Abstract
The unique role of the thymus in the development of T cells was established >4 decades ago. To elucidate how uncommitted lymphoid progenitor cells are instructed to migrate from bone marrow to the thymus to undergo T lymphoid differentiation, we generated and analyzed a genome-wide gene expression profile of CD7+ CD10+ human bone marrow T cell lineage precursors (TLPs) by using the serial analysis of gene expression technique. Unexpectedly, the serial analysis of gene expression profile identified a high number of (pre-) T cell receptor antigen (TCR)-related transcripts in bone marrow TLPs. To determine the configuration of the TCRβ locus in these cells at a quantitative level, we sorted and analyzed bone marrow TLPs from five donors by single-cell PCR. Similar proportions of TLPs harbored TCRβ germ-line alleles, D-J, or V-DJ gene rearrangements. Thus, bone marrow TLPs are heterogenous with respect to TCRβ rearrangement status, suggesting an active recombination machinery that is consistent with the expression of RAG1, RAG2, and TdT in this population. As a hallmark of ongoing TCRβ V-DJ rearrangement, we could amplify broken-ended recombination-signal sequence DNA intermediates from bone marrow TLPs, but not from mature T cells by ligation-mediated PCR. Approximately half of the TCRβ rearrangements were compatible with the expression of a functional pre-TCR, which is in agreement with surface expression of pre-Tα on bone marrow TLPs as shown by confocal laser microscopy and flow cytometry. At a frequency <0.5% of mononucleated cells in human bone marrow, this population is rare, yet it exemplifies T lymphoid differentiation in the human already before immigration into the thymus.
The fundamental role of the thymus in the development of T cell antigen receptor (TCR)αβ+ T cells was established in the mouse >4 decades ago (1) and was also recently demonstrated in humans by the transplantation of thymic tissue, which rescued normal T cell development in children with complete DiGeorge syndrome (2). The lymphoid progenitor cells developing in the bone marrow are thought to include progenitor cells of the T cell lineage, although they are not yet committed to a T cell fate as they still exhibit multilineage differentiation capacity (3, 4). In addition, more recent studies have shown that the earliest T lymphoid progenitor cells within the thymus have at least bipotential T cell and natural killer cell differentiation capacity (5, 6), which suggests that commitment to the T cell lineage occurs only after entry of the precursors into the thymus.
Seemingly in contrast to this notion, several lines of evidence found in mice suggest that T cell lineage commitment can already occur within the bone marrow (5–12). For instance, bone marrow hematopoietic stem cells (HSCs) were able to reconstitute T cell development in thymectomized and irradiated mice (5, 12). In addition, expression of pre-Tα, RAG1, and RAG2 was also found in T cell lineage precursors (TLPs) outside the thymus and even in its absence (6, 8). Notably, two studies identified and further characterized a clonogenic and fully committed TLP in murine bone marrow (10, 12).
Whether the commitment of TLPs can already occur within the bone marrow in humans has not been studied so far. Assuming that also in humans a fraction of lymphoid progenitor cells emigrates from the bone marrow to enter the thymus, we purified TLPs from human bone marrow and generated a quantitative genome-wide gene expression profile of this population by using the serial analysis of gene expression (SAGE) technique. The comparison of SAGE profiles for HSCs and bone marrow TLPs was meant to address (i) how TLPs are directed from the bone marrow to the thymus, and (ii) whether these signals constitute commitment to the T cell lineage.
Materials and Methods
Isolation of Human HSCs, Common Myeloid Progenitor Cells (CMPs), pre-B Cells, and TLPs. CD34+ CD38low HSCs, CD15+ CMPs, and CD10+ CD19+ pre-B cells were purified from human bone marrow as described (13–15). For purification of TLPs, myeloid progenitor cells were depleted from the mononuclear bone marrow cells of five donors (Cambrex, Gaithersburg, MD) by using immunomagnetic anti-CD15 beads (Dynal, Oslo). From the remaining cells, B lineage cells, including plasma cells, natural killer cells, megakaryocytes, and erythroid precursors, were depleted by using magnetic cell sorting (MACS) microbeads specific for CD19, CD138, CD56, CD61, and glycophorin A (Miltenyi Biotec, Bergisch Gladbach, Germany), respectively. Immature lymphoid cells were labeled by using a FITC-conjugated mouse anti-human CD10 IgG1 antibody (CALLA; BD Biosciences, Heidelberg, Germany) and isolated by using anti-mouse IgG1 beads (Miltenyi Biotec). Thereafter, enriched CD10+ cells were labeled with a phycoerythrin (PE)-conjugated anti-CD7 antibody (BD Biosciences) and CD7+ CD10+ cells were sorted by an Epics Elite cell sorter (Coulter) into a vial containing RNAlater solution (Ambion, Austin, TX) for stabilization of RNA (data not shown). Only cell preparations of a purity >95% were included in the SAGE analysis. For verification experiments, T cells were isolated from bone marrow, umbilical cord blood, peripheral blood, and tonsils by using immunomagnetic anti-CD3 beads (Dynal).
Antibodies, Flow Cytometry, and Confocal Laser Microscopy. Anti-CD3 FITC, anti-CD7 PE, anti-CD10 FITC, anti-CD20 PE, rat anti-mouse IgG1 FITC, rat anti-mouse IgG1 PE, rat anti-mouse IgG1 Cy5, and anti-TCRαβ FITC were from BD Biosciences. A mouse anti-human pre-Tα IgG1 antibody, termed K5G3, was prepared as described (16). Flow cytometric analysis of purified cells was carried out on a fluorescence-activated cell sorter (FACSCalibur, BD Biosciences). For confocal laser microscopy, single-cell suspensions of TLPs were triple-stained with antibodies against pre-Tα, CD7, and CD10. As a specificity control, human B cells were purified by using anti-CD19 beads and were double-stained with antibodies against pre-Tα and CD20. Cells were inspected by the use of an LSM 410 confocal laser scanning microscope (Zeiss). A Plan-Neofluar 60-fold oil-immersion objective, numerical aperture 1.25 (Zeiss) was used for the experiments.
SAGE Analysis. cDNA synthesis, SAGE analysis, cloning, and sequencing of SAGE concatemers were carried out as described (17, 18). The UniGene reference database (March 2001) was obtained at www.sagenet.org/SAGEDatabases/unigene.htm. A total of 472,000 SAGE tags were collected for the six SAGE profiles. A total of 106,000 tags were analyzed from the HSC library; 99,000 tags for CMP, 110,000 tags for pre-B cells, 96,500 tags for TLPs, and ≈30,000 tags each for two cases of patients with bone marrow-derived acute lymphoblastic B cell lineage leukemia (B-ALL) carrying a BCR-ABL1 gene rearrangement (F.K., N.F., and M.M., unpublished data). All SAGE libraries were normalized to 100,000 tags.
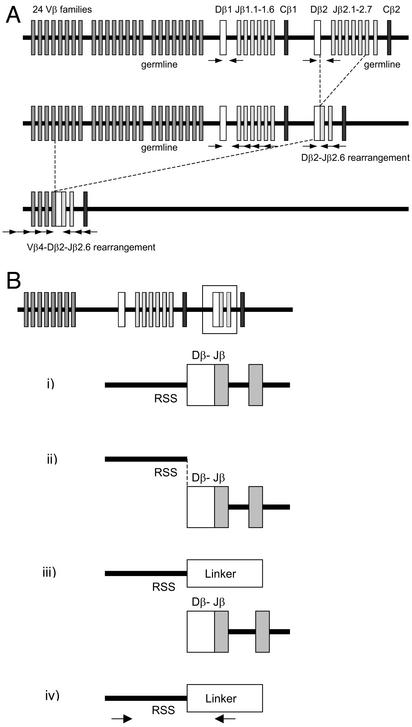
Single-Cell PCR of TCRβ Germ-Line Fragments, D-J, and V-DJ Gene Rearrangements. To analyze individual sorted CD7+ CD10+ TLPs for the presence of germ-line configuration of the TCRβ locus, and TCRβ D-J or V-DJ rearrangements, a whole-genome preamplification step was performed as described (19). This technique, called primer extension preamplification (PEP), multiplies the amount of PCR templates by ≈30-fold in a linear amplification of DNA of the whole genome by using 15-base oligonucleotide random primers (19). Aliquots from these reactions were then subjected to two rounds of seminested PCR amplification (19). For the analysis of the TCRβ loci, the following three PCR strategies were applied to each sorted cell: one that targets germ-line configuration of the TCRβ locus, a second that targets TCRβ D-J gene rearrangements, and a third that detects TCRβ V-DJ gene rearrangements. Amplification of TCRβ VDJ-gene rearrangements was carried out as described by using a panel of 24 Vβ-family-specific primers and two sets of Jβ-gene-specific primers in a seminested approach (Fig. 1A; ref. 19). Germ-line configuration was detected separately for both Cβ loci by using primers binding to intronic sequences flanking the Dβ1- or the Dβ2-gene segment. Dβ–Jβ gene rearrangements were amplified by using the primers specific for intronic sequences in the upstream regions of the Dβ1- and the Dβ2-gene segments, together with primers specific for Jβ gene segments within the Jβ1- or Jβ2-gene clusters, respectively. PCR products were column-purified (Millipore) and were directly sequenced. For sequence analysis of TCRβ V region genes, IMGT V-QUEST software was used at http://imgt.cines.fr/cgi-bin/IMGTdnap.jv?livret=0&Option=humanTcR. The nomenclature of TCRβ V region genes used is according to the work of Rowen et al. (20).
Fig. 1.
Single-cell PCR strategy for the determination of the configuration of TCRβ loci and strategy for ligation-mediated PCR to detect broken-ended TCR Dβ-recombination signal sequence (RSS) DNA intermediates. (A) The organization of the human TCRβ locus and the relative position of PCR primers are depicted for the amplification of specific fragments for germ-line configuration (Top), D-J gene rearrangement (Middle), and V-DJ gene rearrangement (Bottom). (B) The rearrangement of V segments to preexisting DJ joints is initiated by DNA double-strand breaks at the heptamer sequence immediately flanking the D segment (i). DNA double-strand breaks result in blunt ends (ii), which can be ligated (hence “ligation-mediated”) to a blunt-ended linker molecule (iii). By using oligonucleotides specific for sequences upstream of the RSS and the ligated linker sequence, broken-ended Dβ-RSS DNA intermediates can be amplified (iv).
Semiquantitative RT-PCR Analysis. To elucidate the tissue distribution of TLPs, mRNA was extracted from human CD3+ T lymphoid cells from cord blood, bone marrow, peripheral blood, and tonsils. Specific cDNA fragments were amplified for RAG2 by using primers as described (15): 5′-TGCTGCTGGTGGATGGAAAG-3′ and 5′-TCCTGGCTGTAGAAGCCTCT-3′ for pre-Tα; 5′-AGCCCACCCTAAGTTTTCA-3′ and 5′-ACTTCCTGTGCATGATGCT-3′ for RAG1; and 5′-CCTGCTGGATTACATCAAAGCACTG-3′ and 5′-CACCAGCAAGCTTGCGACC-3′ for HPRT, which was used as a standard. Equal amounts of cDNAs were used in 30 and 36 cycles of amplification and separated on a 2% agarose gel.
Amplification of Double-Strand Signal Sequence Breaks in TCRβ V to DJ Recombination by Ligation-Mediated PCR. TLPs were purified from four human bone marrow samples by MACS depletion of non-T lineage cells by using immunomagnetic beads against CD16, CD19, CD56, CD138, and glycophorin A and MACS enrichment by using a mouse anti-human CD10 IgG1 together with anti-mouse IgG1 beads as described above. Mature T cells from peripheral blood were isolated by using anti-CD3 beads. Genomic DNA was isolated from 105 bone marrow TLPs and 105 mature peripheral blood T cells and ligated to a blunt-end linker by using T4 DNA ligase (Invitrogen) at 14°C overnight. The linker was constructed by annealing the oligonucleotides 5′-TTTCTGCTCGAATTCAAGCTTCTAACGATGTACGGGGACATG-3′ and 3′- amino (C7)-GACGAGCTTAAGTTCGAAGATTGCTACATGCCCCT-5′, and protruding 3′ overhangs were removed by 3′→ 5′ exonuclease activity of the Klenow fragment of Escherichia coli DNA polymerase I (Invitrogen). To test whether DNA amounts were comparable for TLPs and mature T cells, a fragment of the MDR1 gene was amplified as described (21). Ligation-mediated PCR was carried out as described (22). In two seminested rounds of amplification (35 and 40 PCR cycles at an annealing temperature of 59°C), recombination signal sequence (RSS)-intermediates with a DNA double-strand break at the 5′ heptamer of the TCR Dβ1 gene segment were amplified by using 5′-GACCAGCCCCTTCGCCAAA-3′ as external forward and 5′-CCTTCGCCAAACAGCCTTA-3′ as internal forward primers, together with 5′-TCCCCGTACATCGTTAGAAG-3′ as reverse primers specific for DNA-ligated linker molecules. To amplify RSS intermediates with a DNA double-strand break at the 5′ heptamer of the TCR Dβ2 gene segment, 5′-TGGTCTGCTCAGGGTGATG-3′ was used as external forward primer and 5′-TCAGGGTGATGCATGTTCCA-3′ as internal forward primer, together with the same linker-specific reverse primer (Fig. 1B).
Results and Discussion
SAGE Analysis of Human Bone Marrow TLPs. TLPs were purified from human bone marrow of five healthy donors, based on the expression of the common acute lymphoblastic leukemia antigen CD10 together with the earliest known T cell lineage marker CD7 (3). Whereas CD7+ CD10+ cells account for <0.5% of mononuclear bone marrow cells, the population was enriched to >95% by a combination of MACS and FACS (not shown). Purified TLPs (6 × 106) were then subjected to SAGE analysis, in which 15,800 unique SAGE tags (i.e., individual transcripts) were identified among 96,500 total SAGE tags collected from this SAGE library. The TLP SAGE profile was compared with already existing libraries for CD34+ HSCs (13), CMPs (14), CD10+ CD19+ pre-B cells (15), and two SAGE libraries for bone marrow-derived B-ALL patients harboring a BCR-ABL1 gene rearrangement (F.K., N.F., and M.M., unpublished work).
In a first survey of the data, SAGE tags were sorted according to the ratio of SAGE tag counts in TLPs to the sum of SAGE tag counts in HSCs, CMPs, pre-B cells, and two B-ALLs. Among the transcripts expressed with highest specificity in TLPs, many molecules related to the (pre-)TCR and (pre-)TCR-associated down-stream signaling were identified (Table 1). The occurrence of such transcripts in bone marrow TLPs was not expected, as the expression of (pre-)TCR components and related signaling molecules, including LCK, ZAP70, CD3 signaling chains, and TRIM were thought to be confined to thymocytes or mature T cells (3). To clarify whether (pre-)TCR-associated transcripts might originate from mature T cells unexpectedly expressing CD7 and CD10, the surface expression of these antigens on peripheral blood mononuclear cells was studied by flow cytometry. Whereas a peripheral blood T cell subset exhibits expression of CD7, no CD10+ mononucleated cells were detected in peripheral blood (data not shown).
Table 1. Distribution of SAGE tags related to the (pre-)TCR and T lineage-specific transcription factors.
| SAGE tag | HSC | CMP | Pre-B | TLP | B-ALL1 | B-ALL2 | UniGene | Gene |
|---|---|---|---|---|---|---|---|---|
| AACAAGTTAG | 125 | 27 | 4 | 4 | 159376 | RAG2 | ||
| TCAAATGTTT | 5 | 75 | 16 | 4 | 73958 | RAG1 | ||
| CTTTATATGA | 1 | 81 | 21 | 272537 | TdT | |||
| TTTATGACTG | 16 | 75 | 18 | 49 | 44 | 272537 | TdT | |
| CATCTGTCAG | 19 | 169002 | preTα | |||||
| GTTTGAAAAA | 1 | 1 | 1 | 9 | 4 | 4 | 74647 | TCRα |
| TCTTTTGCCC | 1 | 4 | 74647 | TCRα | ||||
| AACTGCACTT | 1 | 1 | 74647 | TCRα | ||||
| ACGCTGCGGC | 2 | 4 | 4 | 74647 | TCRα | |||
| AGACTGCCTC | 4 | 74647 | TCRα | |||||
| CCTAAGTGAC | 1 | 1 | 2 | 28 | 9 | 2003 | TCRβ locus | |
| TTCTGTGTGG | 1 | 16 | 4 | 4 | 2003 | TCRβ locus | ||
| AATACTTCTC | 2 | 2 | 2 | 6 | 8 | 2003 | TCRβ locus | |
| GTCAAGAGAA | 1 | 2 | 151 | 274475 | TCR Cβ1 | |||
| TATCCCTTTT | 16 | 247927 | TCRβ V7-8 | |||||
| TATGTCTTGG | 1 | 2 | 2 | 56 | 1765 | LCK | ||
| GCATTCATTG | 1 | 1 | 16 | 1765 | LCK | |||
| CGAGCCTGTT | 1 | 1 | 28 | 4 | 234569 | ZAP70 | ||
| AGACTGGAAG | 1 | 2 | 50 | 4 | 95327 | CD3δ | ||
| GCTTTGGGGT | 1 | 1 | 28 | 211956 | CD3εAP | |||
| GTTTAAAGAT | 1 | 1 | 1 | 9 | 2259 | CD3γ | ||
| GGGTGCTAAG | 1 | 1 | 1 | 44 | 138701 | TRIM | ||
| ATAAGAGCTA | 44 | 82065 | CD130 | |||||
| TCCCCAACTA | 1 | 1 | 1 | 41 | 234434 | HES1 | ||
| CTGGCTCCCT | 1 | 1 | 14 | 234434 | HES1 | |||
| CAGTCCCAGA | 2 | 1 | 21 | 129053 | NOTCH1 | |||
| AAGCCTAAAC | 1 | 1 | 18 | 169946 | GATA3 | |||
| GATTCCAGTT | 1 | 1 | 16 | 233765 | TCF | |||
| AGATGTTTGC | 1 | 1 | 14 | 21704 | HTF4 | |||
| GGCTGATGTT | 1 | 1 | 14 | 21704 | HTF4 | |||
| CTGTGGGACC | 1 | 1 | 1 | 9 | 21704 | HTF4 |
The SAGE profile for bone marrow TLPs also identified a number of T cell lineage-specific transcription factors including HES, NOTCH1, GATA3, TCF, and HTF4 (Table 1). Expression of NOTCH1 IC (23) and oncostatin M receptor/CD130 (24) transgenes induces massive extrathymic T cell development in mice. In this regard, it is notable that both NOTCH1 (21 SAGE tags in TLPs, 1 SAGE tag in CMPs and HSCs) and CD130 (44 SAGE tags in TLPs, no SAGE tags in the other populations) are among the genes expressed with the highest specificity in bone marrow TLPs (Table 1).
As lineage commitment also involves down-regulation of genes belonging to foreign lineages (15), we analyzed lineage-specific transcripts down-regulated in bone marrow TLPs. Silencing of foreign lineage genes primarily involves transcripts associated with MHC molecules and myeloid and B lymphoid differentiation. In the latter case, genes encoding pre-B cell receptor-related molecules (IgH Cμ, CD19, Igα, Igβ, and Vpre-B, and λ5 surrogate light chains), B cell-specific transcription factors (OBF1, PAX5, EBF, and OCT2), and signaling molecules (BLNK, BTK, BLK, BAP29, and IgαBP) were down-regulated. However, also many HSC-specific genes (CD34, CD164, and AML1), myeloid-specific genes (GATA1, CD11C, NF-E2, SET, CD16, CD14, MLL, IL3Rα, and AML1), and erythroid-specific genes (glycophorin C, NF-E2, and GATA1) were down-regulated in bone marrow TLPs (unpublished data). These findings suggest that bone marrow TLPs studied here are committed to the T cell lineage.
As shown in Table 1, we identified many SAGE tags matching to TCR gene segments in the TLP SAGE library. These SAGE tags, however, cannot distinguish between transcripts from TCR gene rearrangements and TCR germ-line transcripts (25). Transcripts from rearranged TCRα genes indicate the presence of mature T cells. In this regard, it is notable, that SAGE tags from TCRβ gene segments were substantially more frequent than SAGE tags derived from the TCRα locus, which were expressed at similar levels in B-ALL cells (Table 1). As a possible indication for TCRβ gene rearrangement, we searched for traces of an active recombination machinery in the TLP library.
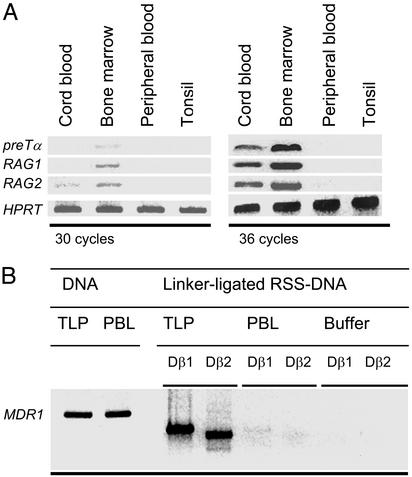
RAG1, RAG2, and TdT are indeed expressed in bone marrow TLPs, albeit at lower levels as compared with bone marrow pre-B cells. Expression of recombination-activating genes, together with pre-Tα in TLPs, was confirmed by semiquantitative RT-PCR (Fig. 2A). These findings, together with the absence of specific expression of mature TCRα chains (Table 1), are consistent with an immature phenotype of the TLP population and argue against a major contamination by mature αβ+ T cells, which also could have been a source of TCRβ gene rearrangements. T cell precursors expressing RAG1, RAG2, and pre-Tα were detected only in human bone marrow and, to a lesser extent, in umbilical cord blood, but not in adult peripheral blood and tonsils (Fig. 2A).
Fig. 2.
Expression of RAGs and ongoing recombination of TCRβ V to DJ segments in bone marrow TLPs. (A) By using immunomagnetic beads, CD3+ T cells from umbilical cord blood, bone marrow, peripheral blood, and tonsillar tissue were isolated and subjected to semiquantitative RT-PCR analysis of mRNA expression of pre-Tα, RAG1, RAG2, and HPRT, which was used for standardization. PCR products after 30 and 36 cycles of amplification are shown. (B) Genomic DNA was isolated from bone marrow TLPs and mature peripheral blood T cells (PBLs). Comparable amounts of DNA (as shown by amplification of a specific fragment of the MDR1 gene) were ligated to a linker molecule and linker-ligated RSS-DNA intermediates carrying double-strand breaks at 5′ heptamers of Dβ1 or Dβ2 gene segments were amplified by ligation-mediated PCR. “Buffer” indicates control without DNA template.
Single-Cell PCR Analysis of Germ-Line and Rearranged TCRβ Alleles in Human Bone Marrow TLPs. To prove that bone marrow TLPs can indeed harbor rearranged TCRβ genes and to quantitate TCRβ germ-line, D-J, and V-DJ alleles in this population, we analyzed the configuration of the TCRβ loci in single sorted CD7+ CD10+ bone marrow TLPs from five donors by single-cell PCR. To this end, single sorted CD7+ CD10+ TLPs were subjected to a whole-genome amplification and aliquots of these reactions were subsequently subjected to three different PCR strategies targeting germ-line fragments, D-J gene, and V-DJ gene rearrangements involving the Cβ1 and the Cβ2 clusters within the TCRβ locus (Fig. 1A). As a positive control, single sorted TCRαβ+ T cells from peripheral blood were used and PCR buffer was used as a negative control. From five healthy donors, a total of 88 bone marrow TLPs were sorted and analyzed. A PCR product was obtained from 69 cells and the remaining 19 cells were considered noninformative (Tables 2 and 3). From 29 cells, only TCRβ germ-line fragments were amplified. These cells were considered to carry nonrearranged alleles of the TCRβ locus. In the absence of polymorphic alleles, however, it is impossible to determine whether TCRβ germ-line PCR products reflect only one or both TCRβ alleles. Assuming that a fraction of cells carry only one TCRβ germ-line allele, whereas a rearrangement on the second allele was missed in the PCR amplification, it is likely that the number of cells in which both TCRβ loci are in the germ-line configuration is somewhat lower. Successful amplification of TCRβ V-DJ gene rearrangements from 21 of 24 sorted TCRαβ+ T cells, however, indicates that not many rearrangements have been missed in the analysis.
Table 2. Summary of single-cell PCR analysis of bone marrow TLPs.
| TCRβ
|
||||
|---|---|---|---|---|
| Germ line | D-J | V-DJ | No product | |
| CD7+ CD10+ TLPs | ||||
| Donor I | 12/40 | 9/40 | 9/40 | 10/40 |
| Donor II | 4/12 | 2/12 | 4/12 | 2/12 |
| Donor III | 5/12 | 2/12 | 2/12 | 3/12 |
| Donor IV | 7/12 | 1/12 | 2/12 | 2/12 |
| Donor V | 1/12 | 5/12 | 4/12 | 2/12 |
| Total | 29/88 | 19/88 | 21/88 | 19/88 |
| TCRαβ+ T cells* | ||||
| With donor I | ND | ND | 10/12 | 2/12 |
| With donors II-V | 0/12 | 1/12 | 11/12 | 1/12 |
| Total | 0/12 | 1/12 | 21/24 | 3/24 |
| Buffer controls* | ||||
| With donor I | 0/8 | 0/8 | 0/8 | 8/8 |
| With donors II-V | 0/12 | 0/12 | 0/12 | 12/12 |
| Total | 0/20 | 0/20 | 0/20 | 20/20 |
ND, not done.
Amplified in parallel as indicated.
Table 3. TCRβ PCR products amplified from single CD7+ CD10+ TLPs.
| Amplified alleles | No. of cells |
|---|---|
| 1 TCRβ germ line | 27 |
| 2 TCRβ germ line | 2 |
| 1 TCRβ D-J | 11 |
| 1 TCRβ D-J, 1 TCRβ germ line | 7 |
| 2 TCRβ D-J | 1 |
| 1 TCRβ V-DJ | 9 |
| 1 TCRβ V-DJ, 1 TCRβ germ line | 5 |
| 1 TCRβ V-DJ, 1 TCRβ D-J | 5 |
| 2 TCRβ V-DJ | 2 |
| No product | 19 |
TCRβ D-J joints in the absence of TCRβ V-DJ gene rearrangements were amplified from 19 of 69 informative TLPs (Tables 2 and 3). Also, in a few of these cells, a TCRβ V-DJ gene rearrangement on the second allele may have been missed. In seven cells, however, either two TCRβ D-J gene rearrangements, or a TCRβ D-J gene rearrangement together with a germ-line allele could be amplified. These cases formally exemplify the presence of T lymphoid bone marrow cells with incomplete or ongoing rearrangement of TCRβ V region genes.
TCRβ V-DJ rearrangements on at least one allele were amplified from 21 of 69 informative cells. To determine whether these TCRβ V-DJ gene rearrangements confer coding capacity for a functional TCRβ chain, which is the prerequisite for the expression of a pre-TCR, we sequenced the amplified TCRβ V region genes. In the sequencing analysis of 21 TCRβ V-DJ junctions from 19 cells, we found 12 potentially functional V region genes and 9 V region genes rearranged out of frame. Thus, in 12 of 69 sorted bone marrow TLPs, the configuration of the TCRβ loci was compatible with the expression of a productive TCRβ chain within a pre-TCR. Also, for the cells harboring a nonproductive TCRβ V region gene, it cannot be excluded that such cells might subsequently acquire coding capacity for a TCRβ chain through successful rearrangement of the second allele. We noted the repetitive usage of Vβ4.1 and Vβ2.1 gene segments within different junctions (Table 4), the significance of which is currently unclear. In summary, the TLP population is heterogenous, with respect to TCRβ rearrangement status and coding capacity.
Table 4. Sequence analysis of TCRβ V-DJ junctions.
| Vβ | Dβ | Jβ | Coding capacity |
|---|---|---|---|
| V1.1 | D1 | J2.2 | + |
| V2.1 | D2 | J1.2 | + |
| V2.1 | D2 | J2.3 | + |
| V2.1 | D1 | J1.2 | + |
| V2.1 | D1 | J2.3 | - |
| V4.1 | D1 | J1.2* | + |
| V4.1 | D1 | J1.2* | + |
| V4.1 | N† | J2.3 | + |
| V4.1 | N† | J2.6 | + |
| V4.1 | N† | J1.3 | - |
| V6.4 | D2 | J1.2 | + |
| V6.4 | N† | J1.3 | - |
| V6.6 | D2 | J1.1 | - |
| V8.1 | D2 | J1.2 | + |
| V8.2 | D2 | J2.3 | - |
| V9.1 | N† | J1.5 | - |
| V10.1 | D2 | J2.3 | + |
| V13.1 | D2 | J2.7 | - |
| V14 | D2 | J2.3 | - |
| V14 | D2 | J1.2 | - |
| V22.1 | N† | J2.3 | + |
Sequences are available at GenBank under nos. AJ549933-AJ549953. TCRβ gene nomenclature used is from Rowen et al. (20).
Junctions differ in N nucleotide sequence.
Rearranged D segments could not be identified.
Ongoing Rearrangement of TCRβ Gene Segments in Human Bone Marrow TLPs. Expression of recombination activating genes (RAGs) (Fig. 2A), together with heterogenous configuration of TCRβ alleles in TLPs, suggests the presence of an active recombination machinery with ongoing rearrangement of TCRβ gene segments. To test this possibility, we amplified broken-ended TCRβ RSS DNA from enriched bone marrow TLPs and mature peripheral blood T cells as a control. During V(D)J recombination, double-strand breaks associated with RSS and covalently closed DNA hairpins associated with coding regions of the TCRβ locus are generated (22). These broken DNA molecules are considered as short-lived reaction intermediates of the recombination process (26). TCRβ genes undergo ordered Dβ to Jβ and Vβ to DβJβ rearrangements, during which Jβ-RSS and, subsequently, Dβ-RSS DNA intermediates are generated. During Dβ to Jβ rearrangement, double-strand breaks are introduced at the 5′ heptamer of the Jβ segment targeted by the recombinase (Jβ-RSS). Subsequent Vβ to Dβ-Jβ rearrangement generates double-strand breaks at the 5′ heptamer of the rearranged Dβ gene segment (Dβ-RSS). Earlier studies (27) have shown that crosslineage rearrangements of the former but not the latter type may also occur in immature B cells at a low frequency. Therefore, we amplified only broken-ended T cell lineage-specific Dβ-RSS DNA intermediates by ligation-mediated PCR. In this case, blunt, 5′ phosphorylated double-strand breaks at the 5′ heptamers of the Dβ1 and Dβ2 gene segments were accessible to ligation to blunt-ended linker molecules (22). Using primers specific for intronic sequences upstream of the Dβ1 and Dβ2 gene segments and the linker molecule, such Dβ1-RSS and Dβ2-RSS, were used, DNA intermediates were amplified from bone marrow TLPs and peripheral blood mature T cells. From bone marrow TLPs, but not from mature T cells and buffer controls, Dβ1-RSS and Dβ2-RSS DNA intermediates were amplified (Fig. 2B).
Based on (i) the expression of RAGs (shown by SAGE and semiquantitative RT-PCR); (ii) a mixed pattern of TCRβ germ-line, D-J, and V-DJ alleles (shown by single-cell PCR); and (iii) specific occurrences of broken-ended Dβ-RSS DNA intermediates (shown by ligation-mediated PCR), we conclude that bone marrow TLPs exhibit ongoing rearrangement of TCRβ gene segments and represent a differentiating T lymphoid population within human bone marrow.
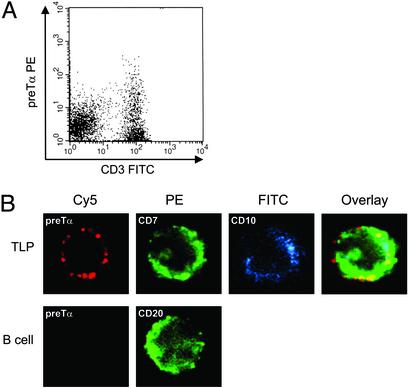
Expression of pre-Tα Within pre-TCR Complexes on the Surface of Bone Marrow TLPs. To determine whether, and at which frequency, human bone marrow TLPs express a pre-TCR on their surface, we used a recently developed antibody to human preTα (16) together with an anti-CD3 antibody. Whereas CD3 is also expressed as constituent of mature TCR signal complexes, the pre-Tα chain is specifically assembled within pre-TCR signal complexes (3). Mononuclear bone marrow cells from two further donors were depleted from myeloid progenitor cells and labeled with antibodies to pre-Tα and CD3. Approximately 5% of the CD3+ T cells coexpressed preTα, whereas cells expressing pre-Tα alone were not detected (Fig. 3A). This finding is consistent with earlier studies, according to which pre-Tα does not reach the cell surface before pairing with a TCRβ chain in complex with CD3 (28). We next studied cellular localization of pre-Tα in single sorted CD7+ CD10+ TLPs by confocal laser microscopy. Some CD7+ CD10+ TLPs also expressed pre-Tα, which was mainly localized to membrane-associated clusters (Fig. 3B). These clusters of pre-Tα expression are reminiscent of the punctate staining pattern observed for pre-Tα expression in murine thymocytes in an earlier study (29). As in murine thymocytes, these clusters may represent preTCRs within glycolipid-enriched membrane domains (rafts) on bone marrow TLPs. We conclude that a small yet appreciable population of human bone marrow TLPs express a pre-TCR on their surface.
Fig. 3.
Expression of pre-Tα within pre-TCR complexes on human bone marrow T lineage precursors. (A) Mononuclear bone marrow cells were depleted from myeloid progenitor cells and were double-stained by using antibodies against CD3 (FITC) and pre-Tα, together with a PE-labeled secondary antibody. (B) Bone marrow TLPs were FACS sorted based on staining for CD7 (PE) and CD10 (FITC) and were subsequently labeled with an anti-pre-Tα antibody, together with a Cy5-conjugated secondary antibody (false-color images). Single cells were analyzed by laser microscopy and staining for CD7 and CD10, and pre-Tα was colocalized in an overlay image (Upper). As a control, single sorted B cells were double-stained by using an antibody against CD20 and pre-Tα, together with a Cy5-conjugated secondary antibody (Lower).
Concluding Remarks
Recent studies in mice suggest that commitment to the TCRαβ+ T cell lineage does not precede colonization of the thymus in most, if not all, cases (4, 30). This conclusion is based on the finding that thymic T lymphoid progenitor cells are capable of generating B, T, and natural killer cells despite mRNA expression of T cell lineage-associated genes, including pre-Tα (30, 28). By using a transgenic reporter mouse strain, it could be formally demonstrated that expression of pre-Tα mRNA in lymphoid bone marrow progenitor cells does not preclude B lymphoid or natural killer cell differentiation (28). Thus, pre-Tα mRNA expression within murine bone marrow would not indicate extrathymic commitment to the T cell lineage and, rather, results from initial opening of the pre-Tα locus in uncommitted lymphoid progenitor cells (28). In the present study, however, we show a differentiating T lymphoid population in human bone marrow, which expresses pre-Tα protein on the cell surface. Unlike pre-Tα transcripts, membrane expression of pre-Tα defines a committed TLP, because pre-Tα reaches the cell membrane only within a constitutively active pre-TCR complex (29). Notably, a recent study (12) demonstrates a clonable committed TLP in murine bone marrow. In agreement with these findings, expression of pre-Tα as part of a pre-TCR complex on human bone marrow TLP, as reported here, shows that committed “extrathymic” T cell progenitors indeed exist in both mice and humans. However, in neither case can it be ruled out that colonization of the bone marrow represents a secondary event after initial commitment within the thymus. To elucidate whether committed bone marrow TLPs are indeed “prethymic,” rather than recirculating from a leaky thymus, we studied configuration and rearrangement of TCRβ alleles. Consistent with the expression of RAG1, RAG2, and TdT, and a heterogenous pattern of cells harboring TCRβ germ-line, D-J, and V-DJ alleles, we amplified broken-ended Dβ-RSS DNA intermediates as a hallmark of ongoing recombination from bone marrow TLPs, but not from mature T cells. We propose that human bone marrow can indeed sustain various stages of T lymphoid differentiation, including committed TLPs carrying TCRβ germ-line alleles, pro-T cells (TCRβ D-J), and pre-T cells (TCRβ V-DJ) expressing a pre-TCR on the surface. Whether bone marrow TLPs can differentiate even beyond the pre-T cell stage independently from the thymus needs further investigation. Although the quantitative contribution of T lymphoid differentiation within the bone marrow to the pool of TCRαβ+ T cells in humans is clearly minor, it will be interesting to compare the developmental checkpoints operative in this alternative pathway to selection processes within the thymus.
Acknowledgments
We thank Klaus Rajewsky and Holger Babbe for critical discussions; Harald von Boehmer, Jos Even, and Samuel Strober for thoughtfully reviewing this manuscript; Stefanie Jauch for excellent technical assistance; Christoph Göttlinger for cell sorting; and Roland Schmitz for sharing his expertise in single-cell PCR. This work was supported by National Institutes of Health Grants CA 78862-01 (to J.D.R.), Deutsche Forschungsgemeinschaft Grant MU1616/2-1 through the Emmy-Noether-Program (to M.M.), a German José Carreras Leukemia Foundation grant (to M.M.), and the Ministry of Science and Research for North Rhine-Westphalia through the Stem Cell Network North Rhine-Westphalia (to M.M.). F.K. is supported by scholarships of the Studienstiftung des Deutschen Volkes and the Köln Fortune Program of the Faculty of Medicine of the University of Cologne. H.W. is supported by the Deutscher Akademischer Austauschdienst Bioscience Program.
Abbreviations: TLP, T lineage precursor; TCR, T cell antigen receptor; HSC, hematopoietic stem cell; CMP, common myeloid progenitor cell; B-ALL, B lineage acute lymphoblastic leukemia; SAGE, serial analysis of gene expression; RSS, recombination signal sequence; RAG, recombination activating gene; PE, phycoerythrin.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AJ549933–AJ549953).
References
- 1.Miller, J. F. A. P. (1961) Lancet ii, 748-749. [DOI] [PubMed] [Google Scholar]
- 2.Markert, M. L., Boeck, A., Hale, L. P., Kloster, A. L., McLaughlin, T. M., Batchvarova, M. N., Douek, D. C., Koup, R. A., Kostyu, D. D., Ward, F. E., et al. (1999) N. Engl. J. Med. 341, 1180-1189. [DOI] [PubMed] [Google Scholar]
- 3.Spits, H. (2002) Nat. Rev. Immunol. 2, 760-772. [DOI] [PubMed] [Google Scholar]
- 4.Blom, B., Res, P., Noteboom, E., Weijer, K. & Spits, H. (1997) J. Immunol. 158, 3571-3577. [PubMed] [Google Scholar]
- 5.Deibakhsh-Jones, S., Jerabek, L., Weissman, I. L. & Strober, S. (1995) J. Immunol. 155, 3338-3344. [PubMed] [Google Scholar]
- 6.Bruno, L., Rocha, L., Rolink, A., von Boehmer, H. & Rodewald, H.-R. (1995) Eur. J. Immunol. 25, 1877-1882. [DOI] [PubMed] [Google Scholar]
- 7.Ktorza, S., Blanc, C., Laurent, C., Sarun, S., Verpilleux, M. P., Debre, P. & Schmitt, C. (1996) J. Immunol. 156, 4120-4127. [PubMed] [Google Scholar]
- 8.Collins, C., Norris, S., McEntee, G., Traynor, O., Bruno, L., von Boehmer, H., Hegarty, J. & O'Farrelly, C. (1996) Eur. J. Immunol. 26, 3114-3118. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Ojeda, M. E., Dejbakhsh-Jones, S., Weissman, I. L. & Strober, S. (1998) J. Exp. Med. 187, 1813-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dejbakhsh-Jones, S. & Strober, S. (1999) Proc. Natl. Acad. Sci. USA 96, 14493-14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsark, E. C., Dao, M. A., Wang, X., Weinberg, K. & Nolta, J. A. (2001) J. Immunol. 166, 170-181. [DOI] [PubMed] [Google Scholar]
- 12.Dejbakhsh-Jones, S., Garcia-Ojeda, M. E., Chatterjea-Matthes, D., Zeng, D. & Strober, S. (2001) Proc. Natl. Acad. Sci. USA 98, 7455-7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou, G., Chen, J., Lee, S., Clark, T., Rowley, J. D. & Wang, S. M. (2001) Proc. Natl. Acad. Sci. USA 98, 13966-13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, S., Zhou, G., Clark, T., Chen, J., Rowley, J. D. & Wang, S. M. (2001) Proc. Natl. Acad. Sci. USA 98, 3340-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müschen, M., Lee, S., Zhou, G., Feldhahn, N., Barath, V. S., Chen, J., Moers, C., Krönke, M., Rowley, J. D. & Wang, S. M. (2002) Proc. Natl. Acad. Sci. USA 99, 10014-10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramiro, A. R., Navarro, M. N., Carreira, A., Carrasco, Y. R., de Yebenes, V. G., Carrillo, G., San Millan, J. L., Rubin, B. & Toribio, M. L. (2001) J. Immunol. 167, 5106-5114. [DOI] [PubMed] [Google Scholar]
- 17.Velculescu, V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. (1995) Science 270, 484-487. [DOI] [PubMed] [Google Scholar]
- 18.Feldhahn, N., Schwering, I., Lee, S., Wartenberg, M., Klein, F., Wang, H., Zhou, G., Wang, S. M., Rowley, J. D., Hescheler, J., et al. (2002) J. Exp. Med. 196, 1291-1305. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Müschen, M., Rajewsky, K., Bräuninger, A., Baur, A. S., Oudejans, J. J., Roers, A., Hansmann, M. L. & Küppers, R. (2000) J. Exp. Med. 191, 387-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowen, L., Koop, B. F. & Hood, L. (1996) Science 272, 1755-1762. [DOI] [PubMed] [Google Scholar]
- 21.Wartenberg, M., Ling, F. C., Müschen, M., Klein, F., Acker, H., Gassmann, M., Petrat, K., Putz, V., Hescheler, J. & Sauer, H. (2003) FASEB J. 17, 503-505. [DOI] [PubMed] [Google Scholar]
- 22.Schlissel, M., Constantinescu, A., Morrow, T., Baxter, M. & Peng, A. (1993) Genes Dev. 7, 2520-2532. [DOI] [PubMed] [Google Scholar]
- 23.Allman, D., Karnell, F. G., Punt, J. A., Bakkour, S., Xu, L., Myung, P., Koretzky, G. A., Pui, J. C., Aster, J. C. & Pear, W. S. (2001) J. Exp. Med. 194, 99-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clegg, C. H., Rulffes, J. T., Wallace, P. M. & Haugen, H. S. (1996) Nature 384, 261-263. [DOI] [PubMed] [Google Scholar]
- 25.Che, F., Rowen, L., Hood, L. & Rothenberg, E. V. (2001) J. Immunol. 166, 1771-1780. [DOI] [PubMed] [Google Scholar]
- 26.Melek, M. & Gellert, M. (2000) Cell 101, 625-633. [DOI] [PubMed] [Google Scholar]
- 27.Ferrier, P., Covey, L. R., Suh, H., Winoto, A., Hood, L. & Alt, F. W. (1989) Int. Immunol. 1, 66-74. [DOI] [PubMed] [Google Scholar]
- 28.Gounari, F., Aifantis, I., Martin, C., Fehling, H. J., Hoeflinger, S., Leder, P., von Boehmer, H. & Reizis, B. (2002) Nat. Immunol. 3, 489-496. [DOI] [PubMed] [Google Scholar]
- 29.Saint-Ruf, C., Panigada, M., Azogui, O., Debey, P., von Boehmer, H. & Grassi, F. (2000) Nature 406, 524-527. [DOI] [PubMed] [Google Scholar]
- 30.Carlyle, J. R. & Zuniga-Pflucker, J. C. (1998) Immunity 9, 187-197. [DOI] [PubMed] [Google Scholar]