masiero, S., Imbriano, C., Ravasio, F., Favaro, R., Pelucchi, N., Gorla, M. S., Mantovani, R., Colombo, L. & Kater, M. M. (2002) J. Biol. Chem. 277, 26429–26435. [DOI] [PubMed] [Google Scholar]
- 44.Ambrose, B. A., Lerner, D. R., Ciceri, P., Padilla, C. M., Yanofsky, M. F. & Schmidt, R. J. (2000) Mol. Cell 5, 569–579. [DOI] [PubMed] [Google Scholar]
- 45.Kang, H.-G., Jeon, J.-S., Lee, S. & An, G. (1998) Plant Mol. Biol. 38, 1021–1029. [DOI] [PubMed] [Google Scholar]
- 46.Moon, Y. H., Jung, J. Y., Kang, H. G. & An, G. (1999) Plant Mol. Biol. 40, 167–177. [DOI] [PubMed] [Google Scholar]
- 47.Kramer, E. M. & Irish, V. F. (1999) Nature 399, 144–148. [DOI] [PubMed] [Google Scholar]
- 48.Dahlgren, R. M. T., Clifford, H. T. & Yeo, P. F. (1985) The Families of the Monocotyledons (Springer, New York).
- 49.Sundstrom, J., Carlsbecker, A., Svenson, M., Svensson, M. E. & Engstrom, P. (1999) Dev. Genet. 25, 253–266. [DOI] [PubMed] [Google Scholar]
- 50.Engstrom, P. (2002) Plant J. 31, 161–169. [DOI] [PubMed] [Google Scholar]
- 51.Mouradov, A., Glassick, T. V., Hamdorf, B. A., Murphy, L. C., Marla, S. S., Yang, Y. & Teasdale, R. D. (1998) Plant Physiol. 117, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mouradov, A., Hamdorf, B., Teasdale, R. D., Kim, J. T., Winter, K. U. & Theissen, G. (1999) Dev. Genet. 25, 245–252. [DOI] [PubMed] [Google Scholar]
- 53.Irish, V. F. (2000) Genome Biol. 1, 1015.1–1015.4. [Google Scholar]
- 54.Ohno, S. (1970) Evolution by Gene Duplication (Springer, New York).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.