Abstract
The 25-year-old debate about the origin of introns between proponents of “introns early” and “introns late” has yielded significant advances, yet important questions remain to be ascertained. One question concerns the density of introns in the last common ancestor of the three multicellular kingdoms. Approaches to this issue thus far have relied on counts of the numbers of identical intron positions across present-day taxa on the assumption that the introns at those sites are orthologous. However, dismissing parallel intron gain for those sites may be unwarranted, because various factors can potentially constrain the site of intron insertion. Demonstrating parallel intron gain is severely handicapped, because intron sequences often evolve exceedingly fast and intron phylogenetic distributions are usually ambiguous, such that alternative loss and gain scenarios cannot be clearly distinguished. We have identified an intron position that was gained independently in animals and plants in the xanthine dehydrogenase gene. The extremely disjointed phylogenetic distribution of the intron argues strongly for separate gain rather than recurrent loss. If the observed phylogenetic pattern had resulted from recurrent loss, all observational support previously gathered for the introns-late theory of intron origins based on the phylogenetic distribution of introns would be invalidated.
Spliceosomal introns are one of the hallmarks of eukaryotic genomes, which are distinctively elusive at providing unmixed clues about their evolutionary origins. Yet, after 25 years of contention (see ref. 1 and references therein), the dispute about the origins of introns between “introns-early” (IE, alternatively known as the exon theory of genes) and “introns-late” (IL) advocates seems to be approaching a synthesis. It is now almost certain that, if the progenote had introns, those could be type II self-splicing introns but never spliceosomal introns (1, 2). Because of the presumably severe constraints imposed on intronic recombination by the role that self-splicing introns play in their own removal, the IE advocates claim that exon shuffling as a factor for the assemblage of primordial genes seems unlikely. However, recent findings in the deeply diverging, putative basal eukaryote Giardia strongly indicate that spliceosomal introns originated in the eukaryotic stem before the diversification of protists, considerably earlier than suggested initially by IL advocates (i.e., around the time of origin of multicellularity; ref. 3). The IL notion that spliceosomal introns as well as the spliceosoma evolved through subfunctionalization of one or more self-splicing group II introns (2, 4, 5) has gained credit. Once released from the constraints of self-splicing, spliceosomal introns may have been instrumental in creating a profusion of new eukaryotic genes by exon shuffling (6). IE supporters now admit that intron insertion is an important process in the evolution of eukaryotic genes, although they persist in asserting that deletion of ancestral introns is the main factor responsible for present-day phylogenetic distributions of introns (7, 8). On their part, IL theorists accept regularities in intron phases and intron genomic distributions, but they explain them, as well as present-day intron phylogenetic distributions, using parsimonious population-genetic arguments that do not demand special evolutionary scenarios (refs. 2 and 9, but see ref. 6). In addition, IL advocates now acknowledge intron sliding as a real evolutionary phenomenon even though it is uncommon (10, 11) and, in most cases, implicates just one nucleotide base-pair slide (12, 13, 14). IL supporters now tend to view spliceosomal introns as genomic parasites that have been co-opted into many essential functions such that few, if any, eukaryotes could survive without them (2).
In this emerging scenario, IE upholders claim that the last common ancestor to all eukaryotes had a genome densely populated with introns, a significant fraction (up to 40%) of which are still conserved in present-day, typically intron-rich multicellular eukaryotes (7, 8). IE theorists base their claim on the observed numbers of intron positions in highly conserved genes, which are identical across animals, fungi, and plants, by assuming that the introns at those sites are all orthologous (and therefore ancestral; ref. 8). But dismissal of parallel intron gain at particular positions seems unjustified if we take into account that the sites of intron insertion may be narrowly confined by, at least, six mutually nonexclusive hypothetical factors: (i) the “protosplice” site (such that intron insertion will occur at the restricted sequence motif MAG│, where M is A or C, R is A or G, and the vertical line represents the site of intron gain) (15, 16); (ii) intron phase, phase 0 being preferred over phase 1 and 2 introns (17); (iii) intron-phase symmetry, favoring symmetrical 0-0, 1-1, and 2-2 insertions over asymmetrically inserted exons (18); (iv) gene–protein structural correlations (such that introns would be best tolerated at linkers or boundary regions between structural and/or functional modules and/or domains) (6, 7); (v) intron spatial distribution (such that even spacing of introns is favored for the proper performance of mRNA surveillance mechanisms) (2); and (vi) a putatively greater potential for nucleosome formation of introns (19). Notably, the operation of factors ii–iv has long been a tenet of the IE theory. To the extent that these requirements invoke adaptive constraints, it would not be surprising that many potentially target sites for intron occupancy have been retained over large phylogenetic distances. In any case, demonstrating convergent intron insertion is critical for the purpose of describing the rate of intron turnover (9).
Detecting parallel intron gain often runs into the difficulties associated with the comparison of intron sequences, which typically evolve exceedingly fast. Alternatively, parallel intron gain can be inferred from clear disjointed intron phylogenetic distributions. Most convincing would be a situation in which two collinear introns show characteristically restricted phylogenetic distributions within far distant lineages (e.g., two different kingdoms), with all intermediate taxa lacking the intron. Thus far no clear instance of such an intron phylogenetic configuration has been reported, perhaps because sequencing efforts continue to be concentrated on entire genomes such that broad phylogenetic sampling of shorter regions remains scarce. Here we focus on one of those few cases, the gene encoding xanthine dehydrogenase (XDH, E.C. 1.1.1.204). The Xdh gene, one of the most intensively investigated housekeeping loci, combines several features that yield it particularly attractive to investigate intron issues: (i) it is ancient, found in prokaryotes and eukaryotes; (ii) in eukaryotes it is very long (>1,300 codons) and can be unambiguously aligned over most of its length across animals, fungi, and plants; (iii) it is present in a single copy in most sequenced genomes; and (iv) its exon/intron structure varies greatly across eukaryotic taxa, ranging from 2 (e.g., Anopheles) to 36 (e.g., mouse and human) exons. A previous survey of the Xdh locus allowed us to identify three of the clearest cases of recent intron gain at a protosplice site because of a highly restricted phylogenetic distribution (1, 16, 20). Intron A, the most phylogenetically circumscribed of all three, was detected only in a cluster of two closely related species of the willistoni group of Drosophila, Drosophila sucinea and Drosophila capricorni, out of many animal and fungal lineages then examined. After 5 years, the growing sequence data have reinforced the picture. Moreover, the first plant Xdh sequences obtained indicate that intron A is ubiquitous throughout this kingdom.
Materials and Methods
The 82 species investigated are shown in Fig. 1. The GenBank accession numbers for the corresponding Xdh nucleotide sequences are given elsewhere (20–25), except for the cases of Anopheles gambiae, Danio rerio, Fugu rubripes, Ciona intestinalis, Caenorhabditis briggsae, Aspergillus fumigatus, Histoplasma capsulatum, Magnaporthe grisea, Dictyostelium discoideum, Oryza sativa, and Chlamydomonas reinhardtii, the hypothetical Xdh sequences of which were obtained by conducting BLAST searches with already-known Xdh amino acid sequences against their genome databases, and unambiguously corroborated by phylogenetic criterion. The data set includes 62 species of drosophilids, specially aimed to accomplish a dense phylogenetic sampling of all main lineages closely related to D. sucinea and D. capricorni (belonging to the bocainensis subgroup of the willistoni group), the two species containing intron A. Twelve of these species are named in Fig. 1, and the other 50 are named in the legend. The 62 species include the bocainensis subgroup (2 species), willistoni subgroup (6 species), Drosophila saltans group (6 species), Drosophila melanogaster and Drosophila obscura groups (13 species), Drosophila subgenus (33 species), Chymomyza genus (1 species), and Scaptodrosophila genus (1 species). At increasingly higher taxonomic levels, the data set comprises four families of dipterans (Drosophilidae, Tephritidae, Calliphoridae, and Culicidae), two orders of insects (dipterans plus lepidopterans), two orders of mammals (primates and rodents), two classes of vertebrates (mammals and bony fishes), two classes of angiosperms (monocots and dicots), four metazoan phyla (arthropods, chordates, nematodes, and urochordates), one phylum of fungi (ascomycetes), two plant phyla (anthophyta and chlorophyta), and three multicellular kingdoms (animals, fungi, and plants). Xdh amino acid sequences were aligned by using CLUSTALX 1.81 (26). In particular, the coding region surrounding the intron A site is well conserved; its alignment did not require further adjustment by eye.
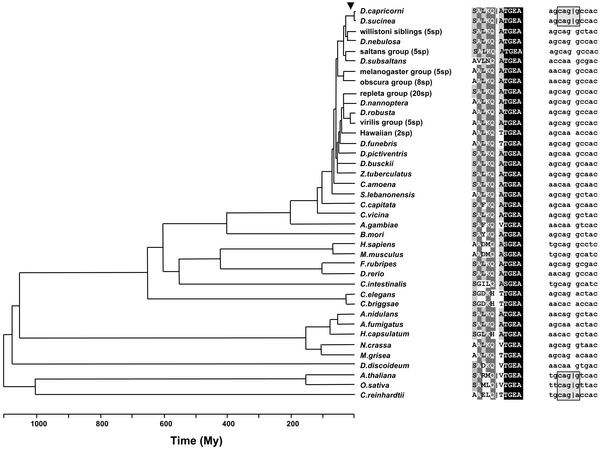
Fig. 1.
Phylogenetic distribution of intron A in Xdh. The time when intron A was gained in animals is indicated by an arrow. The vertical line in the amino acid and nucleotide alignments indicates the site of intron insertion. The two boxes on the nucleotide alignment enclose the protosplice-site motifs. The5 willistoni sibling species are Drosophila equinoxialis, Drosophila paulistorum, Drosophila willistoni, Drosophila tropicalis, and Drosophila insularis; the 5 saltans group species are D. saltans, Drosophila prosaltans, Drosophila neocordata, Drosophila emarginata, and Drosophila sturtevanti; the 5 melanogaster group species are D. melanogaster, Drosophila simulans, Drosophila teissieri, Drosophila erecta, and Drosophila ananassae; the 8 obscura group species are Drosophila pseudoobscura, Drosophila miranda, Drosophila persimilis, Drosophila bifasciata, Drosophila affinis, Drosophila athabasca, Drosophila azteca, and Drosophila subobscura; the 20 repleta group species are Drosophila borborema, Drosophila serido, Drosophila koepferae, Drosophila buzzatii, Drosophila starmeri, Drosophila venezolana, Drosophila uniseta, Drosophila martensis, Drosophila stalkeri, Drosophila richardsoni, Drosophila aldrichi, Drosophila arizonae, Drosophila mojavensis, Drosophila mulleri, Drosophila spenceri, Drosophila leonis, Drosophila eremophila, Drosophila mercatorum, Drosophila eohydei, and D hydei; the 5 virilis group species are Drosophila ezoana, Drosophila lacicola, Drosophila americana, Drosophila lummei, and Drosophila virilis; and the two Hawaiian species are Drosophila gymnobasis and Drosophila mimica.
The phylogenetic hypothesis in Fig. 1 is similar to that adopted by Tarrío et al. (20), except for some changes that ensued from research in our lab concerning the drosophilids (see refs. 21 and 27), and are widely accepted. Tree branches are depicted proportional to the time elapsed as it has been inferred from the fossil record. In any case, alternative phylogenetic rearrangements or branch lengths would not be expected to alter the conclusions of this study.
Results
Fig. 1 displays the phylogenetic distribution of intron A. The distribution is conspicuously disjointed. Intron A shows a markedly restricted distribution in animals, where it has been found exclusively in the dipterans D. sucinea and D. capricorni, two closely related members of the bocainensis subgroup (not >30 million years ago) of the Drosophila willistoni species group. All other animals and fungi in Fig. 1 lack this intron. In plants intron A seems to be common, because it has been found in representatives of the two distantly related phyla anthophyta, including one dicot (Arabidopsis) and one monocot (rice), and chlorophyta (the green algae Chlamydomonas). Intron A is the only Xdh intron known to be shared exclusively between arthropods and plants.
The alignment of the 10 (five on each side) amino acid residues surrounding the site of intron A indicates that the region has been conserved at the protein level across the three multicellular kingdoms, having neither gaps nor ambiguous positions (Fig. 1). Fig. 1 also gives the alignment of the corresponding encoding nucleotides (five on each side). As noted by Logsdon et al. (16), D. sucinea and D. capricorni both conform to the MAG│R protosplice-site model (CAG│G in the two species). This motif was likely already present in their common ancestor before the insertion of the intron, because this motif can be traced back at least to the common ancestor of insects (Fig. 1), much before the site incorporated intron A in the two bocainensis subgroup species. Interestingly, this same protosplice-site motif is also present in the three plant representatives in Fig. 1 (i.e., Arabidopsis, rice, and Chlamydomonas), although in this case the evidence in Fig. 1 does not permit us to decide whether the intron was gained in the last common ancestor of plants or whether it was already present before the split of the three multicellular kingdoms (see below).
Discussion
If intron A would have been present in the ancestor of all species shown in Fig. 1, it would be necessary to invoke a minimum of 14 independent losses to account for the observed disjointed phylogenetic distribution. With 14 losses, the rate of intron turnover at the site of intron A would be 1.33 per billion years (assuming the tree in Fig. 1 cumulatively spans 10.5 billion years). This rate is >400 times greater than the rate of intron turnover at the average site, averaged across Caenorhabditis elegans and D. melanogaster (≈0.0030; the rate ratio increases to ≈800:1 if the computation is circumscribed to animals; see ref. 2). Moreover, 5 of the 14 losses would have occurred in five of the shortest branches of the tree displayed in Fig. 1: the branches representing the last common ancestors of chordata, the melanogaster–obscura stem of Drosophila, the D. saltans species group, the D. willistoni species subgroup, and the subgenus Drosophila (with the two latest branches possibly concealing phyletic radiations). Therefore, the actual number of independent losses that would be necessary to invoke in all likelihood is much greater.
Invoking such a high number of parallel intron losses raises difficulties (9). One would have to postulate that a complex mutation such as the precise reinsertion of a reverse-transcribed spliced Xdh mRNA (probably the most feasible model thus far advanced to account for the precise excision of an intron; refs. 4 and 28) would have happened recurrently in numerous disparate lineages. Moreover, in every case the resulting intronless haplotype would need to rise in frequency until becoming fixed in each species. It seems difficult at this point to identify the reason why specifically intron A (and not other neighboring introns) should exhibit such a marked tendency to be lost in animals (and perhaps in fungi) but not in plants. The rate of intron excision that would be necessary to invoke to account for so many parallel fixations is probably unrealistic. There are not obvious grounds to suspect that intron A carriers might be particularly disfavored in animals. Intron A is a short, phase 0, symmetrical intron placed in a linker region between functional/structural domains (i.e., at the 5′ boundary of the a/b hammerhead domain, pfam01315), without any obvious reason why the intron should be particularly harmful. These difficulties become exacerbated when we take into account that intron A is absent in the vertebrates. This lineage exhibits among the highest known intron densities in eukaryotes (up to 35 introns in Xdh; ref. 16). Moreover, vertebrates are possibly most conservative with regard to intron differences in their genes (33 of 35 Xdh introns are conserved between the puffer fish, F. rubripes, and the mammals). If intron A would have been present in the common ancestor of animals, one would expect to find it much more likely in vertebrates than in an intron-poor lineage such as Drosophila. In conclusion, the disjointed phylogenetic distribution of intron A is explained more parsimoniously by insertion than by deletion.
Several considerations argue in favor of parallel insertion in the two lineages of plants and the Drosophila bocainensis subgroup. In both lineages, intron A occupies a protosplice site (ref. 16; their arguments regarding D. capricorni and D. sucinea apply equally for the Xdh sequences of Arabidopsis, rice, and Chlamydomonas). The coding region embedding the intron site is pretty well conserved in the alignment of Fig. 1, suggesting that the protosplice site has remained basically at the same location since the diversification of multicellular eukaryotes.
Intron A of D. sucinea and D. capricorni probably arose by transposition of a neighboring (92 downstream codons) intron B, because both are significantly similar in sequence (P < 0.05) (20, 29). Intron B was acquired earlier, because it is present in the willistoni as well as in the saltans species groups.
The three dipteran introns described by Tarrío et al. (ref. 20; introns A and B, already mentioned, plus intron C in the Mediterranean fruit fly, Ceratitis capitata) have been acquired in species that exhibit markedly higher AT content than their phylogenetic neighbors (30). In the cases of saltans and willistoni, and possibly analogously in the case of Ceratitis (see ref. 31), the observed bias in nucleotide composition was inferred to have been triggered by a shift toward increased AT content in the pattern of point mutation in the last common ancestor of the two species groups, probably associated with diminished natural selection due to reduced effective population sizes (refs. 23 and 30; see also ref. 25). If this hypothesis is correct, shifting genomic nucleotide composition combined with decreased population numbers may have created a propitious environment for the establishment of new introns. Accurate splicing depends not solely on intron features but also on splicing enhancer and silencer motifs that reside within adjacent exons (32). It is estimated that at least 10% of the total coding DNA of intron-containing genes may typically be involved in the guidance of splicing reactions (2). Hence, increased AT content might have favored the evolution of extra-intronic splicing signals. Moreover, potential intron deleterious effects are more likely to pass undetected to selection in smaller populations. These observations apply to entire genomes, not only to the Xdh region. Therefore, our hypothesis can be tested by finding out whether, on average, the species of the saltans and willistoni groups exhibit a greater number of introns than their relatives.
The phylogenetically disjointed gain of intron A may have come about by either one of two alternative routes: (i) intron A was first gained in the last common ancestor of plants and again much later in the last common ancestor of D. sucinea and D. capricorni (<30 million years ago), or (ii) intron A is as old as the last common ancestor to all multicellular eukaryotes, was lost in the lineage leading to animals and fungi, and regained in the lineage preceding the divergence of D. sucinea and D. capricorni. This second scenario may seem, at present, more likely because the aldehyde oxidase (Ao) gene of the protochordate C. intestinalis also bears an intron at exactly the same location as intron A. The Ao gene arose by duplication of Xdh before the diversification of multicellular eukaryotes. However, the phylogenetic sampling of Ao is too sparse for deciding between intron conservation and convergence.
Convergent intron insertion has been invoked to account for some patchy intron phylogenetic distributions (33–36). Perhaps the strongest case is that reported in several species of Chironomus by Hankeln et al. (ref. 33; see also ref. 1), who found that Chironomus melanotus contains a globin gene, putatively derived from an intronless paralog, with an intron at the same site as one found in plant leghemoglobin genes. Alternatively, that intron could have been ancestral and lost in six intermediate lineages. The parallel intron-gain scenario for the globin genes is sensitive to uncertainties about the phylogenetic relationships of Chironomus adopted by Hankeln et al. (see figure 3 in ref. 33), but their observations together with ours suggest that parallel intron gain is likely to be a real evolutionary phenomenon. If this is so, it raises a “multiple-hits” challenge, which together with the challenge created by parallel intron loss (see ref. 37) should be taken into account for appraising the value of introns as phylogenetic markers and also for obtaining accurate estimates of intron turnover (see ref. 9).
Acknowledgments
R.T. and F.R.-T. received support from Ministerio de Ciencia y Tecnología Contracts Doctor I3P and Ramón y Cajal, respectively. This research was supported by National Institutes of Health Grant GM42397 (to F.J.A.).
Abbreviations: IE, introns early; IL, introns late; XDH, xanthine dehydrogenase.
References
- 1.Logsdon, J. M., Jr. (1998) Curr. Opin. Genet. Dev. 8, 637–648. [DOI] [PubMed] [Google Scholar]
- 2.Lynch, M. & Richardson, A. O. (2002) Curr. Opin. Genet. Dev. 12, 701–710. [DOI] [PubMed] [Google Scholar]
- 3.Nixon, J. E. J., Wang, A., Morrison, H. G., McArthur, A. G., Sogin, M. L., Loftus, B. J. & Samuelson, J. (2002) Proc. Natl. Acad. Sci. USA 99, 3701–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharp, P. A. (1991) Science 254, 663. [DOI] [PubMed] [Google Scholar]
- 5.Stoltzfus, A. (1999) J. Mol. Evol. 49, 169–181. [DOI] [PubMed] [Google Scholar]
- 6.Kaessmann, H., Zöllner, S., Nekrutenko, A. & Li, W.-H. (2002) Genome Res. 12, 1642–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roy, S. W., Nosaka, M., de Souza, S. J. & Gilbert, W. (1999) Gene 238, 85–91. [DOI] [PubMed] [Google Scholar]
- 8.Fedorov, A., Merican, A. F. & Gilbert, W. (2002) Proc. Natl. Acad. Sci. USA 99, 16128–16133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynch, M. (2002) Proc. Natl. Acad. Sci. USA 99, 6118–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoltzfus, A., Logsdon, J. M., Jr., Palmer, J. D. & Doolittle, W. F. (1997) Proc. Natl. Acad. Sci. USA 94, 10739–10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rzhetsky, A., Ayala, F. J., Hsu, L. C., Chang, C. & Yoshida, A. (1997) Proc. Natl. Acad. Sci. USA 94, 6820–6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato, Y., Niimura, Y., Yura, K. & Go, M. (1999) Gene 238, 93–101. [DOI] [PubMed] [Google Scholar]
- 13.Rogozin, I. B., Lyons-Weiler, J. & Koonin, E. V. (2000) Trends Genet. 16, 430–432. [DOI] [PubMed] [Google Scholar]
- 14.Sakharkar, M. K., Tan, T. W. & de Souza, S. J. (2001) Bioinformatics 17, 671–675. [DOI] [PubMed] [Google Scholar]
- 15.Dibb, N. J. & Newman, A. J. (1989) EMBO J. 8, 2015–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Logsdon, J. M., Jr., Stoltzfus, A. & Doolittle, W. F. (1998) Curr. Biol. 8, 560–563. [DOI] [PubMed] [Google Scholar]
- 17.Long, M. & Deutsch, M. (1999) Mol. Biol. Evol. 16, 1528–1534. [DOI] [PubMed] [Google Scholar]
- 18.Patthy, L. (1985) Cell 41, 657–663. [DOI] [PubMed] [Google Scholar]
- 19.Levitsky, V. G., Podkolodnaya, O. A., Kolchanov, N. A. & Podkolodny, N. L. (2001) Bioinformatics 17, 1062–1064. [DOI] [PubMed] [Google Scholar]
- 20.Tarrío, R., Rodríguez-Trelles, F. & Ayala, F. J. (1998) Proc. Natl. Acad. Sci. USA 95, 1658–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarrío, R., Rodríguez-Trelles, F. & Ayala, F. J. (2000) Mol. Phylogenet. Evol. 16, 344–349. [DOI] [PubMed] [Google Scholar]
- 22.Rodríguez-Trelles, F. Alarcón, R. L. & Fontdevila, A. (2000) Mol. Biol. Evol. 17, 1112–1122. [DOI] [PubMed] [Google Scholar]
- 23.Rodríguez-Trelles, F., Tarrío, R. & Ayala, F. J. (2000) Mol. Biol. Evol. 17, 1710–1717. [DOI] [PubMed] [Google Scholar]
- 24.Rodríguez-Trelles, F., Tarrío, R. & Ayala, F. J. (2001) Proc. Natl. Acad. Sci. USA 98, 11405–11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Begun, D. J. & Whitley, P. (2002) Genetics 162, 1725–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1987) Nucleic Acids Res. 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarrío, R., Rodríguez-Trelles, F. & Ayala, F. J. (2001) Mol. Biol. Evol. 18, 1464–1473. [DOI] [PubMed] [Google Scholar]
- 28.Fink, G. R. (1987) Cell 49, 5–6. [DOI] [PubMed] [Google Scholar]
- 29.Rodríguez-Trelles, F., Tarrío, R. & Ayala, F. J. (2000) J. Mol. Evol. 50, 123–130. [DOI] [PubMed] [Google Scholar]
- 30.Rodríguez-Trelles, F., Tarrío, R. & Ayala, F. J. (1999) Genetics 153, 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodríguez-Trelles, F., Tarrío, R. & Ayala, F. J. (2000) J. Mol. Evol. 50, 1–10. [DOI] [PubMed] [Google Scholar]
- 32.Blencowe, B. J. (2000) Trends Biochem. Sci. 25, 106–110. [DOI] [PubMed] [Google Scholar]
- 33.Hankeln, T., Friedl, H., Ebersberger, I., Martin, J. & Schmidt, E. R. (1997) Gene 205, 151–160. [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharya, D., Lutzoni, F., Reeb, V., Simon, D., Nason, J. & Fernandez, F. (2000) Mol. Biol. Evol. 17, 1971–1984. [DOI] [PubMed] [Google Scholar]
- 35.Robertson, H. M. (2000) Genome Res. 10, 192–203. [DOI] [PubMed] [Google Scholar]
- 36.Boudet, N., Aubourg, S., Toffano-Nioche, C., Kreis, M. & Lecharny, A. (2001) Genome Res. 11, 2101–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krzywinski, J. & Besansky, N. J. (2002) Mol. Biol. Evol. 19, 362–366. [DOI] [PubMed] [Google Scholar]