Abstract
During the late Pleistocene, early anatomically modern humans coexisted in Europe with the anatomically archaic Neandertals for some thousand years. Under the recent variants of the multiregional model of human evolution, modern and archaic forms were different but related populations within a single evolving species, and both have contributed to the gene pool of current humans. Conversely, the Out-of-Africa model considers the transition between Neandertals and anatomically modern humans as the result of a demographic replacement, and hence it predicts a genetic discontinuity between them. Following the most stringent current standards for validation of ancient DNA sequences, we typed the mtDNA hypervariable region I of two anatomically modern Homo sapiens sapiens individuals of the Cro-Magnon type dated at about 23 and 25 thousand years ago. Here we show that the mtDNAs of these individuals fall well within the range of variation of today's humans, but differ sharply from the available sequences of the chronologically closer Neandertals. This discontinuity is difficult to reconcile with the hypothesis that both Neandertals and early anatomically modern humans contributed to the current European gene pool.
The origin of anatomically modern humans (a.m.h.) is traditionally explained by two contrasting models. The multiregional model, in its original formulation (1, 2), proposed that modern humans evolved in the last two million years as a single polytypic species, through the independent appearance of modern traits in different areas at different times. Recent adjustments of this model (3–5) retained the concept of a single evolving species, but argue that modern forms expanding from Africa may have mixed, even extensively, with archaic forms, such as Neandertals in Europe. Variants of the multiregional model differ in the extent of predicted admixture between anatomically modern and archaic humans, but all of them assume a dual ancestry, archaic and modern, in the current human gene pool, and therefore some continuity over time between these forms.
The Out-of-Africa model (6), on the other hand, suggests that a.m.h. first arose in Africa some 150,000 years ago and then dispersed replacing archaic forms (Neandertals in Europe). This model does not imply any hypothesis regarding the structure of archaic humans populations, nor that modern humans migrating from Africa were a biologically distinct species. It only assumes that modern traits evolved recently in a single region, Africa, and the vast majority of the genomes of current human populations can be traced back to these African migrants. This hypothesis does not seem to differ radically from Relethford's (4) view of multiregional evolution, whereby the Neandertal's contribution to the modern European gene pool may have been small, yet nonzero.
Analyses of morphological traits, Neandertal ancient DNA and modern DNA (e.g., refs. 7–9), appear to support a recent African origin of all humankind. However, it has been argued that patterns of genetic diversity are not incompatible with a multiregional model (5, 10–12). For instance, Nordborg (10) showed that the differences between Neandertal's and modern mitochondrial sequences are sufficient to rule out random mating between them, but not more complicated models of interbreeding. Those results reflect the existing uncertainties on the European demographic history of the last 30,000 years. Clearly, genetic typing of the earliest a.m.h. in Europe, sometimes referred to as Cro-Magnons or Cro-Magnoid from the site in France where they were first discovered, is a crucial step for solving this question (13–15), because that would allow a genetic comparison between individuals who lived at a much shorter (ideally, zero) time distance. The Out-of Africa model, in fact, predicts genetic discontinuity between Neandertals and early a.m.h. (the former being a separate lineage replaced by the latter) and genetic continuity along the a.m.h. lineages from the Upper Palaeolithic until the present. The multiregional models, on the contrary, predict at least some level of genetic continuity from the archaic Neandertal forms to the almost contemporary Cro-Magnon forms up to today's Europeans.
In this study, we typed the hypervariable region I (HVRI) of the mitochondrial genome (360 bp) from the bones of two early a.m.h. of the Cro-Magnon type from Southern Italy. We validated the sequences obtained through a number of biochemical tests, and we compared them with those of four Neandertal (8, 16–18) specimens and with a large data set of modern human sequences (19, 20).
Materials and Methods
The remains of the two individuals of this study were recovered in the Paglicci cave, Southern Italy. Radioactive carbon determination dated them to 23,000 ± 350 (Paglicci-25) and 24,720 ± 429 (Paglicci-12) years ago, respectively (21, 22). One of us (F.M.) removed from each skeleton two bone fragments, one from a femur and one from a rib.
Authentication Methods: An Overview. Genetic typing of ancient samples is technically challenging, because DNA is generally degraded and present in small amounts in the available specimens. The most stringent standards for authentication of ancient DNA (23, 24, 25) were therefore followed in this study. In particular, using the same order and names of the nine key criteria described in ref. 23, we proceeded as follows.
DNA was extracted in a laboratory room exclusively dedicated to ancient DNA analysis (Physically isolated work area).
For each sample, two independent DNA extractions were performed from fragments of different bones, and PCR controls produced negative results (Control amplifications).
Amplification of large DNA fragments, unusual in ancient DNA analyses, was not observed, and the final consensus sequences make phylogenetic sense, i.e., do not appear to be a combination of different sequences, resulting from contamination of the specimens by exogenous DNA (Appropriate molecular behavior).
All results were identical in two independent extractions and two independent amplifications using four different overlapping primer pairs (Reproducibility).
Ninety-three and 72 clones were analyzed for Paglicci-25 and Paglicci-12, respectively; the average rate of Taq misincorporation across fragments was low (4.3 substitutions every 1,000 bp within the HVRI), with at least 79% of the clones showing the consensus nucleotide at each DNA fragment (Cloning).
A single DNA extraction and amplification of two overlapping fragments was independently repeated in a different laboratory for Paglicci-25; the sequences were consistent across laboratories (Independent replication).
The degree of racemization for three amino acids was low in both samples, suggesting a high probability to obtain intact ancient biomolecules from the specimens (Biochemical preservation).
The estimated copy number of target DNA (between 1,000 and 1,500 in both samples) was larger than the threshold under which sporadic contamination cannot be excluded (Quantitation).
No human sequence was amplified from the horse remains found associated to the Paglicci skeletons using either primers specific for humans or for horse; on the other hand, the DNA sequence obtained from the horse remains using specific primers aligns well with sequences of Equus caballus in GenBank (Associated remains).
The nine key criteria agree in indicating that the authenticity of the sequences we present is supported as much as technically possible at the present. We are not aware of any other published study of ancient DNA considering all of the precautions we used here to exclude contamination.
Authentication Methods: Details. Amino acid racemization. The degree of amino acid racemization was estimated by means of reversephase HPLC (26) using ≈5 mg of bone. The values of the D/L ratio observed for three amino acids are 0.0506 (Asp), 0.0082 (Glu), and 0.0029 (Ala) for Paglicci-25, and 0.0631 (Asp), 0.0167 (Glu), and 0.0055 (Ala) for Paglicci-12.
Quantitation of target DNA. The number of copies of target DNA was estimated by competitive PCR (27). A competitor was used containing a 95-bp deletion (from nucleotide positions 16131–16225 according to ref. 28). PCR components were the same as described below for the amplification of the second HVRI fragment in the Florence laboratory. We included in each amplification a negative control. Thermal cycler conditions consisted of an initial 10-min incubation at 95°C followed by 45 cycles of 50 sec at 94°C, 50 sec at 48°C, and 50 sec at 72°C, with a final extension step at 72°C for 5 min.
DNA extraction. Bones were not washed after recovery to prevent absorption of modern DNA. Each bone surface was brushed and irradiated (1 h under UV light). As described above (point 6), independent extractions, amplifications, and cloning were performed for Paglicci-25 in Florence and in Barcelona. Slightly different protocols were used in these laboratories, and we therefore describe them separately in this and the following two paragraphs. In Florence, DNA was extracted from powdered bone by means of a silica-base protocol (modified from ref. 8). In Barcelona, the powdered sample was decalcified overnight with 10 ml 0.5 M EDTA, incubated over the next night with 1 ml 10% SDS, 0.5 ml 1 M Tris·HCl, and 100 μlof1mg/ml proteinase K, extracted with phenol/chloroform and desalted with Centricon 30 microconcentrators (Amicon).
DNA amplification. In Florence, 2 μl of DNA were amplified with the following profile: 94°C for 10 min and 40 cycles of a denaturation (94°C for 45 sec), annealing (53°C for 1 min), and extension step (72°C for 1 min); the 50-μl reaction mix contained 2 units of AmpliTaq Gold polymerase and 1× reaction buffer (Applied Biosystems), 200 μM of each dNTP, 1.5 mM MgCl2, and 1 μM of each primer (L 15,995-H 16,132, L 16,107-H 16,261, L 16,247-H 16,402, L 16,131-H 16,218). In Barcelona, PCR amplifications were performed in a 25-μl volume containing 1 μl of DNA, 1 unit of TaqDNA polymerase, 1× reaction buffer (EcoGen, Madrid, Spain), 2.5 mM MgCl2, 0.25 mM dNTPs, 2 mg/ml BSA, and 1 μM of each primer (L16,022-H16,218 and L16,185-H16,401); the PCR profile was 40 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with a first denaturation step of 94°C for 5 min. PCR products were visualized in a low-melting point agarose gel, and the appropriate bands were excised from the gel, melted in 150–200 μl of water, and subjected to a second 35 cycles of PCR. The sequences of the primers are available in Fig. 3, which is published as supporting information on the PNAS web site, www.pnas.org.
Cloning and sequencing. PCR products were cloned in Florence by using the TOPO TA Cloning kit (Invitrogen) according to the supplier's instructions. Screening of white recombinant colonies was accomplished by PCR, transferring the colonies into a 30 μl reaction mix with 2 mM MgCl2, 1 μM of each primer (M13 forward and reverse universal primers), 0.125 μM of each dNTP, and 0.75 units of Taq polymerase. After 5 min at 92°C, 30 cycles of PCR (30 sec at 90°C, 1 min at 50°C, 1 min at 72°C) were carried out, and clones with an insert of the expected size were identified by agarose gel electrophoresis. After purification of these PCR products with Microcon PCR devices (Amicon, Beverly, MA), a volume of 1.5 μl was cycle-sequenced following the BigDye Terminator kit (Applied Biosystems) supplier's instructions. The sequence was determined by using an Applied BioSystem 377 DNA sequencer. SureClone Ligation kit (Amersham Pharmacia) was used to clone the PCR products in Barcelona. Cells were grown in LB medium, plated on IPTG/5-bromo-4-chloro-3-indolyl β-D-galactoside (X-Gal) agar plates and incubated overnight at 37°C. White colonies (containing the insert) were transferred to PCRs and sequenced. The rates of Taq misincorporations computed for each fragment as the total number of substitutions every 1,000 bp within the HVRI are: 5.85 for the L15,995-H16,132 fragment, 5.18 for the L16,107-H16,261 fragment, 3.57 for the L16,247-H16,402 fragment, 6.90 for the L16,130-H16,218 fragment, 1.54 for the L16,022-H16,218 fragment, and 2.83 for the L16,185-H16,401 fragment. The sequences of the 165 clones are available in Fig. 3.
Amplification with Neandertal-specific primers. Amplifications of the Paglicci extracts with two pairs of Neandertal-specific primers (L16,022-NH16,139 and NL16,263/264-NH16,400, ref. 8) were also attempted. Five microliters of DNA were amplified with the following profile: 94°C for 10 min and 45 cycles of a denaturation (94°C for 45 sec), annealing (57°C for 1 min for the first couple and 59°C for 1 min for the second couple) and extension step (72°C for 1 min). The 50-μl reaction mix contained 2 units of AmpliTaq Gold polymerase and 1× reaction buffer (Applied Biosystems), 200 μM of each dNTP, 1.5 mM MgCl2, and 1 μM of each primer. No amplification yielded any PCR products.
DNA analysis of faunal remains. The same protocols used for the human remains were used to extract the DNA from a horse bone found in the same layer. A fragment of 155 bp of the mtDNA D-loop was amplified by using the following primers: Equus L 5′-CCCCCACATAACAACATACC-3′ and Equus H 5′-ATGGGGTATGCACGATCAAT-3′. The PCR was performed with 2 μl of DNA, 1 μM of each primer, 200 μM of each dNTPs, 1× reaction buffer (Applied Biosystems), 1.5 mM MgCl2, and 2 units of Taq Gold (Applied Biosystems) in a total volume of 50 μl. Thermocycling was performed as follows: initial denaturation at 94°C for 10 min followed by 40 cycles of 94°C for 1 min, 57°C for 1 min, 72°C for 1 min, final extension at 72°C for 5 min. The PCR product was run on 1.5% agarose gel, excised from the gel and purified with Ultra Free DNA (Amicon, Beverly, MA). Cloning and sequencing of the PCR products was performed as described above (Florence protocol). The sequences of the primers and the 15 clones analyzed are available in Fig. 3. The amplification of DNA from the horse remain by using human specific primers (L 16,107-H 16,261) yielded no PCR products.
Statistical Analysis. Multidimensional scaling (MDS) was used to portray graphically the genetic distances between sequences. In this analysis, sequences are represented by points in a two-dimensional space, and the linear distances between points are proportional to the genetic distances between sequences. The degree of correspondence between the distances among points in the MDS map and the original distances among sequences is measured (inversely) by an index called stress.
The genetic distance between sequences is based on the Tamura–Nei model (29) with a γ distribution for substitution rate heterogeneity. This model is the best fitting model for HVRI when humans, Neandertals, and chimpanzees are considered (12). The γ shape parameter α was set to 0.3. We also considered different values of α (between 0.3 and 0.7) and a different model of nucleotide substitution (Kimura two-parameters with rate heterogeneity), but the graphical representations were not affected by the choice of the model and of the parameters.
Results and Discussion
The D-loop sequence of Paglicci-25 shows no substitutions with respect to the Cambridge reference sequence (28). This sequence is fairly common (14%) in a database of 2,566 today's Europeans and Near Easterners from 35 different populations (updated from ref. 20). The average number of substitutions between Paglicci-25 and these 2,566 sequences is 2.34 (SD = 1.75, range 0–11), whereas the average number of substitutions between today's Europeans is 4.35 (SD = 2.32, range 0–18). The comparison of this 23,000-year-old a.m.h. with four Neandertals dated at ≈29,000, 40,000, 40,000, and 42,000 years ago (8, 16–18) produces very different results: 23, 28, 23, and 24 substitutions separate Paglicci-25 from the Neandertals, respectively. The second individual we typed, Paglicci-12, shows a single difference (a C → T transition at nt position 16,223) from the Cambridge reference sequence, and 3.17 differences on the average from today's Europeans (SD = 1.66, range 0–10). As in the case of Paglicci-25, Paglicci-12 is very different from the four Neandertal sequences: 22, 27, 22, and 23 substitutions, respectively. The consensus sequences of Paglicci-25 and Paglicci-12 are different from the sequences of all scientists who manipulated the bones during the laboratory analyses, thus also excluding the possibility of recent contamination.
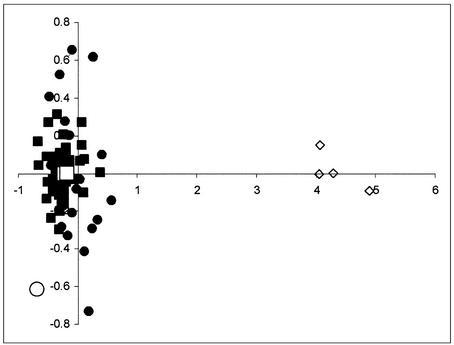
This result is graphically summarized in the multidimensional scaling analysis (MDS), where the plotted points correspond to the Paglicci, Neandertal, and representative today's sequences, and the genetic distances among them are proportional to the linear distances between the points in Fig. 1. Neandertals and a.m.h. form two distinct clouds of points, respectively, with the Paglicci sequences falling well within the range of variation of today's humans. We note that if the sequence from the 40,000 (30) years old sample Lake Mungo 3 (an anatomically modern Australian individual, ref. 31) is added to the MDS analysis, that pattern does not change, and two distinct clusters of data points remain evident. If genuine (25, 32–34), the sequence of Lake Mungo 3 is among the most divergent modern human mtDNAs, but yet it can be unequivocally attributed to the modern cluster of individuals in Fig. 1.
Fig. 1.
MDS of HVRI sequences of 60 modern Europeans (filled squares), 20 modern non-Europeans (filled circles), 4 Neandertals (open diamonds), the Australian Lake Mungo 3 (open circle), and the two early a.m.h. typed in this study (open squares). European and non-European sequences in this figure were selected to represent the most divergent lineages observed in modern individuals. Note that the axes have different scales. The stress value for this analysis was 0.128.
Specific mtDNA sites outside HVRI were also analyzed (by amplification, cloning, and sequencing of the surrounding region) to classify more precisely the ancient sequences within the phylogenetic network of present-time mtDNAs (35, 36). Paglicci-25 has the following motifs: +7,025 AluI, 00073A, 11719G, and 12308A. Therefore, this sequence belongs to either haplogroups HV or pre-HV, two haplogroups rare in general but with a comparatively high frequencies among today's Near-Easterners (35). Paglicci-12 shows the motifs 00073G, 10873C, 10238T, and AACC between nucleotide positions 10397 and 10400, which allows the classification of this sequence into the macrohaplogroupN,containing haplogroups W, X, I, N1a, N1b, N1c, and N*. Following the definition given in ref. 36, the presence of a single mutation in 16,223 within HRVI suggests a classification of Paglicci-12 into the haplogroup N*, which is observed today in several samples from the Near East and, at lower frequencies, in the Caucasus (35). It is difficult to say whether the apparent evolutionary relationship between Paglicci-25 and Paglicci-12 and those populations is more than a coincidence. Indeed, the haplogroups to which the Cro-Magnon type sequences appear to belong are rare among modern samples, and therefore their frequencies are poorly estimated. However, genetic affinities between the first anatomically modern Europeans and current populations of the Near East make sense in the light of the likely routes of Upper Paleolithic human expansions in Europe, as documented in the archaeological record (37).
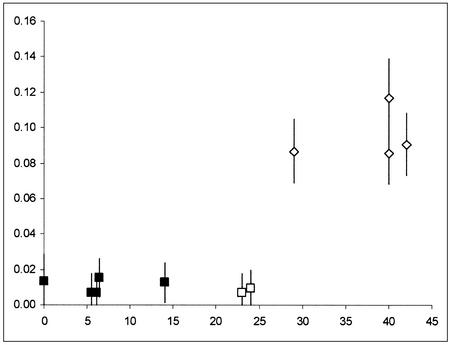
A pattern of genetic distances through time emerges clearly when four additional HVRI sequences from prehistoric anatomically modern Europeans dated between 5,500 and 14,000 years ago (38, 39) are considered (Fig. 2). Going back in time from the present to 25,000 years ago, prehistoric Europeans show an approximately constant number of differences in comparison with today's Europeans, very similar indeed to the number of differences between two randomly chosen modern sequences. Conversely, an abrupt increase of genetic distance from today's Europeans is observed for the Neandertals, even though the most recent of them is separated from the older Paglicci sample by only a few hundreds of generations. Therefore, these results suggest a pattern of genetic continuity in the modern humans' genealogy from the Upper Palaeolithic period to the present, but a clear discontinuity with respect to Neandertals of similar ages.
Fig. 2.
Average genetic distance between ancient and modern samples (2,566 sequences of modern Europeans; y axis), as a function of the samples' age (x axis, in thousands of years). Vertical lines represent two standard deviations above and below the mean. Squares, a.m.h. Diamonds, Neandertals. The Paglicci samples typed in this study are indicated by open squares. The point at 0 years indicates the average pairwise difference between present-day samples.
Even the most stringent available criteria for validating ancient human DNA sequences do not allow one to prove that the sequences determined are authentic. Only if a sequence is radically different from modern ones, as is the case for Neandertals, can one be relatively sure that no contamination has affected the results. Therefore, a certain degree of prudence is necessary before drawing any conclusions from this study. Still, none of the biochemical tests we carried out suggests that different sequences (namely the endogenous one plus some contaminating sequences) were amplified from the 23,000- and 25,000-year-old specimens that we used. In addition, the amino acid racemization test strongly suggests that reasonably well preserved DNA should be present in those specimens. Because DNA from all four Cro-Magnon type bone fragments could be amplified and sequenced only by using primers specific for modern humans, and not for Neandertals, there is little doubt that the mtDNAs of early a.m.h. and of cronologically close Neandertals were, at least, very different.
Under the multiregional model of human origins, Neandertals and modern humans are just one population observed at different times (1, 2). Therefore, when comparing sequences sampled along the single human–Neandertal genealogy, one should not observe any major discontinuity. More recent versions of the multiregional model (3–5) suggest that Neandertals and early a.m.h. were regional populations of the same evolving species connected by gene flow, and both archaic and modern forms contributed, possibly in different proportions, to the present day human gene pool. Under this updated multiregional model, the absence of Neandertal mtDNA lineages in living humans is regarded as a consequence of a random drift or a selection process of lineage extinction since the disappearance of Neandertals. In this case, unless the extinction of Neandertal lineages was almost instantaneous, the probability of finding such lineages in early a.m.h. should not be too low. All these expectations are inconsistent with the data and the analysis here presented. Two a.m.h. dated between 23,000 and 25,000 years ago appear to have HVRI sequences fully compatible with the variation observed both in contemporary and in ancient samples of a.m.h., and certainly they do not show any special relationships with the almost contemporary Neandertals. These results are at odds with the view whereby Neandertals were genetically related with the anatomically modern ancestors of current Europeans or contributed to the present day human gene pool. Although only six HVRI sequences of ancient a.m.h and four sequences of Neandertals are available to date, the sharp differentiation among them represents a problem for any model regarding the transition from archaic to modern humans as a process taking place within a single evolving human lineage.
Supplementary Material
Acknowledgments
We thank A. Torroni and L. Castrì for their help in understanding the classification of mtDNA haplogroups; E. Cappellini, S. Sanna, C. Conti, and E. Pecchioli for their technical contribution; and J. Moggi-Cecchi for his critical reading of the manuscript. This research was supported by funds from the University of Florence, the University of Ferrara, the Italian National Research Council (CNR) “Progetto Beni Culturali,” the Italian CNR Eurocores Project “The Origin of Man, Language, and Languages,” the “Fondazione Dino Terra,” and the Departament d'Universitats, Recerca i Societat de la Informació de la Generalitat de Catalunya.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: a.m.h., anatomically modern human(s); HVRI, hypervariable region I; MDS, multidimensional scaling.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY283027 and AY283028).
References
- 1.Wolpoff, M. H., Wu, X. & Thorne, A. (1984) in The Origins of Modern Humans: A World Survey of the Fossil Evidence, eds. Smith, F. & Spencer, F. (Liss, New York), pp. 411–483.
- 2.Wolpoff, M. H., Thorne, A. G., Smith, F. H., Frayer D. W. & Pope, G. G. (1994) in Origins of Anatomically Modern Humans, eds. Nitecki, M. H. & Nitecki, D. V. (Plenum, New York), pp. 175–199.
- 3.Wolpoff, M. H. & Caspari, R. (1997) Race and Human Evolution: A Fatal Attraction (Simon & Schuster, New York).
- 4.Relethford, J. H. (2001) Am. J. Phys. Anthropol. 115, 95–98. [DOI] [PubMed] [Google Scholar]
- 5.Templeton, A. R. (2002) Nature 416, 45–51. [DOI] [PubMed] [Google Scholar]
- 6.Stringer, C. B. & Andrews, P. (1988) Science 239, 1263–1268. [DOI] [PubMed] [Google Scholar]
- 7.Sokal, R. R., Oden, N. L., Walker, J. & Waddle, D. M. (1997) J. Hum. Evol. 32, 501–522. [DOI] [PubMed] [Google Scholar]
- 8.Krings, M., Stone, A., Schmitz, R. W., Krainitzki, H., Stoneking, M. & Pääbo, S. (1997) Cell 90, 19–30. [DOI] [PubMed] [Google Scholar]
- 9.Underhill, P. A., Shen, P., Lin, A. A., Jin, L., Passarino, G., Yang, W. H., Kauffman, E., Bonne-Tamir, B., Bertranpetit, J., Francalacci, P., et al. (2000) Nat. Genet. 26, 358–361. [DOI] [PubMed] [Google Scholar]
- 10.Nordborg, M. (1998) Am. J. Hum. Genet. 63, 1237–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawks, J., Hunley, K., Lee, S. H. & Wolpoff, M. H. (2000) Mol. Biol. Evol. 17, 2–22. [DOI] [PubMed] [Google Scholar]
- 12.Gutierrez, G., Sanchez, D. & Marin, A. (2002) Mol. Biol. Evol. 19, 1359–1366. [DOI] [PubMed] [Google Scholar]
- 13.Relethford, J. H. (2001) Proc. Natl. Acad. Sci. USA 98, 390–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stringer, C. B. & Davies, W. (2001) Nature 413, 791–792. [DOI] [PubMed] [Google Scholar]
- 15.Gibbons, A. (2001) Science 291, 1726. [Google Scholar]
- 16.Ovchinnikov, I. V., Gotherstrom A., Romanova, G. P., Kharitonov, V. M., Liden, K. & Goodwin, W. (2000) Nature 404, 490–493. [DOI] [PubMed] [Google Scholar]
- 17.Krings, M., Capelli, C., Tschentscher, F., Geisert, H., Meyer, S., von Haeseler, A., Grossschmidt, K., Possnert, G., Paunovic, M. & Pääbo, S. (2000) Nat. Genet. 26, 144–146. [DOI] [PubMed] [Google Scholar]
- 18.Schmitz, R. W., Serre, D., Bonani, G., Feine, S., Hillgruber, F., Krainitzki, H., Pääbo, S. & Smith, F. H. (2002) Proc. Natl. Acad. Sci. USA 99, 13342–13347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Excoffier, L. & Schneider, S. (1999) Proc. Natl. Acad. Sci. USA 96, 10597–10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simoni, L., Calafell, F., Pettener, D., Bertranpetit, J. & Barbujani, G. (2000) Am. J. Hum. Genet. 66, 262–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallegni, F., Bertoldi, F. & Manolis, S. K. (1999) Homo 50 (2), 127–148. [Google Scholar]
- 22.Mallegni, F. & Palma di Cesnola, A. (1994) Anthropologie 32, 45–47. [Google Scholar]
- 23.Cooper, A. & Poinar, H. N. (2000) Science 289, 1139. [DOI] [PubMed] [Google Scholar]
- 24.Hofreiter, H., Serre, D., Poinar, H. N., Kuch, M. & Pääbo, S. (2001) Nat. Rev. Genet. 2, 353–359. [DOI] [PubMed] [Google Scholar]
- 25.Cooper, A., Rambaut, A., Macaulay, V., Willerslev, E., Hansen, A. J. & Stringer, C. (2001) Science 292, 1655–1656. [DOI] [PubMed] [Google Scholar]
- 26.Poinar, H. N., Hoss, M., Bada, J. L. & Pääbo, S. (1996) Science 272, 864–866. [DOI] [PubMed] [Google Scholar]
- 27.Handt, O., Krings, M., Ward, R. H. & Pääbo, S. (1996) Am. J. Hum. Genet. 59, 368–376. [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson, S., Bankier, A. T., Barrell, B. G., de Bruijn, M. H., Coulson, A. R., Drouin, J., Eperon, I. C., Nierlich, D. P., Roe, B. A., Sanger, F., et al. (1981) Nature 290, 457–465. [DOI] [PubMed] [Google Scholar]
- 29.Tamura, K. & Nei, M. (1993) Mol. Biol. Evol. 10, 512–526. [DOI] [PubMed] [Google Scholar]
- 30.Bowler, J. M., Johnston, H., Olley, J. M., Prescott, J. R., Roberts, R. G., Shawcross, W. & Spooner, N. A. (2003) Nature 421, 837–840. [DOI] [PubMed] [Google Scholar]
- 31.Adcock, G. J., Dennis, E. S., Easteal, S., Huttley, G. A., Jermiin, L. S., Peacock, W. J. & Thorne A. (2001) Proc. Natl. Acad. Sci. USA 98, 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trueman, J. W. H. (2001) Archaeol. Oceania 36, 163–165. [Google Scholar]
- 33.Groves, C. (2001) Archaeol. Oceania 36, 166–167. [Google Scholar]
- 34.Colgan, D. (2001) Archaeol. Oceania 36, 168–169. [Google Scholar]
- 35.Richards M., Macaulay, V., Hickey, E., Vega, E., Sykes, B., Guida, V., Rengo, C., Sellitto, D., Cruciani, F., Kivisild, T., et al. (2000) Am. J. Hum. Genet. 67, 1251–1276. [PMC free article] [PubMed] [Google Scholar]
- 36.Finnila, S., Lehtonen, M. S. & Majamaa, K. (2001) Am. J. Hum. Genet. 68, 1475–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mellars, P. A. (1992) Philos. Trans. R. Soc. London B 337, 225–234. [DOI] [PubMed] [Google Scholar]
- 38.Handt, O., Richards, M., Trommsdorff, M., Kilger, C., Simanainen, J., Georgiev, O., Bauer, K., Stone, A., Hedges, R., Schaffner, W., et al. (1994) Science 264, 1775–1778. [DOI] [PubMed] [Google Scholar]
- 39.Di Benedetto, G., Nasidze, I. S., Stenico, M., Nigro, L., Krings, M., Lanzinger, M., Vigilant, L., Stoneking, M., Pääbo, S. & Barbujani, G. (2000) Eur. J. Hum. Genet. 8, 669–677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.