Abstract
We have found previously that during tumor growth intact human chromosome 3 transferred into tumor cells regularly looses certain 3p regions, among them the ≈1.4-Mb common eliminated region 1 (CER1) at 3p21.3. Fluorescence in situ hybridization analysis of 12 mouse orthologous loci revealed that CER1 splits into two segments in mouse and therefore contains a murine/human conservation breakpoint region (CBR). Several breaks occurred in tumors within the region surrounding the CBR, and this sequence has features that characterize unstable chromosomal regions: deletions in yeast artificial chromosome clones, late replication, gene and segment duplications, and pseudogene insertions. Sequence analysis of the entire 3p12-22 revealed that other cancer-associated deletions (regions eliminated from monochromosomal hybrids carrying an intact chromosome 3 during tumor growth and homozygous deletions found in human tumors) colocalized nonrandomly with murine/human CBRs and were characterized by an increased number of local gene duplications and murine/human conservation mismatches (single genes that do not match into the conserved chromosomal segment). The CBR within CER1 contains a simple tandem TATAGA repeat capable of forming a 40-bp-long secondary hairpin-like structure. This repeat is nonrandomly localized within the other tumor-associated deletions and in the vicinity of 3p12-22 CBRs.
Solid tumors are characterized by complex karyotypes. Numerical and structural chromosomal alterations result in multiple genomic imbalances with nonrandom patterns of segmental losses and gains. The loss of the short arm of chromosome (chr) 3 is among the most frequent genomic changes in human solid tumors (1, 2). The development of high-resolution genetic analysis has led to the discovery of numerous regions on 3p, where losses occurred at a high frequency (3, 4, 5). Such regions may harbor multiple tumor-suppressor gene candidates (2, 6). High frequency of chromosome rearrangements, the occurrence of multiple deletion breakpoints within small segments (deletion “hot spots”) (4, 5), and changes affecting groups of genes suggested that yet-unknown unstable sites may play an important role in tumor-associated chromosome instability as it was demonstrated for known fragile sites, virus integration sites, and pericentromeric regions (7–10). Genetic instability may participate in evolutionary genome rearrangements as shown for pericentromeric chromosomal regions (11), and it has been suggested that the hot spots of tumor-related deletions on 3p may coincide with the evolutionary chromosome breakpoints (1). The availability of more precise data about hot-spot locations together with newly released human and mouse genome sequences have permitted us to examine this hypothesis.
We previously developed an assay named the “elimination test” for the functional definition of regions that contain genes capable of antagonizing tumor growth (12). Earlier findings showed that fusion of normal and malignant cells leads to the suppression of tumorigenicity as long as the full hybrid chromosome complement is maintained (13). Single normal cell-derived chr 3 transferred by microcell fusion can also suppress tumorigenicity (14). Reappearance of the tumorigenic phenotype was associated with the loss of the transferred chromosome or its parts. To analyze these losses we have generated monochromosomal human chr 3 hybrids by using a tumorigenic mouse fibrosarcoma or a human nonpapillary renal cell carcinoma as recipients. We found that an ≈1.4-Mb segment, designated as the common eliminated region (CER)1, was missing from all derived tumors (15, 16).
Analyzing CER1 orthologous sequences in the mouse, we found that it corresponds to two conserved segments on chr 9. We also showed that some instability features characterize sequences surrounding the conservation breakpoint region (CBR).
Two more regions identified in our system (16, 17) and four well characterized deletion hot-spot regions found in tumor biopsies and cell lines (6, 7, 18, 19) are located at 3p12-22 (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). Comparing human and mouse genomic sequences assembled by the Celera Discovery System, we found that CBRs localize preferentially within the tumor-associated deletions in this chromosomal segment.
Methods
Fluorescence in Situ Hybridization (FISH). DNA from P1 artificial chromosome (PAC) and bacterial artificial chromosome (BAC) clones was prepared by using Qiagen (Valencia, CA) columns. Expand long-template PCR system (Roche Molecular Biochemicals) was used to PCR-amplify the DNA segments between primers MLztf l1.F (5′-ACGGTAGATTCCTGCTTTCA-3′) and MLztfl1.R (5′-TTGACTTCTCATCCAGAGCA-3′) for Lztfl1, MLtf.F (5′-CATCAGGTTATTGACCATGCCT-3′) and MLtf.R (5′-CTTTGAGGCTATCACATCCTGC-3′) for Ltf, and MTdgf1.F (5′-GAGTTGAGGACCCGGAAGAA-3′) and MTdgf1.R (5′-GTGAGGGTCTTGCCATT-3′) for Tdgf1 from mouse genomic DNA. The obtained fragments (>6 kb) were isolated in low-melting-point agarose and purified. Probes were labeled with either biotin-dUTP (Bionick labeling system, BRL) or digoxigenin-dUTP (DIG-Nick translation mix, Roche Molecular Biochemicals). Two- and three-color FISH was performed on metaphase chromosomes and nuclei were prepared from BALB/c embryonic fibroblasts as described (20). A fluorescence microscope (Leitz-DMRB, Leica, Heidelberg) equipped with a Hamamatsu (Herrsching, Germany) C 4800 cooled charge-coupled device camera and PHOTOSHOP 5.5 (Adobe Systems, Mountain View, CA) were used for the analysis of FISH results.
FISH-Based Replication Timing. FISH using human PAC probes was performed on nuclei obtained from the human KH39 cell line. The interpretation of results was done basically as described (21, 22). We counted the percentage of nuclei with double-hybridization dots (doublets), which reflects the percentage of cells that have undergone replication. Relative difference in replication time of different parts of CER1 was estimated by using comparison of results for different PACs, hybridized by pairs in two-color FISH experiments on the nuclei prepared from the same cell suspension.
Bioinformatics. The information about human–mouse orthologous gene pairs and the probable paralogs was obtained from the Celera Discovery System (http://cds.celera.com).
Detailed information about positions of sequence-tagged site markers, genomic clones, known genes, repeat masker, and simple tandem repeats was taken from University of California, Santa Cruz (UCSC) Genome Bioinformatics (http://genome.cse.ucsc.edu) and R EPEATMASKER WEB (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker). For comparison of identities between human and mouse sequences, the sequences that were identified in the Celera Discovery System were submitted to the PIPMAKER server (http://nog.cse.psu.edu/pipmaker) (23).
To define the palindrome (hairpin) structures in the analyzed sequences we used the PALINDROME server (http://bioweb.pasteur.fr/seqanal/interfaces/palindrome.html).
To determine whether the localization of TATAGA repeats and CBRs within tumor-related deletions on the 3p12-22 segment is random, we used CHITEST (Microsoft EXCEL), comparing the values corresponding to the number of repeats (CBRs) found in each deletion (actual range) to the number expected if the distribution would be random (expected range). The expected number = DS × TN/S, where DS is deletion size in Mb, TN is the total number of repeats (CBRs) within the analyzed region, and S is the size of analyzed region.
Results and Discussion
Human CER1 Contains a Murine/Human CBR. Initially, we analyzed the representation of human CER1 in the mouse genome. We performed FISH with 12 mouse probes corresponding to human genes located within CER1 and flanking regions. We assigned all probes by FISH to mouse chr 9 (9F). The results of FISH probe ordering suggest that CER1 is represented in two distinct segments on mouse chr 9 (Fig. 1).
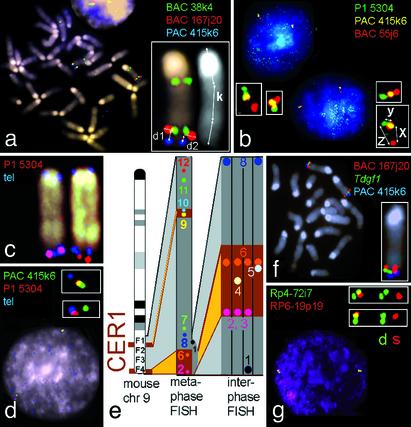
Fig. 1.
FISH for mapping of probes on mouse chr 9 (a–f) and for replication-timing analysis (g). Red signal, biotin-labeled probe detected by Cy3-conjugated avidin; green signal, digoxigenin-labeled probe detected by FITC-conjugated antidigoxigenin antibodies; yellow signal, probe labeled with both biotin and digoxigenin; blue signal, artificial color given to the signal of the probe used in a subsequent hybridization on the same metaphase spread. (a) Metaphase measurements. Distance between two probes (PAC 415k6 and BAC 167j20) = average of d/k (minimum of 20 measurements). Order: cen, BAC 38k4, BAC 167j20, PAC 415k6. Note that the BAC 38k4 containing Rbm6 gene produces additional signals at 9A and 1G, reflecting duplication of this genomic segment in mouse. (b) Interphase measurements. The average values (minimum of 20 measurements) of distances between probes (x, y, and z) show the order of probes: BAC55j6, PAC 415k6, P1 5304. The average ratio of distances (x/y) yields the relative position of the PAC 415k6 between two others. (c) The probe P1 5304 signal was detected in close vicinity but often distal to the telomere-specific probe signal on the metaphase chromosomes, reflecting a structure specificity of metaphase chromosome-terminal regions. (d) Interphase measurements in the vicinity of the closest telomere signal to clarify the probe order. (e) Summary of metaphase and interphase measurements. Probe 1, telomere PNA kit/FITC (DAKO); probes 2, 3, 8, 11, and 12, P1 5304, P1 5203, BACs 55j6, 167j20, and 38k04, correspondingly (Incyte Genomics, St. Louis); probes 4, 9, and 10, amplified probes for Lztfl1, Ltf, and Tdgf1 genes, correspondingly (see Methods); probes 5 and 6, PACs 443e19 and 415k6, correspondingly (37); probe 7, p12698 (a kind gift of G. Eggertsen, Karolinska Institute; ref. 38). The interphase FISH part of the picture summarizes results of four experiments with different three-probe combinations and shows the probe positions in the vicinity of the chr 9 telomere. Two brown boxes on mouse 9F indicate that the segment, which corresponds to human CER1, is divided into two distinct regions. (f) Tdgf1 FISH probe, which was made by long-range PCR, is located between two large-size genomic fragments. (g) Two-color FISH-based replication-timing experiments in human cells. Magnified images show, separately, green and red signals: singlet “s” pattern (region not yet replicated) for RP6-19p19 and doublet pattern “d” (region already replicated) for RP4-721i7.
We compared our FISH-derived map to the human and mouse genome sequence-based maps, which are available from the Celera Discovery System and UCSC Genome Bioinformatics (Fig. 2). Our results were in agreement with these sequence-based maps throughout the entire region analyzed. In conclusion, the human CER1 belongs to two conserved chromosomal segments (CCSs), in which the distances between genes and their order are similar in man and mouse, and a CBR lies between CCR5 and LTF genes.
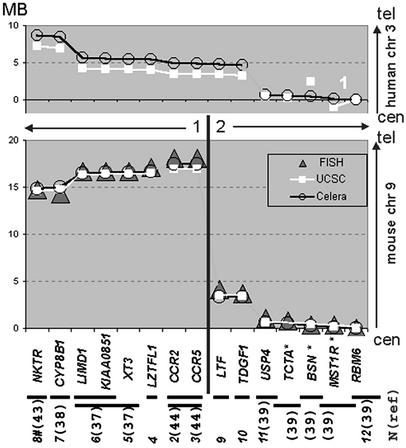
Fig. 2.
Comparison of mapping data for human genes located within CER1 and its flanking regions as well as their mouse orthologs. (Bottom) Mouse FISH probes (horizontal lines), whose numbers (N) correspond to Fig. 1, contain orthologs of the listed human genes (reference). #, This BAC contains marker D9Mit152, which is adjacent to gene Nktr on chr 9. Genes located in one sequenced contig in human and mouse genome assemblies in the Celera Discovery System (July 2002) and in UCSC Genome Bioinformatics (June 2002) are connected by lines. The Celera sequence, which contained fewer gaps, was used to establish human gene order. In the available human UCSC assembly, the contig indicated by number 1 is inverted and gene BSN is in the wrong position. The relative positions of mouse FISH probes (Fig. 1e) between probes 3 and 12 are shown as triangles. Genes marked by * are within BACs from an ≈1-Mb contig, flanked by BACs numbers 11 and 12. Our mouse FISH map, in agreement with both sequence-based maps, suggested that the analyzed region consists of two CCSs (1 and 2) and a CBR (black vertical line) is located between CCR5 and LTF. The numbers in parentheses indicate reference numbers.
Characteristics of CBR Within CER1 Are Consistent With Knowledge About Unstable Chromosomal Sites. Knowing that CER1 contains a CBR and that there are several breakpoints of tumor-related chr 3 rearrangements in the region of 300 kb surrounding CBR (Fig. 3a, black vertical arrows), we searched for further support of the notion that the CBR-containing part of human CER1 is genetically unstable.
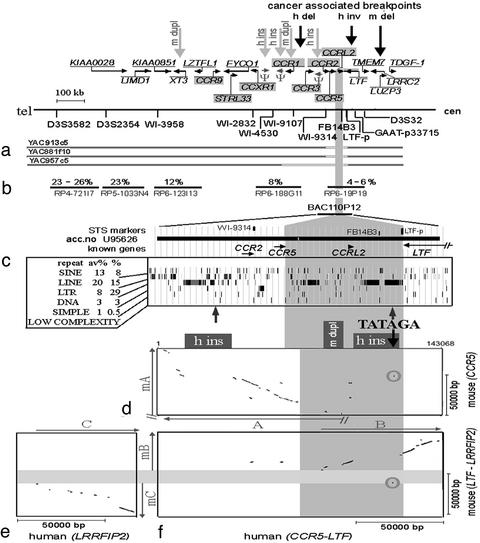
Fig. 3.
Detailed analysis of CER1 sequence containing CBR (gray area from top to bottom). (a) Simplified transcriptional map of CER1 (40). CCR genes are highlighted in gray. Ψ, pseudogene. The gray arrows point to the positions of mouse gene duplications (m dupl) and human pseudogene insertions (h ins) (41), and the black arrows point to cancer-associated breakpoints (deletions and inversion in human and mouse tumors: h del, h inv, and m del) (16, 40). At the bottom, positions of sequence-tagged site markers used for the analysis of YACs covering CER1 are shown. Interstitial deletions (unfilled boxes) were detected in three YACs. (b) Replication-timing analysis of PAC clones along CER1. The percentage of nuclei with double-hybridization dots, counted in one or several FISH experiments (Fig. 1g), is given above the PACs. The lowest percentage corresponds to the latest replication time. (c) Description of the BAC110P12 sequence according to UCSC Genome Bioinformatics (November 2002). Repeat content (%) was determined by using repeatmasker and compared with the average in the human genome (av%) (42). The two gray arrows show the sequences with potential to form the hairpin palindromic structures as determined by using palindrome. The length of the TATAGA-involving palindrome is 40 bp with 14 mismatches. (d–f) Sequence conservation between human (horizontal axis) and mouse (vertical axis) computed by pipmaker. Two dot plots (d and f) show identities between human BAC sequence, surrounding CCR5 and LTF genes and two mouse segments derived from the Celera Discovery System containing the orthologs of CCR5 (d) and LTF and LRRFIP2 (f) genes. (e) Comparison of the “mouse (LTF–LRRFIP2)” segment with human sequence “human (LRRFIP2)” derived from the Celera Discovery System. The human regions (A–C) showing identities to mouse (mA–mC) are indicated by arrows. // indicates that the forward region could not be analyzed, because the mouse sequence contained a gap in this region. The gray area from left to right highlights the segment corresponding to CBR within mouse (LTF–LRRFIP2). h-ins and m-dupl in d refer indicate human insertion and mouse duplication, respectively, and the arrow indicates the position of the TATAGA repeat. On the plots (d) and two dots (f), areas highlighted by gray circles correspond to human–mouse sequence identity in the regions containing (TATAGA)n.
Deletions in yeast artificial chromosomes (YACs). Large genomic fragments, cloned in YACs, often show instability. The sequences unstable in YACs are not randomly distributed in the genome and may represent somatically unstable chromosomal segments (24, 25). CER1 was covered previously by nine YACs, and none of them seemed to bridge fully the segment between CCR2/CCR5 and LTF genes (15). We reanalyzed six of these YACs, 913c5, 881f10, 957c5, 930h5, 870h5, and 743h10, using 11 sequence-tagged site markers. None of these YACs contained the marker FB14B3, localized at CBR. Three of the YACs span the whole CER1 but contain interstitial deletions of various sizes, surrounding FB14B3 (Fig. 3a). Yet another YAC (743h10) was reported to contain this marker. After reanalysis using PCR, we found that this YAC lost all analyzed CER1 markers. FISH, however, showed hybridization to 3p23 and 3p21 (data not shown), indicating that a deletion in CER1 appeared after yeast culture.
Gene and segment duplications. Many recently duplicated segments are located in hot-spot regions of chromosomal and/or evolutionary instability, indicating that there may be a link between the chromosomal rearrangements and gene duplications (11, 26). Eight of 19 characterized human genes of the chemokine receptor (CCR) family form the largest cluster within CER1 (Figs. 3 and 4). This family seems to have evolved recently, because it does not contain any Drosophila or other invertebrate genes (data not shown). According to the phylogenetic tree of this family (Fig. 4), the latest evolutionary duplications could be those two forming CCR1–CCR3 and CCR2–CCR5 pairs. These genes are very similar and are located within CER1 immediately adjacent to the CBR. Some of the closest relatives (CCR9 and STRL33) of the above-mentioned four genes are also located within CER1 or close to it on 3p (CX3CR1, CCR8, and CCR4). The evolutionarily more distant genes either remained within CER1 or were spread to different chromosomal locations. Such gradient of sequence similarity between duplicated segments was described also for unstable pericentromeric regions, with a possible explanation that the duplication events occur within a certain region, and then the original sequence may be dislocated by new duplications and by chromosomal rearrangements such as inversions (27, 28). The location of the most recent duplications near the CBR suggests involvement of it in the duplication process and its evolutionary instability. Human-specific processed pseudogene insertions nearby (Fig. 3a) and enrichment of the CBR with LTRs (Fig. 3c) reflect increased transposition capacity, which is also characteristic for unstable regions. Moreover, comparison of mouse and human CER1 sequences revealed additional duplications in mouse, and they involved not only CCR gene but also an other gene, XT3 ortholog (Fig. 3a, gray vertical arrows), and even a gene empty segment located at the CBR (Fig. 3 c, d, and f). This suggests that the duplication process characterizes not the particular gene cluster but the genomic segment.
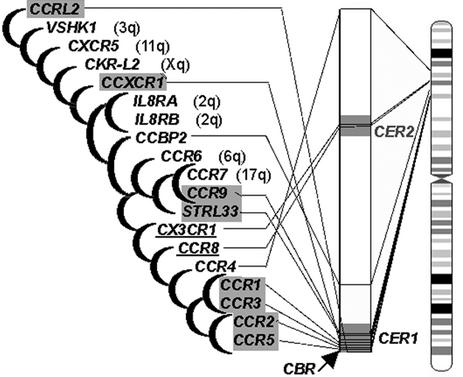
Fig. 4.
Phylogenetic tree of 19 human CCR-related proteins, drawn according to Celera. Shown at the right are locations of genes on 3p21-23. Black arrow, CBR within CER1. The genes located within CER1 and CER2 regions are highlighted in gray boxes and underlined, respectively. Seven genes are located outside 3p, and their chromosomal assignments are indicated in parentheses.
Late replication. The best known unstable chromosomal regions potentially involved in tumorigenesis are fragile sites (9). Their instability is often associated with late replication (22, 29, 30, 31, 32). We performed FISH-based replication-timing analysis within CER1 (Fig. 1g) and found that the CBR region replicated latest as compared with the other studied sites (Fig. 3b), suggesting that a mechanism of instability within CBR may be similar to that in fragile sites.
A TATAGA repeat capable of forming hairpin-like secondary structure colocalizes with CBR. Various repeats were found within fragile sites and within unstable loci responsible for neurodegenerative diseases. These repeats are often capable of forming secondary hairpin-like structures (29). Hairpin formation may play a destabilizing role in eukaryotic genomes (33). There is also strong evidence for genomic instability of inverted repeats, which form long palindromes in prokaryotes. These repeats are deleted at a very high rate in Escherichia coli (34). The most common human constitutional translocation breakpoint t(11;22) is located on the tip of hairpins formed on both chrs 11 and 22, and these inverted repeats, when cloned in different vectors such as YACs, PACs, and BACs, are also unstable (35).
We identified two sequences with the potential to form palindromic, hairpin-like structures in the vicinity of CBR within CER1 (Fig. 3c). One of these, (TA)28, being widely distributed in the genome, is not likely to be associated with regional instability. The other simple tandem repeat, (TATAGA)11, was located within the CBR and is rare in the human genome (see below). The conserved sequences containing this repeat were found only close to the border of the CCS within the analyzed mouse segments surrounding the CCR5 ortholog (Fig. 3d) and at the CBR within the 0.5-Mb mouse sequence spanning LTF–LRRFIP2 orthologs (Fig. 3 e and f).
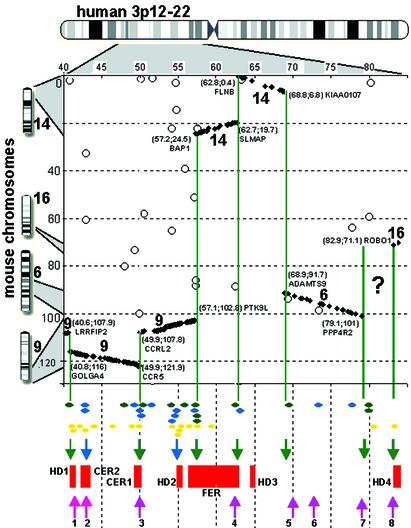
Conservation of Human 3p12-22 in the Mouse. We tested our hypothesis about the role of regional instability in the cancer-associated chromosomal rearrangements, extending our analysis to a larger chr 3 segment. Seven regions were identified on the chr 3 as being preferentially involved in tumor growth-associated deletions (see Table 1). These regions are located within 3p12-22. To analyze murine/human conservation over this chromosomal segment we used the Celera Discovery System computation data about mouse orthologs of 303 human genes located between Mb positions 40 and 85 (Fig. 5). We found that 278 genes match into 7 CCSs. Mouse orthologs of 25 genes were localized outside the corresponding CCSs (conservation mismatches). Analysis of probable paralogs of these mouse genes has shown that for 13 of these 25, one of their paralogs was located within the corresponding CCS (Fig. 5, blue ovals). It is likely that the miscomputation of the orthologous pair took place because of the high similarity of the paralogs and the low similarity between the real orthologs, suggesting that these loci represent sites of recent gene duplications (similarity of paralogs) and unusually strong sequence divergence between orthologs. These 13 mismatches, as well as the other 12 that may represent sites of additional chromosomal rearrangements or transpositions (Fig. 5, green ovals), surround the CBRs. This may reflect the evolutionary instability/plasticity of these chromosomal regions (Fig. 5, green arrows). We discussed above that such instability also may be characteristic for the regions enriched with local duplications (see text about CCR-related genes within CER1 and Fig. 4). Therefore we considered the presence of the groups of two and more paralogs within the same chromosomal locus (Fig. 5, yellow ovals) as an additional indication of instability. These local duplications were distributed along the analyzed region in a very similar manner as the conservation mismatches. Based on this we propose additionally two unstable zones at which mismatches and local duplications are preferentially located (Fig. 5, blue arrows).
Fig. 5.
Dot-plot comparison of the chromosomal positions of the human and mouse orthologous genes according to Celera. Each dot coordinate corresponds to Mb position of a particular gene on human chr 3 (horizontal axis) and on mouse chromosomes (vertical axis). A gene that matches the CCS is displayed as a black rhombus. CCSs are labeled with numbers that correspond to their localization on mouse chrs 9, 14, 6, and 16 (shown at the left of the plot). In the 79.1- to 82.9-Mb segment seven genes were determined as pseudogenes, two have orthologs located on mouse chrs X and 16, and for the other 14 genes mouse orthologs were not identified. Therefore, we could not define any CCS in this region (“?” on the plot). The coordinates of the CCS borders and the closely located genes are indicated. CBRs are shown as green lines. Circles correspond to genes that by their mouse position do not match the CCSs (conservation mismatches). Below the plot, mismatches on 3p12-22 are shown in different colors indicating that the mouse ortholog may (blue) or may not (green) have a paralog located within the corresponding CCS. CBRs are surrounded by conservation mismatches that may reflect the evolutionary instability of these regions (green arrows). We show also that sites containing two or more paralogous genes located in the vicinity of each other (yellow ovals) are distributed similarly to the conservation mismatches. We propose two additional regions of evolutionary instability (blue arrows) where both mismatches and paralogous gene clusters are preferentially located. The evolutionarily unstable regions (green and blue arrows) often colocalize with TATAGA repeats (pink arrows), and both are pointing into the tumor-associated deletion regions (red).
Tumor-Associated Deletions on 3p Colocalize with Evolutionarily Unstable Regions and TATAGA Repeats. We found that these evolutionarily unstable regions (Fig. 5, blue and green arrows) colocalize with the tumor-associated deletions (Fig. 5, red boxes). Four regions of deletions contain CBRs (Fig. 5, green arrows; nonrandom event, P = 0.99; see Methods). Two others do not but contain conservation mismatches and gene duplications (Fig. 5, blue arrows). Within CER2, closely related paralogous genes CCR8 and CX3CR1 have been most likely derived from the same locus as their closest relatives (on the family tree and on the chromosome), namely CCR1, CCR2, CCR3, and CCR5 on CER1 (Fig. 4). This suggests that CER2 was involved in evolutionary rearrangements before the murine/human divergence. HD2 contains three groups of paralogous genes. We detected the duplication of a large mouse segment corresponding to its telomeric part (BAC 38k4) by FISH (Fig. 1a). This region was also found to be unstable in YACs (24). In contrast, HD3 does not show any signs of evolutionary instability, but it harbors the most common fragile site, FRA3B (7). It has to be noted that the coincidence of the synteny breakpoints with tumor-associated deletions means not the colocalization of breakpoints in one spot but the involvement of the same, probably unstable region (up to 1 Mb in size) in both evolutionary and tumor-related chromosomal alterations.
Tandem-Repeat Finder (36) at UCSC Genome Bioinformatics defined eight sites of TATAGA repeats on the 3p12-22 (see Table 2, which is published as supporting information on the PNAS web site). These sites (Fig. 5, pink arrows) were located nonrandomly within the sites of tumor-associated deletions (P = 0.95; see Methods) and in the vicinity of CBRs but not precisely at the breakpoints similarly to fragile sites, where the breaks occur not necessarily very close to repeat (8). Moreover the repeat numbers 2, 3, and 8, identified with the highest scores, were located in two of our CERs (CER1 and CER2) and close to the overlapping region of three homozygous deletions in HD4. It is conceivable that the length of this repeat is polymorphic in the human population. Such variation might constitute a predisposing factor for certain types of cancers. Repeat expansion might also participate in rearrangements during evolution.
In conclusion, the coincidence of synteny breakpoints and evolutionary duplications with malignancy-related deletions, the presence of TATAGA repeats at these sites, and the instability features characterizing the breakpoint region within CER1 suggest that regional instability plays an important role in both cancer-associated and evolutionary chromosomal rearrangements.
Supplementary Material
Acknowledgments
This work was supported by grants from the Swedish Cancer Society, the Swedish Medical Research Council, the Cancer Society in Stockholm, the Cancer Research Institute/Concern Foundation (New York and Los Angeles), the Karolinska Hospital, and the Karolinska Institute.
Abbreviations: chr, chromosome; CER, common eliminated region; UCSC, University of California, Santa Cruz; CBR, conservation breakpoint region; FISH, fluorescence in situ hybridization; PAC, P1 artificial chromosome; BAC, bacterial artificial chromosome; CCS, conserved chromosomal segment; YAC, yeast artificial chromosome; CCR, chemokine receptor.
References
- 1.Kok, K., Naylor, S. L. & Buys, C. H. (1997) Adv. Cancer Res. 71, 27–92. [DOI] [PubMed] [Google Scholar]
- 2.Zabarovsky, E. R., Lerman, M. I. & Minna, J. D. (2002) Oncogene 21, 6915–6935. [DOI] [PubMed] [Google Scholar]
- 3.Alimov, A., Kost-Alimova, M., Liu, J., Li, C., Bergerheim, U., Imreh, S., Klein, G. & Zabarovsky, E. R. (2000) Oncogene 19, 1392–1399. [DOI] [PubMed] [Google Scholar]
- 4.Wistuba, I. I., Behrens, C., Virmani, A. K., Mele, G., Milchgrub, S., Girard, L., Fondon, J. W., III, Garner, H. R., McKay, B., Latif, F., et al. (2000) Cancer Res. 60, 1949–1960. [PubMed] [Google Scholar]
- 5.Maitra, A., Wistuba, I. I., Washington, C., Virmani, A. K., Ashfaq, R., Milchgrub, S., Gazdar, A. F. & Minna, J. D. (2001) Am. J. Pathol. 159, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lerman, M. I. & Minna, J. D. (2000) Cancer Res. 60, 6116–6133. [PubMed] [Google Scholar]
- 7.Huebner, K., Druck, T., Siprashvili, Z., Croce, C. M., Kovatich, A. & McCue, P. A. (1998) Recent Res. Cancer Res. 154, 200–215. [DOI] [PubMed] [Google Scholar]
- 8.Auer, R. L., Jones, C., Mullenbach, R. A., Syndercombe-Court, D., Milligan, D. W., Fegan, C. D. & Cotter, F. E. (2001) Blood 97, 509–515. [DOI] [PubMed] [Google Scholar]
- 9.Richards, R. I. (2001) Trends Genet. 17, 339–345. [DOI] [PubMed] [Google Scholar]
- 10.Padilla-Nash, H. M., Heselmeyer-Haddad, K., Wangsa, D., Zhang, H., Ghadimi, B. M., Macville, M., Augustus, M., Schrock, E., Hilgenfeld, E. & Ried, T. (2001) Genes Chromosomes Cancer 30, 349–363. [DOI] [PubMed] [Google Scholar]
- 11.Jackson, M. S., Rocchi, M., Thompson, G., Hearn, T., Crosier, M., Guy, J., Kirk, D., Mulligan, L., Ricco, A., Piccininni, S., et al. (1999) Hum. Mol. Genet. 8, 205–215. [DOI] [PubMed] [Google Scholar]
- 12.Imreh, S., Kholodnyuk, I., Allikmetts, R., Stanbridge, E. J., Zabarovsky, E. R. & Klein, G. (1994) Genes Chromosomes Cancer 11, 237–245. [DOI] [PubMed] [Google Scholar]
- 13.Harris, H., Miller, O. J., Klein, G., Worst, P. & Tachibana, T. (1969) Nature 223, 363–368. [DOI] [PubMed] [Google Scholar]
- 14.Anderson, M. J. & Stanbridge, E. J. (1993) FASEB J. 7, 826–833. [DOI] [PubMed] [Google Scholar]
- 15.Yang, Y., Kiss, H., Kost-Alimova, M., Kedra, D., Fransson, I., Seroussi, E., Li, J., Szeles, A., Kholodnyuk, I., Imreh, M. P., et al. (1999) Genomics 62, 147–155. [DOI] [PubMed] [Google Scholar]
- 16.Yang, Y., Kost-Alimova, M., Ingvarsson, S., Qianhui, Q., Kiss, H., Szeles, A., Kholodnyuk, I., Cuthbert, A., Klein, G. & Imreh, S. (2001) Proc. Natl. Acad. Sci. USA 98, 1136–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kholodnyuk, I. D., Kost-Alimova, M., Yang, Y., Kiss, H., Fedorova, L., Klein, G. & Imreh, S. (2002) Genes Chromosomes Cancer 34, 341–344. [DOI] [PubMed] [Google Scholar]
- 18.Daigo, Y., Nishiwaki, T., Kawasoe, T., Tamari, M., Tsuchiya, E. & Nakamura, Y. (1999) Cancer Res. 59, 1966–1972. [PubMed] [Google Scholar]
- 19.Sundaresan, V., Chung, G., Heppell-Parton, A., Xiong, J., Grundy, C., Roberts, I., James, L., Cahn, A., Bench, A., Douglas, J., et al. (1998) Oncogene 17, 1723–1729. [DOI] [PubMed] [Google Scholar]
- 20.Fedorova, L., Kost-Alimova, M., Gizatullin, R. Z., Alimov, A., Zabarovska, V. I., Szeles, A., Protopopov, A. I., Vorobieva, N. V., Kashuba, V. I., Klein, G., et al. (1997) Eur. J. Hum. Genet. 5, 110–116. [PubMed] [Google Scholar]
- 21.Amiel, A., Litmanovich, T., Gaber, E., Lishner, M., Avivi, L. & Fejgin, M. D. (1997) Hum. Genet. 101, 219–222. [DOI] [PubMed] [Google Scholar]
- 22.Hellman, A., Rahat, A., Scherer, S. W., Darvasi, A., Tsui, L. C. & Kerem, B. (2000) Mol. Cell. Biol. 20, 4420–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz, S., Zhang, Z., Frazer, K. A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R. & Miller, W. (2000) Genome Res. 10, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kok, K., van den Berg, A., Veldhuis, P. M., van der Veen, A. Y., Franke, M., Schoenmakers, E. F., Hulsbeek, M. M., van der Hout, A. H., de Leij, L., van de Ven, W., et al. (1994) Cancer Res. 54, 4183–4187. [PubMed] [Google Scholar]
- 25.Collins, J. E., Cole, C. G., Smink, L. J., Garrett, C. L., Leversha, M. A., Soderlund, C. A., Maslen, G. L., Everett, L. A., Rice, K. M., Coffey, A. J., et al. (1995) Nature 377, 367–379. [DOI] [PubMed] [Google Scholar]
- 26.Samonte, R. V. & Eichler, E. E. (2002) Nat. Rev. Genet. 3, 65–72. [DOI] [PubMed] [Google Scholar]
- 27.Crosier, M., Viggiano, L., Guy, J., Misceo, D., Stones, R., Wei, W., Hearn, T., Ventura, M., Archidiacono, N., Rocchi, M. & Jackson, M. S. (2002) Genome Res. 12, 67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey, J. A., Yavor, A. M., Viggiano, L., Misceo, D., Horvath, J. E., Archidiacono, N., Schwartz, S., Rocchi, M. & Eichler, E. E. (2002) Am. J. Hum. Genet. 70, 83–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Handt, O., Sutherland, G. R. & Richards, R. I. (2000) Mol. Genet. Metab. 70, 99–105. [DOI] [PubMed] [Google Scholar]
- 30.Wang, L., Darling, J., Zhang, J. S., Huang, H., Liu, W. & Smith, D. I. (1999) Hum. Mol. Genet. 8, 431–437. [DOI] [PubMed] [Google Scholar]
- 31.Le Beau, M. M., Rassool, F. V., Neilly, M. E., Espinosa, R., III, Glover, T. W., Smith, D. I. & McKeithan, T. W. (1998) Hum. Mol. Genet. 7, 755–761. [DOI] [PubMed] [Google Scholar]
- 32.Hansen, R. S., Canfield, T. K., Fjeld, A. D., Mumm, S., Laird, C. D. & Gartler, S. M. (1997) Proc. Natl. Acad. Sci. USA 94, 4587–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sinden, R. R. (2001) Nature 411, 757–758. [DOI] [PubMed] [Google Scholar]
- 34.Leach, D. R. (1994) BioEssays 16, 893–900. [DOI] [PubMed] [Google Scholar]
- 35.Tapia-Paez, I., Kost-Alimova, M., Hu, P., Roe, B. A., Blennow, E., Fedorova, L., Imreh, S. & Dumanski, J. P. (2001) Hum. Genet. 109, 167–177. [DOI] [PubMed] [Google Scholar]
- 36.Benson, G. (1999) Nucleic Acids Res. 27, 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiss, H., Kedra, D., Kiss, C., Kost-Alimova, M., Yang, Y., Klein, G., Imreh, S. & Dumanski, J. P. (2001) Genomics 73, 10–19. [DOI] [PubMed] [Google Scholar]
- 38.Gafvels, M., Olin, M., Chowdhary, B. P., Raudsepp, T., Andersson, U., Persson, B., Jansson, M., Bjorkhem, I. & Eggertsen, G. (1999) Genomics 56, 184–196. [DOI] [PubMed] [Google Scholar]
- 39.Persons, D. A., Paulson, R. F., Loyd, M. R., Herley, M. T., Bodner, S. M., Bernstein, A., Correll, P. H. & Ney, P. A. (1999) Nat. Genet. 23, 159–165. [DOI] [PubMed] [Google Scholar]
- 40.Kiss, H., Yang, Y., Kiss, C., Andersson, K., Klein, G., Imreh, S. & Dumanski, J. P. (2002) Eur. J. Hum. Genet. 10, 52–61. [DOI] [PubMed] [Google Scholar]
- 41.Kiss, H., Darai, E., Kiss, C., Kost-Alimova, M., Klein, G., Dumanski, J. P. & Imreh, S. (2002) Mamm. Genome 13, 646–655. [DOI] [PubMed] [Google Scholar]
- 42.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W. (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 43.Korenberg, J. R., Chen, X. N., Sun, Z., Shi, Z. Y., Ma, S., Vataru, E., Yimlamai, D., Weissenbach, J. S., Shizuya, H., Simon, M. I., et al. (1999) Genome Res. 9, 994–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boring, L., Gosling, J., Monteclaro, F. S., Lusis, A. J., Tsou, C. L. & Charo, I. F. (1996) J. Biol. Chem. 271, 7551–7558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.