Abstract
The mouse DBA/2 (D2) strain is very susceptible to infection with virulent Mycobacterium tuberculosis, whereas C57BL/6 (B6) is much more resistant. Infection of D2 and B6 mice with M. tuberculosis H37Rv by the respiratory route is biphasic: during the first 3 weeks, there is rapid bacterial growth in the lung of both strains, whereas beyond this point replication stops in B6 but continues in D2, causing rapidly fatal pulmonary disease. To identify the genes regulating growth of M. tuberculosis in the lungs of these two strains, 98 informative (B6 × D2) F2 mice were infected by the respiratory route with M. tuberculosis H37Rv (2 × 102 colony-forming units), and the extent of bacterial replication in the lungs at 90 days was used as a quantitative measure of susceptibility in a whole-genome scan. Quantitative trait locus mapping identified a major locus on chromosome 19 (Tuberculosis resistance locus-4, Trl-4; logarithm of odds 5.6), which regulated pulmonary replication of M. tuberculosis and accounted for 25% of the phenotypic variance. B6 alleles at Trl-4 were inherited in an incompletely dominant fashion and associated with reduced bacterial replication. An additional effect of a locus (Trl-3), previously shown to affect survival to i.v. infection with M. tuberculosis, was also noted. F2 mice homozygous for B6 alleles at both Trl-3 and Trl-4 were as resistant as B6 parents, whereas mice homozygous for D2 alleles were as susceptible as D2 parents. These results suggest a strong genetic interaction between Trl-3 and Trl-4 in regulating pulmonary replication of M. tuberculosis.
The global prevalence of Mycobacterium tuberculosis infection was recently estimated at 32% (1.9 billion people), with an evaluated 8 million new cases of active tuberculosis (TB) and 2 million deaths that year (1). Only a small proportion of individuals that come in contact with M. tuberculosis develop active TB, and a wide clinical spectrum of disease severity is observed in such individuals. Appearance of full-blown disease is determined in part by microbial virulence determinants (2) and by environmental and host factors, such as social conditions and immune status, most critically by the presence of concomitant HIV infection (3). An important genetic component of vulnerability to TB in humans affecting susceptibility per se, disease progression, and ultimate outcome has been well documented (4). This genetic component is supported by epidemiological data pointing to sex (5, 6) and racial differences in susceptibility (7), as well as geographical distribution and familial aggregation (4). In addition, studies of first-contact epidemics in isolated populations with no ancestral experience of this infection (Yanomami, Qu'Appelle Indians) (8, 9), survival data from accidental injection of virulent M. tuberculosis during a bacillus Calmette–Guérin vaccination trial (Lubeck disaster) (10), and studies in twins showing higher concordance rates of TB in monozygotic vs. dizygotic twins (11–13) provide compelling evidence that host genes affect the outcome of M. tuberculosis infection.
To date, inherited mutations in gp91/phox (14), IL-12 (15), and in the IFN-γ receptor genes (16) have been found in a few rare familial cases of infantile TB or of disseminated Mycobacterium bovis (bacillus Calmette–Guérin) infection (4, 17). However, such Mendelian disorders are exceedingly rare. The genetic component of TB susceptibility has been investigated in population- and family-based studies. Case control studies in areas of endemic disease have pointed to several gene variants contributing to TB risk, including the HLA (4), the natural resistance-associated macrophage protein NRAMP1 (18–22), the vitamin D receptor (23, 24), and the mannose-binding protein (25). A strong association [logarithm of odds (lod) 3.8] of NRAMP1alleles on 2q35 with susceptibility to TB was independently found by linkage analysis in a large Aboriginal Canadian pedigree in the outbreak situation (26). Major gene effects were recently investigated by whole-genome scan by using 173 affected sib pairs from The Gambia and South Africa; this analysis identified suggestive linkages (lod ≈2) on chromosomes (Chr.) 15q and Xq (27, 28). These studies suggest that the genetic control of susceptibility to TB in humans is complex.
Such complex genetic traits can be studied in mouse models of disease, where environmental and genetic components can be best controlled, and where single gene effects may have become fixed in inbred, recombinant inbred, and recombinant congenic strains of mice (29). Genetic control of susceptibility to TB is complex in mouse and is influenced by the M. tuberculosis isolate, the route and dose of infection, the mouse strains used, and the phenotypic measure of susceptibility (30–35). Using survival time after i.v. injection of 1 × 105 M. tuberculosis, inbred mouse strains are classified (36) into either highly susceptible (CBA, C3H, DBA/2, 129svJ) or highly resistant (C57BL/6, BALB/c). Susceptibility of DBA/2 (D2) and resistance of C57BL/6 (B6) and BALB/c strains, as measured by mean survival time, are also observed after airborne infection with 102 bacilli (37). Using informative backcross and F2 mice issued from C3HeB/Fe and B6 parents and infected i.v. with 106 M. tuberculosis Erdman, Kramnik et al. (38) have mapped a locus on distal Chr.1 (sst1; position 49–58 cM) that controls the rate of bacterial replication and granuloma formation in the lung.
Susceptibility of D2 mice to M. tuberculosis infection is characterized by (i) progressive bacterial replication in the lung; (ii) extended neutrophil-dominated lung pathology, including large numbers of acid-fast bacilli and areas of necrosis; and (iii) early death (36–39). We have previously mapped three quantitative trait loci (QTL) on distal Chr. 1 (Trl-1; lod 4.8), proximal Chr. 7 (Trl-3; lod 4.7), and proximal Chr. 3 (Trl-2; lod 3.9) that affect survival time of (B6 × D2) F2 mice after i.v. infection with 1 × 105 M. tuberculosis H37Rv (39). In this report, the genetic analysis of D2 susceptibility to pulmonary TB was expanded by using an infection protocol more closely related to the human situation than the previously used i.v. model (35, 38, 39). (B6 × D2) F2 mice were infected by the respiratory route with 2 × 102 M. tuberculosis H37Rv, and pulmonary bacterial load at 90 days was used as a quantitative measure of susceptibility. A whole-genome scan revealed a major locus on Chr. 19 (designated Trl-4) regulating replication of M. tuberculosis in the lung.
Materials and Methods
Animals. Inbred pathogen-free 8-week-old C57BL/6J (B6) and DBA/2J (D2) mice were purchased from the Trudeau Institute Animal Breeding Facility. Ninety-eight (B6 × D2) F2 progeny were bred by systematic brother–sister mating of (B6 × D2) F1.
Mycobacteria. M. tuberculosis strain H37Rv (TMC no. 102) was obtained from the Trudeau Mycobacterial Culture Collection as a frozen (-70°C) log phase dispersed culture in Proskauer and Beck medium (Difco) containing 0.01% Tween 80. For each experiment, a vial was thawed, subjected to 5-s ultrasound to break up aggregates, and diluted appropriately in PBS containing 0.01% Tween 80. Mice (8–10 weeks of age) were inoculated with 2 × 102 colony-forming units (cfu) by aerosol in a Middle-brook airborne infection apparatus (Tri Instruments, Jamaica, NY). Bacilli were enumerated in the lungs of infected mice 90 days postinfection by preparing lung homogenates in PBS containing 0.05% Tween 80 and by plating 10-fold serial dilutions of the homogenates on enriched agar (Middlebrook 7H11; Difco). cfu were enumerated after 3–4 weeks of incubation at 37°C, and the data are presented as log10 of total cfu count per lung.
Genotyping. Before infection, tail biopsies were obtained, and genomic DNA was prepared (39). A total of 151 microsatellite markers distributed over all Chr. except Chr. Y (≈10 cM coverage) were selected (www-genome.wi.mit.edu) and purchased from Research Genetics (Huntsville, AL). Genotyping was performed by standard PCR-based method by using trace amounts of [32P]α-dATP, followed by separation on denaturing polyacrylamide gels, exactly as described (39). Some markers were genotyped by using primer pairs fluorescently labeled, either commercially available or custom-synthesized by Applied Biosystems. In this case, one of the primers was synthesized and labeled with FAM, HEX, or NED phosphoramides. Products were analyzed by capillary separation by using an Applied Biosystems Prism 3700 automated DNA sequencer.
Statistical Analysis. Genome-wide interval mapping analysis between lung cfus (log10) and genetic markers for the identification of QTLs was performed by using MAPMAKER/EXP Ver. 3.0 and MAPMAKER/QTL Ver. 1.1 (40). lod scores were calculated as χ2/2ln (10). Permutations of the phenotypes in this sample were conducted by using linear regression in QTL CARTOGRAPHER (41, 42) to obtain empirical significance levels at each locus (10,000 iterates, unless noted otherwise). Genome-wide significance levels were also obtained by using QTL CARTOGRAPHER, providing thresholds appropriate for this particular study. Initial linkage analyses were conducted by using a “free” model involving codominance and dominance effects, yielding χ2 statistics and lod scores with 2 df. Tests of specific genetic submodels were conducted by fitting each 1-df model (dominant, recessive, and codominant) and comparing the likelihood ratios [χ2(1) = 2[LL(free) - LL(nested)] (LL, log likelihood). For significant linkage regions, similar likelihood-ratio tests were conducted to test whether apparent linkage results were due to mean trait differences between sexes: heterogeneity test for sex-specific effects, χ2 = 2[LL(combined)-(LL(male) + LL(female))]. Significant results in this test indicate that the linkage results differ between males and females; nonsignificant results indicate that apparent differences in lod scores are consistent with random variation.
The distribution of lung cfus (log10) in (B6 × D2) F2 mice closely followed a normal distribution (see Fig. 1). Significant differences were noted between mean cfu counts in female vs. male mice, (Table 1; male mean = 7.11, female mean = 6.57; two-tailed t = 6.92, P < 0.001) (43). Linkage to Chr. X was initially detected (lod 4.56), suggesting a possible sex effect. However, further analysis indicated no significant linkage to Chr. X in either males or females when analyzed independently (lod of 0.25 and 0.81, respectively), suggesting that the original linkage simply reflects mean trait differences between genders instead of the influence of any Chr. X loci. To compensate for sex effects, the lung cfu counts (log10) were then adjusted by subtracting the gender-specific mean from each individual to create a “sex-adjusted cfu” value. This measure allows the male and female cfu data to be analyzed jointly on the same scale, free of gender-specific influences.
Fig. 1.
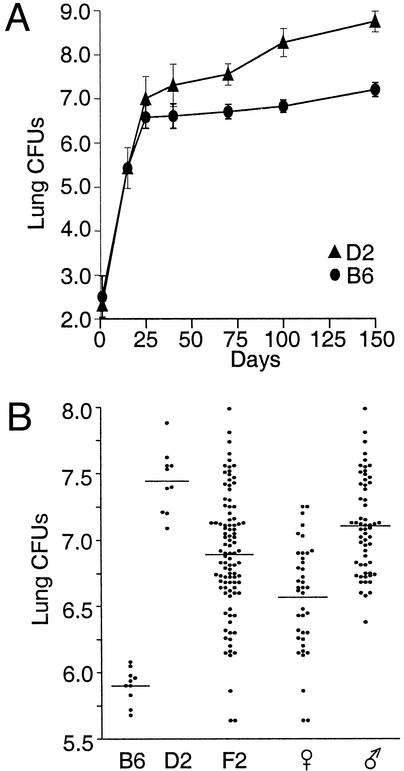
Replication of M. tuberculosis in the lungs of B6, D2, and (B6 × D2) F2 mice. Resistant B6 and susceptible D2 control mice were infected via the aerosol route with 2 × 102 M. tuberculosis H37Rv. (A) The number of M. tuberculosis present in the lungs was determined (log10 cfu) at preset time points, where each data point corresponds to the mean cfu counts for groups of five mice. (B) Lung cfus (log10) of B6 and D2 controls and of individual (B6 × D2) F2 mice 90 days after aerosol infection with 2 × 102 M. tuberculosis H37Rv. Horizontal bars represent means of cfus for each group.
Table 1. Distribution of the cfu counts phenotype in the 98 (B6 × D2) F2 population.
| Trait | n | Mean | SD | Skewness | Kurtosis | Normality test, D* | Normality test, P* |
|---|---|---|---|---|---|---|---|
| cfu counts | 98 | 6.89 | 0.46 | -0.25 | 0.12 | 0.068 | >0.15 |
| Female cfu counts | 39 | 6.57 | 0.40 | -0.37 | -0.35 | 0.095 | >0.15 |
| Male cfu counts | 59 | 7.11 | 0.35 | 0.30 | -0.62 | 0.100 | >0.14 |
| Sex adjusted cfu counts | 98 | 0.00 | 0.37 | -0.03 | -0.43 | 0.068 | >0.15 |
Normality tests conducted by using the Kolmogorov-Smirnov test, as implemented in the SAS computer package (43).
Results
Groups of DBA/2 (D2) and C57BL/6 (B6) mice were infected with 2 × 102 cfu of highly virulent M. tuberculosis H37Rv by the aerosol route, and at predicted times (0–150 days) the number of cfus recovered from infected lungs was determined (Fig. 1A). Pulmonary replication of M. tuberculosis in the two mouse strains was biphasic. In the early phase (up to 3–4 weeks), the pathogen grew to a similar extent in B6 and D2 lungs. In the second phase (4 weeks to 5 months), the infection was held stationary in B6 mice with only a 2-fold increase in the lung cfus between day 25 (5 × 106) and day 150 (1 × 107). Histological analysis has shown that limited growth of M. tuberculosis in the lungs of B6 mice late in infection is associated with pathology dominated by macrophages in proximity of large aggregates of lymphoid cells (39). B6 mice ultimately succumb to infection with a mean survival time of 239 days (range: days 147–292). In contrast, there is progressive replication of M. tuberculosis in the lungs of D2 mice between days 25 and 150, with an ≈50- to 75-fold increase in lung bacterial load and some associated mortality. Histological examination shows large diffuse lesions, dominated by neutrophils and containing areas of tissue necrosis (37, 39, 44). This severe lung pathology leads to early and uniform death in this group with a mean survival time of 102 days (range: days 88–126) (data not shown).
The genetic control of differential lung replication in the late phase of infection was investigated. For these studies, 98 informative male and female (B6 × D2) F2 animals as well as D2 and B6 controls were infected by the respiratory route with 2 × 102 live M. tuberculosis H37Rv, and 90 days later the extent of bacterial replication in the lung was determined (Fig. 1B). The 90-day time point was chosen because (i) parental strains show clear differences in lung cfus at this time, and (ii) D2 mice begin dying beyond 90 days. In this experiment, there was a highly significant 50- to 100-fold difference in cfu counts recovered from susceptible D2 (X = 7.45; range 7.1–7.9) when compared with resistant B6 controls (X = 5.9; range 5.6–6.1). cfu counts in (B6 × D2) F2 showed a continuous distribution (log10 cfus 5.55–8.0) between that of resistant B6 and susceptible D2 parents (Fig. 1B) with minor deviations from normality (Table 1) (43). The mean log10 cfus in the F2 was at 6.89, a value closer to susceptible D2 than to resistant B6 controls, suggesting that susceptibility does not segregate as a recessive trait in this cross. Comparison of cfu counts in male and female (B6 × D2) F2 mice showed a clear gender effect, with females more resistant to M. tuberculosis replication (X = 6.57; range 5.55–7.25) than males (X = 7.11; range 6.35–8.0). No significant deviations from normality were observed for the full or gender-specific distributions of lung cfus (Table 1).
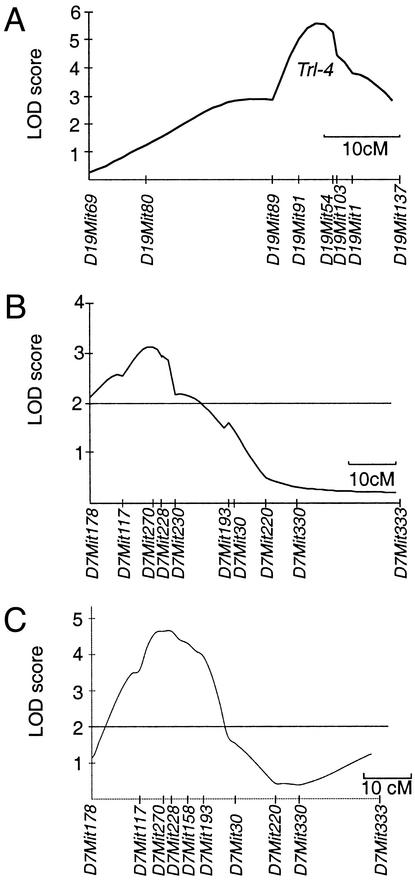
The raw cfu data for the total F2 cross behaves as a quantitative trait amenable to study by QTL analysis. QTL mapping was carried out by a whole-genome scan approach, using a total of 151 polymorphic markers providing an average coverage of ≈10 cM along each chromosome (www-genome.wi.mit.edu/cgi-bin/mouse/index) (Table 3, which is published as supporting information on the PNAS web site, www.pnas.org). The largest gap was estimated at ≈18.5 cM for Chr. 7 and X. Genome-wide multipoint linkage analysis was performed by using MAPMAKER/EXP Ver. 3.0, MAPMAKER/QTL 1.1 (40), and QTL CARTOGRAPHER (42). Results of this analysis are shown as multipoint lod score traces in Fig. 2 A and B, and numerical data for individual intervals are shown in Table 2. Using raw cfu (log10) data as a quantitative trait, one highly significant linkage was identified on the distal portion of Chr. 19, with maximal lod score obtained for the interval defined by markers D19Mit91 and D19Mit54 (χ2 = 25.74; lod 5.59, P = 0.000005). This level of significance was not observed one time in 50,000 permutations of the data. This QTL overlaps ≈10 cM on Chr. 19, explains 24.1% of the total phenotypic variance in the F2 cross, and was given the designation Trl-4 (for Tuberculosis resistance locus-4). Several weaker linkages were also detected on proximal (D5Mit254; χ2 = 17.42; lod 3.78, empirical P = 0.0002) and distal (D5Mit216; χ2 = 15.77; lod 3.42, empirical P = 0.04) portions of Chr. 5 and Chr. 7 (D7Mit270; χ2 = 14.44; lod 3.14, empirical P = 0.0005), and Chr. 10 (D10Mit194; χ2 = 15.83; lod 3.4, empirical P = 0.02). However, none of these linkages reached genome-wide statistical significance at the 0.01 level.
Fig. 2.
Linkage analysis by whole-genome scan of susceptibility to infection with M. tuberculosis. lod score traces along Chr. 19 (A) and 7 (B), for which highly significant (lod >5.0) and suggestive (lod >3) QTL controlling bacterial replication of M. tuberculosis H37Rv after aerosol infection with 2 × 102 cfus in (B6 × D2) F2 mice was detected. For comparison, the lod score plot for Chr. 7 from an independent genome scan for a significant QTL (Trl-3) that controls survival of (B6 × D2) F2 mice after i.v. infection with 1 × 105 cfus of M. tuberculosis H37Rv is shown [C reproduced with permission from ref. 39 (copyright 2000, Nature Publishing Group)]. The map positions of microsatellite markers used are indicated, and chromosomal lengths are shown to scale.
Table 2. Statistical values for linkages obtained from the (B6 × D2) F2 population.
| Marker | cM | lod | χ2 | % variance explained |
|---|---|---|---|---|
| cfu counts | ||||
| D5Mit297 | 17.5 | 3.37 | 15.50 | 15.7 |
| D5Mit254 | 25.1 | 3.78 | 17.42 | 17.6 |
| D5Mit201 | 28.4 | 3.24 | 14.90 | 14.2 |
| D5Mit95 | 57.9 | 2.11 | 9.71 | 9.5 |
| D5Mit216 | 62.3 | 3.42 | 15.77 | 14.8 |
| D5Mit168 | 66.7 | 3.12 | 14.35 | 14.0 |
| D7Mit117 | 10.9 | 2.61 | 12.01 | 12.8 |
| D7Mit270 | 15.3 | 3.14 | 14.44 | 16.2 |
| D7Mit228 | 16.4 | 2.85 | 13.11 | 12.5 |
| D10Mit194 | 18.5 | 3.40 | 15.83 | 18.5 |
| D10Mit186 | 36.1 | 2.58 | 11.87 | 13.1 |
| D19Mit89 | 28.4 | 2.85 | 13.11 | 13.2 |
| D19Mit91 | 35.0 | 5.06 | 23.28 | 21.6 |
| D19Mit54 | 39.3 | 5.59 | 25.74 | 24.1 |
| D19Mit103 | 40.4 | 5.44 | 25.02 | 22.6 |
| D19Mit1 | 43.7 | 4.64 | 21.34 | 19.6 |
| Sex adjusted | ||||
| D5Mit297 | 19.7 | 2.66 | 12.24 | 11.8 |
| D5Mit254 | 25.1 | 3.10 | 14.26 | 13.5 |
| D5Mit201 | 28.4 | 2.89 | 13.29 | 12.7 |
| D19Mit91 | 35.0 | 3.59 | 16.51 | 15.8 |
| D19Mit54 | 39.3 | 4.00 | 18.40 | 18.0 |
| D19Mit103 | 40.4 | 3.88 | 17.85 | 16.7 |
| D19Mit1 | 43.7 | 3.29 | 15.13 | 14.3 |
Possible linkage was initially detected to all Chr. X markers tested (lod 3.3–4.6). However, further analyses failed to reveal significant linkage to Chr. X in either males or females alone when analyzed independently (lod of 0.25 and 0.81, respectively), suggesting that the original linkage reflects mean differences between genders instead of any Chr. X loci. In this gender-specific analysis, evidence for linkage to Trl-4 was stronger in males (n = 59; lod 3.8, empirical P = 0.0002) than in females (n = 39; lod 1.59, empirical P = 0.02). A likelihood-ratio heterogeneity test (see Materials and Methods) indicated that these apparent differences were not statistically significant [χ2(2) = 2(25.7-(17.54 + 7.37) = 1.66, P = 0.44], suggesting that the differences in lod scores between males and females may be due to differences in sample size and random variation. To further explore gender specificity in linkage to Chr. 19 QTL (Trl-4), residual cfu values for F2 mice were examined after controlling for sex effects (Table 1; see Materials and Methods). This sex-adjusted cfu count transformation provided a means to retain all data, whereas it eliminated the effects of baseline sex differences in bacterial replication. Analysis of the sex-adjusted cfus retained solely the Chr.19 hit as significant (χ2 = 18.40; lod 4.00; explaining 18% of the phenotypic variance). A result this large was not observed in 50,000 permutations of the data. Conditioning on the genotypes at each of the Chr. X markers genotyped revealed no evidence for improved linkage at any autosomal loci (45). Thus, the linkage to Trl-4 did not appear to be sex-specific.
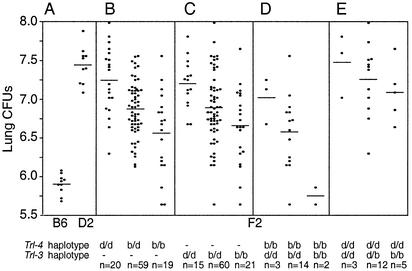
To visualize the effect of parental alleles (D2, d; B6, b) at Trl-4 on lung cfus, F2 animals were separated according to their genotype at D19Mit54 (Fig. 3B). B6 alleles were associated with reduced bacterial replication and were inherited in an incompletely dominant fashion, with mean cfu counts (log10) of 7.25 (“d/d” homozygotes), 6.85 (“b/d” heterozygotes), and 6.55 (“b/b” homozygotes) (Fig. 3B). We have previously detected a QTL on Chr. 7 (Trl-3) that affects survival of (B6 × D2) F2 mice after i.v. injection of 1 × 105 M. tuberculosis (D7Mit270; χ2 = 21.4; lod 4.66) (39). In the present study, D7Mit270 shows suggestive linkage to lung cfus after aerosol infection (Table 2; lod 3.14; 16.2% of the variance), and alignment of lod score plots from both studies (Fig. 2 B vs. C) suggests that Trl-3 may indeed affect both phenotypes. Thus, F2 animals were also separated according to their genotype for D7Mit270 (Trl-3). Results in Fig. 3C also suggest that “b” alleles at the Chr. 7 linkage are associated with reduced M. tuberculosis replication in the lung and are inherited in a codominant fashion as previously noted for their effect on survival (39).
Fig. 3.
Effect of haplotype combination at individual QTLs on M. tuberculosis replication in the lungs. The effect of parental allele combinations at Trl-4 (B, D19Mit54), Trl-3 (C, D7Mit228), as well as Trl-3 and Trl-4 in combination (D and E) on bacterial replication (log10 cfu at day 90 postinfection) is shown. The D2 and B6 parental alleles are identified as d and b, respectively, and the number of animals (n) in each group is shown beneath the figure. Each data point represents a single mouse, and horizontal bars indicate mean cfus in each group. The log10 cfus in the parental B6 and D2 groups are shown in A for comparison.
Analysis of combined effects of Chr. 19 and 7 loci on lung cfus suggested an additive and very strong effect of the two QTLs. Two-loci linkage analysis yielded a lod of 10.09 for the combined QTLs, explaining 38% of the variation in raw cfus. Regression tests of lung cfus on both D7Mit270 and D19Mit54 did not indicate a significant interaction between the loci (t =-1.46, P = 0.15). However, although few animals were available for analysis, mice homozygous for “b/b” alleles at both loci were completely resistant and showed cfu counts (5.55, 5.80) in the range of B6 controls (range 5.6–6.1), whereas mice homozygous for “d/d” alleles were completely susceptible and showed cfu counts (7.0, 7.6, and 7.8) similar to those seen in the susceptible D2 controls (range 7.1–7.9) (Fig. 3 D and E). This effect was specific and was not seen when Chr. 19 haplotypes were analyzed in conjunction with any of the weaker linkages (Chr. 5 and 10) detected in this study (data not shown). Finally, the presence of permissive homozygous “d/d” haplotypes at Trl-4 largely dissipated the protective effect of “b” alleles at Trl-3.
Discussion
Compared with other inbred strains, D2 is uniquely sensitive to infection with virulent isolates of human M. tuberculosis H37Rv, with a concomitant pathology and outcome that resemble M. tuberculosis infection in many AIDS patients. This susceptibility is characterized by unrestricted pulmonary microbial replication, massive inflammatory response in situ, and early death. Differential pulmonary growth of M. tuberculosis in B6 and D2 mice is associated with dramatic differences in histopathology starting 3 weeks postinfection. Whereas B6 lungs show well formed granulomas containing M. tuberculosis-infected epithelioid macrophages in close proximity to aggregates of lymphocytes, D2 lungs show extensive diffuse lesions containing a large number of degenerating neutrophils replete with acid-fast bacilli (37, 39, 44). Thus, the genetic control appears to regulate aspects of lung-specific host immune response that are triggered at 3–4 weeks (46, 47). This mechanism may be impaired in D2 (and replaced by a pronounced inflammatory response), possibly through impairment of mononuclear, polymorphonuclear, and/or lymphocytic lineages.
To examine genetic control of susceptibility of D2 mice, we have carried out independent genome scans in informative (B6 × D2) F2 mice. To sample different aspects of host response to infection that may be under unique or common genetic controls, we used different doses (2 × 102, 1 × 105) and routes of infection (aerosol, i.v.) and monitored different quantitative measures of susceptibility (bacterial replication in the lung, overall survival time). The major conclusions of these experiments are that (i) the genetic control of susceptibility in D2 is complex; (ii) individual QTLs affecting this trait can be mapped in this experimental setting, with four significant loci mapped to date (Trl-1 to Trl-4); and (iii) different infection models reveal different gene effects with little overlap between the different experimental protocols. In a first genome scan (39), we infected (B6 × D2) F2 mice with 1 × 105 M. tuberculosis H37Rv by the i.v. route and used time of survival (log10) as a quantitative measure of susceptibility. The genetic control was found to be complex, with two significant linkages mapping on distal Chr. 1 (Trl-1; lod 4.8) and proximal Chr. 7 (Trl-3; lod 4.7), each accounting for 21% of the phenotypic variance. A third suggestive linkage was mapped to proximal Chr. 3 (Trl-2; lod 3.9) (39). In the present study, M. tuberculosis was introduced by the respiratory route (which closely resembles the mode of infection in humans), and the extent of pulmonary replication (log10 cfus at day 90) was used as a quantitative measure of susceptibility. QTL mapping using either raw or sex adjusted cfu counts revealed a highly significant linkage on Chr. 19 (Trl-4; D19Mit91 and D19Mit54; χ2 = 25.67; lod 5.58, P = 0.000005), which explains 24.1% of the total phenotypic variance in the F2 cross. The Trl-4 QTL is distinct from other QTLs previously mapped in murine models of M. tuberculosis infection, including the sst1 locus (susceptibility to tuberculosis 1) (38) originally mapped on Chr. 1 (D1Mit49) in a (C3H × C57BL/6) F2 cross, and which regulates pulmonary growth, inflammatory response, and overall survival after i.v. infection with high dose (1 × 106 i.v.) of M. tuberculosis. Trl-4 is also distinct from the QTLs mapped by Lavebratt et al. (35) on Chrs. 3, 5, 9, and 10 that regulate in a gender-specific fashion differential body weight loss after i.v. infection with high doses of M. tuberculosis (>106 cfu). Importantly, Trl-4 was identified as a locus that regulates replication of M. tuberculosis in the lung after aerosol infection with small numbers of the pathogen.
The interval for Trl-4 on Chr. 19 is ≈10 cM and contains an estimated 70 transcription units (data not shown). Several of these may be potential candidates by virtue of their established role in host immune and inflammatory responses. Trl-4 maps to a QTL designated Pgia12 (48), previously shown to control onset of arthritis induced by injection of human cartilage-derived proteoglycan (PGIA), a known mouse model of human rheumatoid arthritis. The relationship between Trl-4 and Pgia12 is unknown, but it is interesting to note that both QTLs seem to affect host inflammatory responses. The Trl-4 interval also contains the NF-κB (position 45.8 cM) and Iκκα (chuk; position 45 cM) genes. NF-κB (p52) is a subunit of the NF-κB factors, a group of transcription factors implicated in the induction of numerous genes in response to inflammatory stimuli, as well as pathogen-derived or stress signals (lipopolysaccharide, IL-1, or tumor necrosis factor-α) (49). Mouse mutants lacking functional NF-κB (p52) show absence of B cell follicles in secondary lymphoid organs and cannot produce antibodies to T dependent antigens (50). NF-κB mutant mice become susceptible to Leishmania major infection, which is associated with uncontrolled parasite replication, nonhealing lesions, and failure to develop an IFN-γ response (51). NF-κB mutant mice are also susceptible to Toxoplasma gondii (52). Iκκα is one of two known kinases that phosphorylate IkBs (inhibitor of κ B kinase) and thus acts as a regulatory subunit of NF-κB factors. Iκκα mutant mice (studied in chimeras) show a phenotype similar to NF-κB mutants, with respect to impaired B cell function (53). Finally, the Trl-4 region also contains the α chain of the cell surface receptor for granulocyte/macrophage colony-stimulating factor (GM-CSF), known as GM-CSFRα (CSF2rα; position 51 cM). GM-CSF, IL-3, and IL-5 are related cytokines that bind to cell surface receptors composed of a cytokine-specific α chain and a β chain common to the three receptors (54). GM-CSF acts as a growth factor for macrophages and granulocytes, and GM-CSF mutant mice show increased susceptibility to pneumonia caused by Pasteurella pneumotropica, group B Streptococcus, Penumocytis carinii, and others (55–58). They also display reduced pathogen killing by isolated alveolar macrophages (59), which is concomitant to exaggerated inflammatory response in the lungs (57). Importantly, the only “constitutive” phenotype displayed by GM-CSF and GM-CSFRβ mutant mice is “pulmonary alveolar proteinosis” (PAP), a condition characterized by abnormal catabolism of lung surfactant by lung epithelial cells and by alveolar macrophages (54). Together with the recent discovery of a GM-CSFRβ mutation in a human PAP patient (60), these findings suggest that GM-CSF plays a major role in lung surfactant homeostasis by these cells. Interestingly, both alveolar type II epithelial cells and alveolar macrophages are invaded in vivo by M. tuberculosis (61) and are key hosts to this pathogen early in infection.
In two genome scans conducted to date in (B6 × D2) F2 mice (scan 1: i.v. infection, survival; scan 2: aerosol, lung replication), Trl-4 constitutes the strongest linkage identified to date with a lod ≈5.6. Interestingly, the Trl-4 linkage was detected only in scan 2, whereas the Chr. 1 (Trl-1) and Chr. 3 (Trl-2) QTLs were detected only in scan 1. This observation suggests that Trl-4 may affect pulmonary replication per se (after aerosol infection), whereas Trl-1/Trl-2 may influence time of death in the presence of high lung bacterial load. Alternatively, Trl-1/2/4 may be statistical accidents that await validation in larger groups of mice of the same cross or a different cross. This explanation is unlikely for Trl-4, which reaches a very high degree of significance and explains a large proportion of the phenotypic variance in the cross. Thus, we believe that Trl-4 is a major determinant of M. tuberculosis replication in the lungs, after infection by the respiratory route. The Trl-3 QTL was the only QTL detected in both scans 1 (39) and 2 (this study). The observation that in both scans the Trl-3 alleles of B6 are protective and inherited in a codominant fashion suggests that the gene effect is real and thus this QTL affects both the extent of M. tuberculosis replication (scan 2) and the survival to infection (scan 1). Further-more, a major additive effect of Trl-3 and Trl-4 on pulmonary replication of M. tuberculosis (lod 10.09; ≈40% of variance) was detected in scan 2 (this study). Remarkably, and although only a few animals were available for study, mice homozygous for B6 alleles at Trl-3/Trl-4 were completely resistant to infection and phenotypically undistinguishable from B6 parents. Also, mice homozygous for D2 alleles at both loci were completely susceptible and similar to susceptible D2 controls (Fig. 3 D and E). The independent and combined contribution of Trl-3 and Trl-4 to regulation of M. tuberculosis replication in the lung is currently being investigated in congenic mice.
The Chr. 19 Trl-4 region is syntenic with human 10q, whereas the Chr.7 Trl-3 is syntenic with human 19q13. A possible association of these chromosomal regions with susceptibility to TB in humans can now be tested in population studies from areas where the disease is endemic.
Supplementary Material
Acknowledgments
P.G. is a Distinguished Scientist from the Canadian Institutes of Health Research. L.R.C. is supported by the Wellcome Trust. L.-M.M. is supported by a studentship from the Fonds de Recherche en Santé du Québec. This work was supported by research grants to P.G. (RO1 AI35237) and R.J.N. (AI-37844 and HL-64565) from the National Institutes of Health. The genotyping core at McGill University was supported by a donation from the J.-L. Lévesque Foundation.
Abbreviations: Chr., chromosome; lod, logarithm of odds; cfu, colony-forming unit; QTL, quantitative trait locus; GM-CSF, granulocyte/macrophage colony-stimulating factor.
References
- 1.Dye, C., Scheele, S., Dolin, P., Pathania, V. & Raviglione, M. C. (1999) J. Am. Med. Assoc. 282, 677–868. [DOI] [PubMed] [Google Scholar]
- 2.Young, D. B. & Duncan, K. (1995) Annu. Rev. Microbiol. 49, 47–55. [DOI] [PubMed] [Google Scholar]
- 3.Lienhardt, C. (2001) Epidemiol. Rev. 23, 288–301. [DOI] [PubMed] [Google Scholar]
- 4.Casanova, J.-L. & Abel, L. (2002) Annu. Rev. Immunol. 20, 581–620. [DOI] [PubMed] [Google Scholar]
- 5.Hinman, A. R., Judd, J. M., Kolnick, J. P. & Daitch, P. B. (1976) Am. J. Epidemiol. 103, 486–497. [DOI] [PubMed] [Google Scholar]
- 6.Rieder, H. L., Kelly, G. D., Bloch, A. B., Cauthen, G. M. & Snider, D. E. (1991) Chest 100, 678–681. [DOI] [PubMed] [Google Scholar]
- 7.Stead, W. W., Sener, J. W., Reddick, W. T. & Lofgren, J. P. (1990) N. Engl. J. Med. 322, 422–427. [DOI] [PubMed] [Google Scholar]
- 8.Motulsky, A. G. (1960) Hum. Biol. 32, 28–62. [PubMed] [Google Scholar]
- 9.Sousa, A. O, Salem, J. I., Lee, F. K., Vercosa, M. C., Cruaud, P., Bloom, B. R., Lagrange, P. H. & David, H. L. (1997) Proc. Natl. Acad. Sci. USA 94, 13227–13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubos, R. & Dubos, J. (1952) in The White Plague: Tuberculosis, Man and Society (Little, Brown, Boston), pp. 1–207.
- 11.Kallmann, F. J. & Reisner, D. (1942) Am. Rev. Tuberc. 47, 549–574. [Google Scholar]
- 12.Simonds, B. (1963) Tuberculosis in Twins (Pitman Medical Pub. Co., London), pp. 1–81.
- 13.Comstock, G. W. (1978) Am. Rev. Respir. Dis. 117, 621–624. [DOI] [PubMed] [Google Scholar]
- 14.Reichenbach, J., Rosenzweig, S., Doffinger, R., Dupuis, S., Holland, S. M. & Casanova, J.-L. (2001) Curr. Opin. Clin. Immunol. Allergy 1, 503–511. [DOI] [PubMed] [Google Scholar]
- 15.Picard, C., Fieschi, C., Altare, F., Al-Jumaah, S., Al-Hajjar, S., Feinberg, J., Dupuis, S., Soudais, C., Al-Mohsen, I. Z., Genin, E., et al. (2002) Am. J. Hum. Genet. 70, 336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jouanguy, E., Lamhamedi-Cherradi, S., Lammas, D., Dorman, S. E., Fondaneche, M. C., Dupuis, S., Doffinger, R., Altare, F., Girdlestone, J., Emile, J. F., et al. (1999) Nat. Genet. 21, 370–378. [DOI] [PubMed] [Google Scholar]
- 17.Doffinger, R., Dupuis, S., Picard, C., Fieschi, C., Feinberg, J., Barcenas-Morales, G. & Casanova, J-L. (2002) Mol. Immunol. 38, 903–909. [DOI] [PubMed] [Google Scholar]
- 18.Bellamy, R., Ruwende, C., Corah, T., McAdam, K. P. W. J., Whittle, H. C. & Hill, A. V. (1998) N. Engl. J. Med. 338, 640–644. [DOI] [PubMed] [Google Scholar]
- 19.Gao, P. S., Fujishima, S., Mao, X. Q., Remus, N., Kanda, M., Enomoto, T., Dake, Y., Bottini, N., Tabuchi, M., Hasegawa, N., et al. (2000) Clin. Genet. 58, 74–76. [DOI] [PubMed] [Google Scholar]
- 20.Ryu, S., Park, Y. K., Bai, G. H., Kim, S. J., Park, S. N. & Kang, S. (2000) Int. J. Tuberc. Lung. Dis. 4, 577–580. [PubMed] [Google Scholar]
- 21.Cervino, A. C., Lakiss, S., Sow, O. & Hill, A. V. (2000) Ann. Hum. Genet. 64, 507–512. [DOI] [PubMed] [Google Scholar]
- 22.Mesiner, S. J., Mucklow, S., Warner, G., Sow, S. O., Lienhardt, C. & Hill, A. V. (2001) Am. J. Trop. Med. Hyg. 65, 733–735. [DOI] [PubMed] [Google Scholar]
- 23.Bellamy, R., Ruwende, C., Corrah, T., McAdam, K. P., Thursz, M., Whittle, H. C. & Hill A. V. (1999) J. Infect. Dis. 179, 721–724. [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson, R. J., Llewelyn, M., Toossi, Z., Patel, P., Pasvol, G., Lalvani, A., Wright, D., Latif, M. & Davidson, R. N. (2000) Lancet 355, 618–621. [DOI] [PubMed] [Google Scholar]
- 25.Selvaraj, P., Narayanan, P. R. & Reetha, A. M. (1999) Tuberc. Lung Dis. 79, 221–227. [DOI] [PubMed] [Google Scholar]
- 26.Greenwood C. M., Fujiwara, T. M., Boothroyd, L. J., Miller, M. A., Frappier, D., Fanning, E. A., Schurr, E. & Morgan, K. (2000) Am. J. Hum. Genet. 67, 405–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellamy, R., Beyers, N., McAdam, K. P., Ruwende, C., Gie, R., Samaai, P., Bester, D., Meyer, M., Corrah, T., Collin, M., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 8005–8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cervino, A. C., Lakiss, S., Sow, O., Bellamy, R., Beyers, N., Hoal-Van Helden, E., Van Helden, P., McAdam, K. P. & Hill, A. V. (2002) Hum. Mol. Genet. 11, 1599–1603. [DOI] [PubMed] [Google Scholar]
- 29.Fortin, A., Stevenson, M. M. & Gros, P. (2002) Genes Immun. 3, 177–186. [DOI] [PubMed] [Google Scholar]
- 30.Pierce, C., Dubos, R. J. & Middlebrook, G. (1947) J. Exp. Med. 86, 159–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donovick, R., McKee, C. M., Jambor, W. P. & Rake, G. (1949) Am. Rev. Tub. 60, 109–120. [DOI] [PubMed] [Google Scholar]
- 32.Lynch, C. J., Pierce-Chase, C. H. & Dubos, R. (1965) J. Exp. Med. 121, 1051–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musa, S. A., Kim, Y., Hashim, R., Wang, G. Z., Dimmer, C. & Smith, D. W. (1987) Infect. Immun. 55, 1862–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brett, S., Orrell, J. M., Beck, S. & Ivanyi, J. I. (1992) Immunology 76, 129–132. [PMC free article] [PubMed] [Google Scholar]
- 35.Lavebratt, C., Apt, A. S., Nikonenko, B. V., Schalling, M. & Schurr, E. (1999) J. Infect. Dis. 180, 150–155. [DOI] [PubMed] [Google Scholar]
- 36.Medina, E. & North, R. J. (1998) Immunology 93, 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medina, E. & North, R. J. (1996) J. Exp. Med. 183, 1045–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kramnik, I., Dietrich, W. F., Demant, P. & Bloom, B. R. (2000) Proc. Natl. Acad. Sci. USA 97, 8560–8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitsos, L-M., Cardon, L. R., Fortin, A., Ryan, L., LaCourse, R., North, R. J. & Gros, P. (2000) Genes Immun. 1, 467–477. [DOI] [PubMed] [Google Scholar]
- 40.Lander, E., Green, P.,Abrahamson, J., Barlow, A., Daley, M., Lincoln, S. & Newburg, L. (1987) Genomics 1, 174–181. [DOI] [PubMed] [Google Scholar]
- 41.Basten, C. J., Weir, B. S. & Zeng, Z.-B. (1994) in Proceedings of the 5th World Congress on Genetics Applied to Livestock Production: Computing Strategies and Software, eds. C. Smith, C., Gavora, J. S., Benkel, B., Chesnais, J., Fairfull, W., Gibson, J. P., Kennedy, B. W. & Burnside, E. B. (University of Guelph, Guelph, Canada), pp. 65–66.
- 42.Basten, C. J., Weir, B. S. & Zeng, Z.-B. (2002) QTL Cartographer (Department of Statistics, North Carolina State University, Raleigh, NC), Ver. 1.16.
- 43.SAS Institute (1990) SAS/STAT User's Guide (SAS Inst., Cary, NC), Ver. 6, 4th Ed.
- 44.Medina, E. & North, R. J. (1999) Immunology 96, 16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez, F., Radaeva, T. V., Nikonenko, B. V., Persson, A-S., Sengul, S., Schalling, M., Schurr, E., Apt, A. S. & Lavebratt, C. (2003) Infect. Immun. 71, 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mogues, T., Goodrich, M. E., Ryan, L., LaCourse, R. & North, R. J. (2001) J. Exp. Med. 193, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung, Y-J., Lacourse, R., Ryan, L. & North, R. J. (2002) Infect. Immun. 70, 6436–6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Otto, J. M., Cs-Szabo, G., Gallagher, J., Velins, S., Mikecz, K., Buzas, E. I., Enders, J. T., Li, Y., Olson, B. R. & Glant, T. (1999) Arthritis Rheum. 42, 2524–2531. [DOI] [PubMed] [Google Scholar]
- 49.Baeuerle, P. A. & Henkel, T. (1994) Annu. Rev. Immunol. 12, 141–179. [DOI] [PubMed] [Google Scholar]
- 50.Franzoso, G., Carlson, L., Poljak, L., Shores, E. W., Epstein, S., Leonardi, A., Grinberg, A., Tran, T., Scharton-Kersten, T., Anver, M., et al. (1998) J. Exp. Med. 187, 147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Speirs, K., Caamano, J., Goldschmidt, M. H., Hunter, C. A. & Scott, P. (2002) J. Immunol. 168, 4406–4413. [DOI] [PubMed] [Google Scholar]
- 52.Caamano, J., Tato, C., Cai, G., Villegas, E. N., Speirs, K., Craig, L., Alexander, J. & Hunter, C. A. (2000) J. Immunol. 165, 5720–5728. [DOI] [PubMed] [Google Scholar]
- 53.Kaisho, T., Takeda, K., Tsujimura, T., Kawai, T., Nomura, F., Terada, N. & Akira, S. (2001) J. Exp. Med. 193, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell, B. C. & Whitsett, J. A. (2002) Annu. Rev. Physiol. 64, 775–802. [DOI] [PubMed] [Google Scholar]
- 55.Stanley, E., Lieschke, G. J., Grail, D., Metcalf, D., Hodgson, G., Gall, J. A., Maher, D. W., Cebon, J., Sinickas, V. & Dunn, A. R. (1994) Proc. Natl. Acad. Sci. USA 91, 5592–5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.LeVine, A. M., Reed, J. A., Kurak, K. E., Cianciolo, E. & Whitsett, J. A. (1999) J. Clin. Invest. 103, 563–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paine, R., 3rd, Preston, A. M., Wilcoxen, S., Jin, H., Siu, B. B., Morris, S. B., Reed, J. A., Ross, G., Whitsett, J. A. & Beck, J. M. (2000) J. Immunol. 164, 2602–2609. [DOI] [PubMed] [Google Scholar]
- 58.Zhan, Y., Lieschke, G. J., Grail, D., Dunn, A. R. & Cheers, C. (1998) Blood 91, 863–869. [PubMed] [Google Scholar]
- 59.Shibata, Y., Berclaz, P. Y., Chroneos, Z. C., Yoshida, M., Whitsett, J. A. & Trapnell, B. C. (2001) Immunity 15, 557–567. [DOI] [PubMed] [Google Scholar]
- 60.Dirksen, U., Nishinakamura, R., Groneck, P., Hattenhorst, U., Nogee, L., Murray, R. & Burdach, S. (1997) J. Clin. Invest. 100, 2211–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sato, K., Tomioka, H., Shimizu, T., Gonda, T., Ota, F. & Sano, C. (2002) J. Infect. Dis. 185, 1139–1147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.