Abstract
Cholesterol, a lipid not normally found in prokaryotes, was identified in purified Chlamydia trachomatis elementary bodies and in the chlamydial parasitophorous vacuole (inclusion) membrane of infected HeLa cells. Chlamydiae obtained eukaryotic host cell cholesterol both from de novo synthesis or low-density lipoprotein. Acquisition of either de novo-synthesized cholesterol or low-density lipoprotein-derived cholesterol was microtubule-dependent and brefeldin A-sensitive, indicating a requirement for the Golgi apparatus. Transport also required chlamydial protein synthesis, indicative of a pathogen-directed process. The cholesterol trafficking pathway appears to coincide with a previously characterized delivery of sphingomyelin to the inclusion in that similar pharmacological treatments inhibited transport of both sphingomyelin and cholesterol. These results support the hypothesis that sphingomyelin and cholesterol may be cotransported via a Golgi-dependent pathway and that the chlamydial inclusion receives cholesterol preferentially from a brefeldin A-sensitive pathway of cholesterol trafficking from the Golgi apparatus to the plasma membrane.
Chlamydia trachomatis is the etiologic agent of the most common form of sexually transmitted disease in the United States and preventable blindness worldwide (1). Chlamydiae are obligate intracellular bacteria with a biphasic life cycle consisting of an extracellular infectious form and an intracellular replicative form. Infection is initiated by the metabolically inert elementary body (EB), which develops into the noninfectious but metabolically active reticulate body (RB). RBs differentiate back to EBs before release from the infected cell (2). Chlamydiae replicate within a specialized parasitophorous vacuole, termed an inclusion, that is neither acidified nor fusogenic with lysosomes (3, 4). Rather than interacting with the endocytic pathway, the chlamydial inclusion is fusogenic with exocytic vesicles containing sphingomyelin en route from the Golgi apparatus to the plasma membrane (5, 6). Acquisition of sphingomyelin requires chlamydial de novo transcription and translation of proteins that are thought to modify the inclusion membrane (7).
Because sphingomyelin is delivered to the chlamydial inclusion and EBs contain both cholesterol and sphingomyelin (5, 8, 9), we asked whether cholesterol may be similarly regulated and transported to the inclusion. Cholesterol is an essential component of eukaryotic membranes and is enriched in the plasma membrane. Although synthesis occurs in the endoplasmic reticulum (ER), the role of the Golgi apparatus in the transport of cholesterol to the plasma membrane has been controversial. Dissection of intracellular cholesterol transport is complicated by the fact that there are a number of possible sources of this lipid. Eukaryotic cells acquire cholesterol from either de novo synthesis or from the extracellular media via the low-density lipoprotein (LDL) pathway (10–12). We find that C. trachomatis has the ability to obtain host cholesterol from either pathway. Similar pharmacological agents blocked delivery of both cholesterol and sphingomyelin to chlamydiae, suggesting that the two lipids are cotransported to the chlamydial inclusion, with the Golgi apparatus playing a critical role. The chlamydial inclusion thus offers a target organelle to characterize Golgi-dependent mechanisms of intracellular cholesterol trafficking.
Materials and Methods
Cell Lines and Chlamydial Strains. C. trachomatis serovar L2 was grown in HeLa 229 cells and purified by Renografin density gradient centrifugation as described (13). HeLa cells were propagated in RPMI medium 1640 supplemented with 10% FBS or CPSR-1 low-lipid formula (Sigma) in an atmosphere of 5% CO2 in humidified air.
Reagents. Brefeldin A (BFA), chloramphenicol, nocodazole, non-hydroxy fatty acid ceramide from bovine brain, cholesterol (chromatography grade), squalene, filipin III from Streptomyces filipinensis, methyl-β-cyclodextrin, 4-Methylumbelliferyl-α-D-mannopyranoside, sodium thymidine 5′-monophosphate, p-nitrophenyl ester, peroxidase-conjugated goat anti-mouse IgG antibody, and 4-chloro-1-naphthol tablets were purchased from Sigma. N-(4,4-dif luoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoyl)sphingosine (BODIPY FL C5-ceramide), 6-{[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]hexanoyl}sphingosine (NBD C6-ceramide), and N-{[6-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]hexanoyl}sphingosyl phosphocholine (NBD C6-SM) were purchased from Molecular Probes. [4,8,12,13,17,21-3H]Squalene (10–30 Ci/mmol; 1 Ci = 37 GBq), [1,5,9,14,20,24-14C]Squalene (100–200 mCi/mmol), D-erythro-3-[3H]Sphingosine (15–30 Ci/mmol), [choline methyl-14C]Sphingomyelin (50–60 mCi/mmol), and N-1-[14C]oleoy-D-sphingosine (50–60 mCi/mmol) were purchased from American Radiolabeled Chemicals (St. Louis). [1,2,6,7-3H(N)]Cholesteryl linoleate (60–100 Ci/mmol) was purchased from NEN. GRP78/BiP mAb was purchased from Transduction Laboratories (San Diego).
Analysis of Lipids Associated with EBs. C. trachomatis-infected HeLa 229 cells in RPMI medium 1640 supplemented with 10% CPSR-1 low-lipid formula serum (Sigma) were labeled with 5 μCi of N-[14C]oleoyl-d-sphingosine (50–60 mCi/mmol) or 2 μCi of [14C]squalene (10–30 Ci/mmol) for 30 h postinfection and the EBs were purified by density gradient centrifugation. Lipids were extracted from purified EBs by the Folch extraction method (14), dried under a stream of N2, and redissolved in a 1:1 (vol/vol) mixture of MeOH:CHCl3. A 25-μl aliquot was subjected to HPLC by using a Hewlett-Packard Model 1090 liquid chromatograph with an Alltech adsorbosphere 250 × 4.6-mm, 5-μm silica particle column. Initial solvent conditions were as follows: 0.5% double-distilled H2O (ddH2O), 99.5% MeCN, and 0.0% MeOH followed by a gradient of: 5 min, 0.5% ddH2O, 99.5% MeCN, and 0% MeOH; 15 min, 8.0% ddH2O, 59.0% MeCN, and 33.0% MeOH; and 35 min, 8.0% ddH2O, 59.0% MeCN, and 33.0% MeOH, with a flow rate of 1.0 ml/min. Radioactively labeled lipids were detected by using an IN/US (Tampa, FL) β-RAM flow-through scintillation detector.
Inhibition of Cholesterol and Sphingomyelin Transport. At 24 h postinfection, cultures were preincubated with BFA (1 μg/ml) or nocodazole (10 μg/ml) for 2 h. The infected cells were labeled with either 10 μCi of N-oleoyl[14C]-D-sphingosine (50–60 mCi/mmol), 6 μCi of [14C]squalene (100–300 mCi/mmol), or [3H]Cholesteryl linoleate-labeled LDL (15) for 1 h at 15°C in the presence of the inhibitors. The labeling medium was removed, cells were rinsed twice with cold HBSS, and incubated in chase medium [RPMI medium 1640 with 10% FCS or 10% CPSR-1 serum, and either 5 mg/ml ceramides derived from bovine brain or 40 μM squalene, BFA (1 μg/ml) or nocodazole (10 μg/ml)]. The cells were incubated at 37°C for 1 h in chase medium and EBs were purified. Scintillation counting was performed with a Beckman LS 6000LL.
Effects of Chloramphenicol Treatment. One set of T150 flasks with HeLa 229 cell monolayer was infected with L2 EBs as described above. At 24 h postinfection, chloramphenicol (40 μg/ml) was added, while a second set of T150 flasks was infected with L2. At 24 h postinfection (relative to the second set of flasks), radioactive labeling was performed as above to both chloramphenicol-treated and untreated flasks. Chloramphenicol was continuously present in the treated group throughout the labeling and chase procedures. EBs were purified, total lipid extracted as described above, and were subjected to thin layer chromatography. A 10-μl aliquot from each flask was subjected to Western blotting and probed with a mouse mAb (L2 I-45) to the major outer membrane protein of L2, to determine the relative number of EBs isolated from the chloramphenicol-treated and untreated flasks.
Fluorescence Microscopy. HeLa 229 cells growing on 12-mm glass coverslips (no. 1 thickness) were infected with C. trachomatis L2 EBs. At 24 h postinfection, cells were stained with filipin as described (6, 16).
Results
Cholesterol and Sphingomyelin Are Present in Chlamydial EBs and the Inclusion Membrane. Analysis of total lipids from C. trachomatis L2 EBs by HPLC revealed that cholesterol and sphingomyelin represent ≈6.47 ± 0.08% and 3.69 ± 0.02% of the total lipid content. These values are consistent with previous observations (8, 9, 17). Steady-state labeling of C. trachomatis EBs was accomplished by incubation of infected cells for 30 h in the presence of [14C]squalene or [14C]ceramide to uniformly label endogenously synthesized cholesterol and sphingomyelin, respectively. EBs were density gradient-purified at 30 h postinfection and total lipids extracted for HPLC analysis. Cholesterol was the singular product observed after [14C]squalene labeling whereas four peaks were observed after [14C]ceramide labeling (data not shown). Thin layer chromatography was thus included in subsequent experiments to specifically analyze sphingomyelin synthesis. Approximately 6% of the total cellular cholesterol and 12% of the total sphingomyelin was associated with EBs. These values are likely underestimates of the total cholesterol and sphingomyelin trafficked to the inclusion as RBs and fragments of the inclusion membrane would be lost during the purification protocol. To exclude the possibility of contamination of the EB preparations by cellular debris, uninfected control cells similarly labeled with [14C]squalene or [14C]ceramide were mixed with an equivalent number of unlabeled EBs and subjected to the same purification scheme. In this experiment, <0.2% of the total cholesterol and 0.08% of the total sphingomyelin were associated with the purified EBs. Additional controls were performed to further rule out nonspecific contamination of EB preps by cellular membranes. Renografin density gradient-purified EBs are typically free of host debris as determined by electron microscopy. Furthermore, contamination of purified EBs by host plasma membrane or inclusion membrane was minimal, as shown by the absence of host plasma membrane proteins or the chlamydial inclusion membrane protein IncG, respectively (see Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org). Analysis of specific radioactive products in purified EBs thus appears to be a useful measure of transport to the chlamydial inclusion, from which they are acquired by the intracellular chlamydiae.
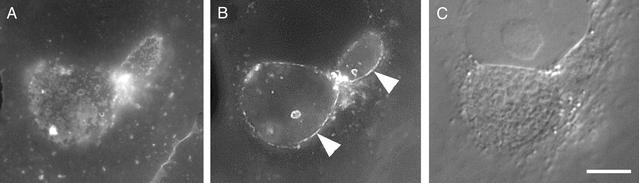
The distribution of cholesterol in C. trachomatis-infected cells was visualized by using filipin, a fluorescent polyene antibiotic that binds the 3′ hydroxyl group of steroids. Intense filipin staining is observed in the plasma membrane as well as in the inclusion membrane (Fig. 1). Staining of the intracellular chlamydial developmental forms was also observed although the intensity was less than that of the inclusion membrane or plasma membrane. Furthermore, filipin staining of chlamydiae was best observed when the focal plane was adjusted to the top or bottom of the inclusion. Because filipin staining depends on cholesterol concentration, it is likely that the inclusion membrane has a higher cholesterol content than the chlamydiae. However, the relative lack of staining of the chlamydial developmental forms could occur because of either an enrichment of cholesterol in the inclusion membrane or poor accessibility of the fluorescent probe to the lumen of the inclusion.
Fig. 1.
Filipin staining of the chlamydial inclusion. At 20 h p.i., HeLa cells were gently fixed with paraformaldehyde and incubated with filipin for 30 min for visualization of cholesterol. Images were acquired at different focal planes to allow visualization of the chlamydial developmental forms (A) and the inclusion membrane (B). The inclusion membrane typically stains much brighter than the chlamydiae. Images of RBs were therefore acquired first so as to compensate for the effects of photobleaching. Arrowheads indicate the inclusion membrane. (C) Nomarski differential interference contrast image of the same field. Scale bar, 1 μm.
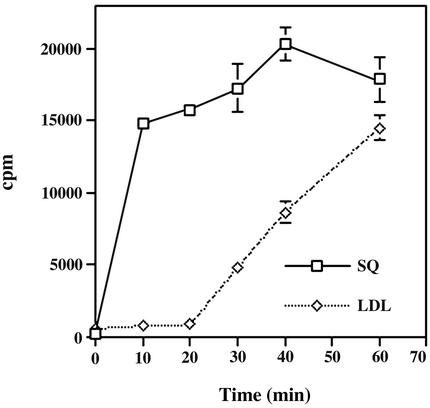
De Novo-Synthesized and LDL-Derived Cholesterol Are Trafficked to the Chlamydial Inclusion. The ability of chlamydiae to acquire cholesterol synthesized from [14C]squalene indicates that the biosynthetic pathway is one possible source of cholesterol for Chlamydia. Thus, the kinetics of cholesterol acquisition from the biosynthetic pathway were determined. L2-infected HeLa cells grown in lipid-deficient serum for 24 h were labeled for 1 h with [14C]squalene at 15°C, then temperature-shifted to 37°C, and chased with nonradioactive squalene. EBs were purified by Renografin density gradient centrifugation at 10-min intervals to monitor associated cholesterol (Fig. 2). Radioactive cholesterol was detected in purified EBs within 10 min after temperature shift, thus newly synthesized cholesterol appeared to be rapidly transported to the inclusion.
Fig. 2.
Kinetics of transport to the chlamydial inclusion of cholesterol derived from de novo synthesis or LDL uptake. Infected HeLa cells were incubated with [14C]squalene or [3H]cholesterol oleate/LDL and radioactive cholesterol content from purified EBs monitored as a function of time. Data are expressed as mean ± SD.
The ability of the LDL pathway to contribute to the cholesterol pool in the inclusion was also investigated. HeLa cells grown in lipid-deficient serum for 24 h were labeled with LDL-[3H]cholesteryl linoleate. HeLa cells were incubated with progesterone during the labeling period to arrest the mobilization of LDL-derived cholesterol from the lysosomes (18). The appearance of radioactive cholesterol in density gradient-purified EBs was monitored at 10-min intervals after removal of progesterone. Association of LDL-derived cholesterol with EBs was not observed until 30 min after progesterone removal (Fig. 2). To verify that the cholesterol found in EBs indeed came from the LDL uptake pathway, infected cells were treated with NH4Cl to prevent acidification of lysosomes (19). NH4Cl treatment resulted in an 80-fold reduction of radioactive cholesterol associated with EBs (data not shown). Thus, the LDL uptake pathway also provided a source of cholesterol for the chlamydial inclusion.
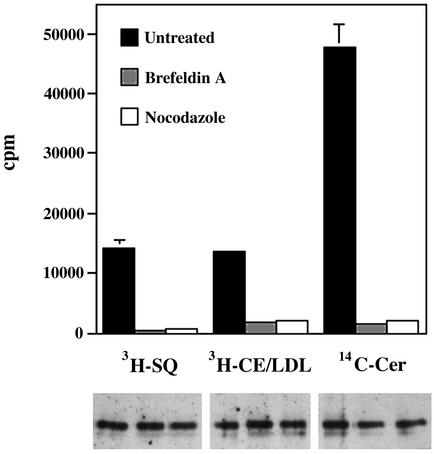
Transport of Cholesterol from Different Sources to the Chlamydial Inclusion Is Golgi- and Microtubule-Dependent. To determine whether the same pharmacological agents (BFA and nocodazole) that block sphingomyelin delivery to the inclusion similarly inhibited cholesterol trafficking to the inclusion, infected HeLa cells were treated with either BFA or nocodazole during a 1-h pulse-labeling with [14C]squalene or [3H]cholesteryl linoleate/LDL. EBs were then purified by Renografin density gradient centrifugation before determination of associated radioactivity. Regardless of the source of radioactive cholesterol, BFA and nocodazole markedly reduced the amount of cholesterol associated with purified EBs (Fig. 3). Consistent with previous results using fluorescent analogs (6, 20), [14C]sphingomyelin transport to the inclusion was also inhibited by BFA and nocodazole treatments. Although both BFA and nocadazole decrease the rate of cholesterol and sphingomyelin delivery to EBs, the continuous presence of the inhibitors throughout the chlamydial developmental cycle does not reduce the yield of progeny EBs (ref. 6 and unpublished observations).
Fig. 3.
The Golgi apparatus and microtubules are required for the transport of cholesterol and sphingomyelin to the chlamydial inclusion. Infected HeLa cells were incubated with [14C]squalene, [3H]cholesterol oleate/LDL, or [14C]ceramide in the presence of BFA or nocodazole to inhibit Golgi and microtubule functions, respectively. Radioactive cholesterol or sphingomyelin associated with EBs were monitored. Data are expressed as mean ± SD. (Lower) A representative immunoblot against the chlamydial major outer membrane protein (MOMP) from density gradient-purified EBs to show equivalence in the samples used in the analysis of incorporated isotope. Lanes correspond to the samples directly above.
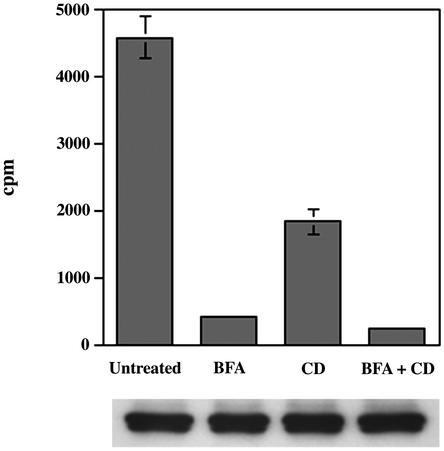
Typically, cholesterol originating from the lysosome after LDL degradation is trafficked to the plasma membrane (10–12, 16, 21). There have been reports of Golgi-dependent and Golgi-independent pathways of transport of LDL-derived cholesterol to the plasma membrane (10, 16, 22). To test the possibility of the plasma membrane being an intermediate in the transport of cholesterol to the inclusion, infected cells were incubated with [3H]cholesteryl linoleate-LDL at 37°C in the presence of progesterone and BFA with or without cyclodextrin added to the medium to remove plasma membrane cholesterol. The samples were washed to remove unbound LDL and initiate cholesterol mobilization from the lysosomes. EBs were density gradient-purified 1 h later for determination of incorporated cholesterol (Fig. 4). Cyclodextrin alone led to a moderate reduction in the amount of EB-associated cholesterol, suggesting that some proportion of the cholesterol originating from LDL was exposed at the plasma membrane. However, treatment with BFA alone was comparable to BFA plus cyclodextrin in inhibiting the amount of LDL-derived cholesterol associated with the EBs and was much more effective than cyclodextrin alone. Thus, even cholesterol that cycles through the plasma membrane must transit the Golgi apparatus before delivery to the inclusion. These data indicate that the transport to the chlamydial inclusion of cholesterol originating from the biosynthetic or LDL-uptake pathways is directly from the Golgi apparatus and confirms a role for this organelle in the trafficking of cholesterol.
Fig. 4.
LDL-derived cholesterol transits to the Golgi apparatus and the plasma membrane before arriving to the chlamydial inclusion. Infected HeLa cells were incubated with [3H]cholesterol oleate/LDL in the presence of BFA and/or cyclodextrin to inhibit Golgi functions and/or sequester cholesterol at the plasma membrane, respectively. Radioactive cholesterol associated with EBs was monitored. Data are expressed as mean ± SD. (Lower) Representative immunoblots against the MOMP from density gradient-purified EBs used in the analysis of incorporated isotope. Lanes correspond to the samples directly above.
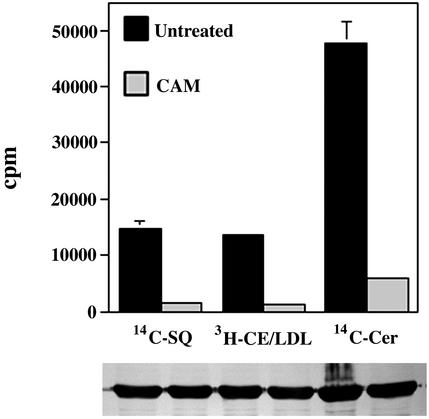
Chlamydial Protein Synthesis Is Required for the Acquisition of Cholesterol. Chlamydial protein synthesis is required for the initiation and maintenance of fusogenicity with sphingomyelin-containing vesicles (7). To determine whether the transport of cholesterol to the chlamydial inclusion is similarly pathogen-directed, chlamydial protein synthesis in mature 24-h inclusions was inhibited by chloramphenicol for 18 h before addition of [14C]squalene, [3H]cholesteryl linoleate/LDL, or [14C]ceramide (Fig. 5). A dramatic decrease in the amount of radioactive cholesterol associated with EBs in the presence of chloramphenicol is observed regardless of whether the cholesterol was synthesized endogenously from squalene or obtained exogenously from LDL. The data suggest that trafficking of cholesterol to the inclusion requires chlamydial protein synthesis for modification of the fusion competence of the inclusion.
Fig. 5.
Acquisition of cholesterol and sphingomyelin requires chlamydial protein synthesis. The amount of radioactive cholesterol and sphingomyelin associated with EBs harvested from untreated and chloramphenicol (CAM)-treated cultures were compared. The significant decrease in radioactivity associated with chloramphenicol-treated EBs indicates an active role of Chlamydia in acquiring cholesterol and sphingomyelin from the host. Data are expressed as mean ± SD. (Lower) Representative immunoblots against the MOMP from density gradient-purified EBs used in the analysis of incorporated isotope. Lanes correspond to the samples directly above.
A similar effect of chloramphenicol on [14C]sphingomyelin trafficking to the chlamydial inclusion was observed. The amount of [14C] sphingomyelin found associated with EBs in the presence of chloramphenicol was significantly lower (15-fold, P < 0.001) when compared with the untreated culture. This result is consistent with previous observations using fluorescent analogs of sphingomyelin (5, 6, 7). Thus the trafficking of both cholesterol and sphingomyelin to the chlamydial inclusion appear to require de novo protein synthesis by chlamydiae.
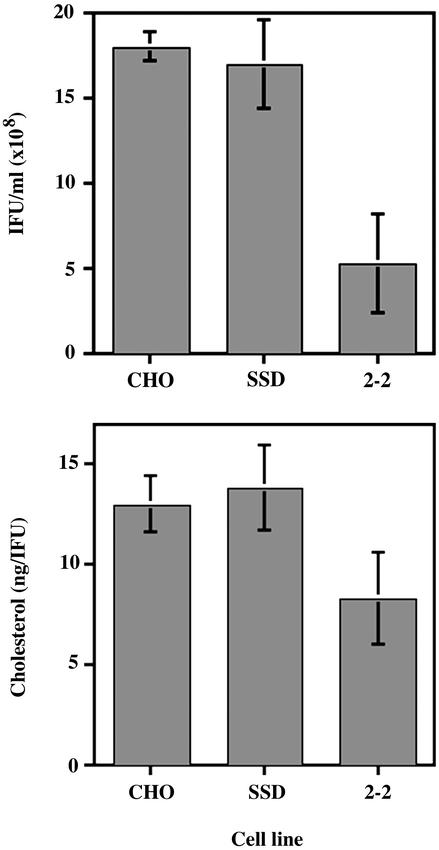
Chlamydial Multiplication in 2–2 and SSD Chinese Hamster Ovary (CHO) Cell Mutants Defective in Cholesterol Transport and Synthesis. To determine the biological significance of cholesterol in chlamydial infection, two mutant CHO cell lines defective in transport, 2–2 (23), or synthesis, SSD (24), of cholesterol were infected and the yield of infectious forming units at 30 h postinfection were compared with that of wild-type CHO-K1 cells (Fig. 6). SSD cells produced approximately the same number of infectious EBs as the wild-type CHO-K1 cells, whereas the 2–2 cells yielded ≈70% fewer EBs than the wild-type or SSD cells. Long-term (30 h) growth in media in which both sources of cholesterol were depleted (2–2 + lovastatin, SSD + lipid-deficient serum) was detrimental to both mutants. Because chlamydiae may acquire cholesterol from either source, the marginal reduction in inclusion-forming unit (IFU) yield was likely due to compensatory activity of the alternate pathway(s) in the 2–2 and SSD mutant cell lines. The presence of cholesterol in EBs harvested from 2–2 or SSD cells was confirmed by cholesterol determination (Fig. 6).
Fig. 6.
Multiplication of C. trachomatis in SSD and 2–2 CHO cell mutants defective in cholesterol synthesis and LDL uptake. (Upper) Infectious progeny EBs from control CHO-K1 cells, SSD, and 2–2 cells lysed and plated for IFUs at 30 h postinfection. (Lower) Relative amount of cholesterol in density gradient-purified EBs from the parent and mutant cell lines. Data are expressed as mean ± SD.
Discussion
The cell walls of Chlamydia trachomatis contain the eukaryotic lipids cholesterol and sphingomyelin (refs. 8, 9, 17, and references therein). Chlamydiae do not encode the enzymatic capacity for synthesis of these lipids (25, 26), which are not typically found in prokaryotes, thus they are likely acquired from the host cell. The transport of endogenously synthesized sphingomyelin from the Golgi apparatus to the chlamydial inclusion has been characterized by using a fluorescent precursor of sphingomyelin, C6-NBD-ceramide (5, 6), and its delivery was confirmed here by using radioactive precursors. Because sphingomyelin and cholesterol are frequently associated in membrane microdomains (27) and may share transport pathways from the Golgi apparatus to the plasma membrane (28), we initiated studies of cholesterol delivery to the chlamydial inclusion to determine whether these lipids might be transported by means of the same mechanisms. C. trachomatis obtained both de novo-synthesized or LDL-derived cholesterol from the host cell. Acquisition of cholesterol from either source was inhibited by BFA and nocodazole. Thus, the same pharmacological inhibitors of Golgi or microtubule function that inhibit sphingomyelin delivery to the inclusion also block cholesterol trafficking, implying that the Golgi apparatus is a necessary intermediate in the transport of cholesterol to the chlamydial inclusion. The ability to acquire sphingomyelin and cholesterol requires chlamydial protein synthesis indicative of a pathogen-directed process and suggests that sphingomyelin and cholesterol may be cotransported to the chlamydial inclusion. Infection of mutant CHO cell lines defective in either the LDL pathways or biosynthetic pathways resulted in little to no reduction in the yield of progeny EBs, although compensatory activity of alternative pathways apparently supplied cholesterol, suggesting that either cholesterol or the activity of this vesicular transport pathway is necessary for chlamydial growth.
The Golgi apparatus has been thought to play only a minor role in transport of cholesterol from the ER to the plasma membrane (21) although a gradient of increasing cholesterol concentration exists through the cis-, medial, and trans-Golgi (29). Transport of de novo-synthesized cholesterol to the plasma membrane is energy- and temperature-dependent and thought to be mediated via lipid-rich transport intermediates (11, 30–32). Inhibitors of glycoprotein export, such as BFA, result in only modest reductions in cholesterol export, suggesting that the majority of cholesterol transported to the plasma membrane bypasses the Golgi apparatus in BFA-treated cells (28). Because of possible compensatory activity of BFA-insensitive pathways in the presence of the inhibitor, it is difficult to estimate the relative activity of the BFA-sensitive and BFA-insensitive pathways in normal cells. Analysis of cholesterol delivery to the chlamydial inclusion, as measured by cholesterol content of purified EBs, provides an organelle target membrane that may aid in the dissection of distinct lipid transport pathways obscured in intact cells.
Sphingolipid synthesis is initiated in the ER, but the transfer of the phosphorylcholine headgroup to ceramide in sphingomyelin synthesis occurs at the cis- or medial Golgi (33). Sphingomyelin transits the trans-Golgi before vesicular transport to the plasma membrane. Cholesterol appears to associate with sphingomyelin in detergent insoluble lipid rafts in the Golgi apparatus before distribution to the plasma membrane (28, 34, 35). Sub-cellular fractionation of cells labeled at 15°C has identified cholesterol primarily in the ER and lipid-rich vesicular fractions, although a percentage was found in the Golgi-enriched fractions (36, 37). Our results provide additional evidence for cholesterol/sphingomyelin association intracellularly (28).
The properties of chlamydial cholesterol acquisition are quite different from that of other bacterial and protozoan parasites. Like C. trachomatis, Toxoplasma gondii occupies a parasitophorous vacuole that is nonfusogenic with endocytic compartments (38–41). T. gondii membranes contain cholesterol although these parasites also lack the enzymatic capacity for de novo sterol synthesis and therefore must acquire it from the host cell. In contrast to C. trachomatis, T. gondii derives cholesterol exclusively from LDL via a pathway that transits host lysosomes, but is Golgi- and microtubule-independent (42). Consistent with the inability to acquire Golgi-derived cholesterol and also distinct from chlamydial inclusions, T. gondii parasitophorous vacuoles do not acquire sphingomyelin endogenously synthesized from ceramide (40). Cholesterol is also found in the parasitophorous vacuolar membrane surrounding intracellular Plasmodium falciparum (43) and Salmonella typhimurium (44); however, the source of cholesterol is believed to be the plasma membrane via a tubulovesicular network or interactions with early endosomes, respectively.
Another chlamydial species, Chlamydia pneumoniae, has been implicated in the etiology of atherosclerosis (45), thus there is interest in possible effects of C. pneumoniae infection on lipid metabolism as a contributing factor in coronary artery disease. C. pneumoniae, or its purified lipopolysaccharide (46), induces cellular oxidation of LDL (47) and foam cell formation in macrophages (48). LDL oxidation may involve indirect mechanisms requiring induction of inflammatory cytokines by macrophages and subsequent uptake by scavenger receptors (49). The relationship of these events to the Golgi-dependent trafficking of cholesterol to the C. trachomatis inclusion described here is unclear. Whether chlamydiae productively infect the relevant cell types to disrupt cholesterol homeostasis and how this may contribute to foam cell formation and LDL oxidation are questions in need of additional study.
The chlamydial inclusion intersects a secretory pathway that intercepts sphingomyelin en route from the Golgi apparatus to the plasma membrane (50). This property is unique to chlamydiae and shared by all species within the genus (3, 5, 20). Delivery of sphingomyelin is directly from the Golgi apparatus (50) and requires chlamydial transcription and translation (7), presumably to modify the inclusion membrane (51). Cholesterol delivery to the C. trachomatis inclusion exhibits similar properties in that it is inhibited by BFA, nocodazole, and reduced temperature and requires chlamydial protein synthesis. These observations support the hypothesis that the chlamydial inclusion intersects on a subset of vesicles containing sphingomyelin and cholesterol from the Golgi apparatus. This pathway does not coincide with the export of model glycoproteins (52). That chlamydial protein synthesis is required for intersection of this pathway suggests that inclusion-membrane-associated proteins may subvert the cellular machinery controlling this poorly understood pathway and thus offers a potential means of identifying requisite host factors.
Supplementary Material
Acknowledgments
We thank Dr. Laura Liscum (Tufts University Medical School, Boston) and Dr. Robert Simoni (Stanford University, Stanford, CA) for their generosity and their gifts of mutant 2-2 and SSD cell lines, respectively; Isabelle Coppens (Yale University, New Haven, CT) for suggestions on LDL purification and labeling; Dr. H. Caldwell, Dr. O. Steele-Mortimer, and members of the Hackstadt lab for reviewing the manuscript; and Janet Sager for technical assistance.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: EB, elementary body; LDL, low-density lipoprotein; BFA, brefeldin A; NBD, 7-nitrobenz-2-oxa-1,3-diazol-4-yl; CHO, Chinese hamster ovary.
References
- 1.Schachter, J. (1999) in Chlamydia: Intracellular Biology, Pathogenesis, and Immunity, ed. Stephens, R. S. (Am. Soc. Microbiol., Washington, DC), pp. 139-169.
- 2.Moulder, J. W. (1991) Microbiol. Rev. 55, 143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinzen, R. A., Scidmore, M. A., Rockey, D. D. & Hackstadt, T. (1996) Infect. Immun. 64, 796-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schramm, N., Bagnell, C. R. & Wyrick, P. B. (1996) Infect. Immun. 64, 1208-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hackstadt, T., Scidmore, M. A. & Rockey, D. D. (1995) Proc. Natl. Acad. Sci. USA 92, 4877-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hackstadt, T., Rockey, D. D., Heinzen, R. A. & Scidmore, M. A. (1996) EMBO J. 15, 964-977. [PMC free article] [PubMed] [Google Scholar]
- 7.Scidmore, M. A., Rockey, D. D., Fischer, E. R., Heinzen, R. A. & Hackstadt, T. (1996) Infect. Immun. 64, 5366-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatch, G. M. & McClarty, G. (1998) Infect. Immun. 66, 3727-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wylie, J. L., Hatch, G. M. & McClarty, G. (1997) J. Bacteriol. 179, 7233-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liscum, L. & Faust, J. R. (1989) J. Biol. Chem. 264, 11796-11806. [PubMed] [Google Scholar]
- 11.Liscum, L. & Dahl, N. K. (1992) J. Lipid Res. 33, 1239-1254. [PubMed] [Google Scholar]
- 12.Liscum, L. & Underwood, K. W. (1995) J. Biol. Chem. 270, 15443-15446. [DOI] [PubMed] [Google Scholar]
- 13.Caldwell, H. D., Kromhout, J. & Schachter, J. (1981) Infect. Immun. 31, 1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folch, J. (1957) J. Biol. Chem. 266, 497-509. [PubMed] [Google Scholar]
- 15.Poumay, Y. & Ronveaux-Dupal, M. F. (1985) J. Lipid Res. 26, 1476-1480. [PubMed] [Google Scholar]
- 16.Neufeld, E. B., Cooney, A. M., Pitha, J., Dawidowicz, E. A., Dwyer, N. K., Pentchev, P. G. & Blanchette-Mackie, E. J. (1996) J. Biol. Chem. 271, 21604-21613. [DOI] [PubMed] [Google Scholar]
- 17.Newhall, W. J. (1988) in Microbiology of Chlamydia, ed. Barron, A. L. (CRC, Boca Raton, FL), pp. 47-70.
- 18.Butler, J. D., Blanchette-Mackie, J., Goldin, E., O'Neill, R. R., Carstea, G., Roff, C. F., Patterson, M. C., Patel, S., Comly, M. E., Cooney, A., et al. (1992) J. Biol. Chem. 267, 23797-23805. [PubMed] [Google Scholar]
- 19.Grupping, A. Y., Cnop, M., Van Schravendijk, C. F., Hannaert, J. C., Van Berkel, T. J. & Pipeleers, D. G. (1997) Endocrinology 138, 4064-4068. [DOI] [PubMed] [Google Scholar]
- 20.Wolf, K. & Hackstadt, T. (2001) Cell Microbiol. 3, 145-152. [DOI] [PubMed] [Google Scholar]
- 21.Liscum, L. & Munn, N. J. (1999) Biochim. Biophys. Acta 1438, 19-37. [DOI] [PubMed] [Google Scholar]
- 22.Underwood, K. W., Andemariam, B., McWilliams, G. L. & Liscum, L. (1996) J. Lipid Res. 37, 1556-1568. [PubMed] [Google Scholar]
- 23.Dahl, N. K., Gutheil, W. G. & Liscum, L. (1993) J. Biol. Chem. 268, 16979-16986. [PubMed] [Google Scholar]
- 24.Bradfute, D. L., Silva, C. J. & Simoni, R. D. (1992) J. Biol. Chem. 267, 18308-18314. [PubMed] [Google Scholar]
- 25.Read, T. D., Brunham, R. C., Shen, C., Gill, S. R., Heidelberg, J. F., White, O., Hickey, E. K., Peterson, J., Utterback, T., Berry, K., et al. (2000) Nucleic Acids Res. 28, 1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens, R. S., Kalman, S., Lammel, C., Fan, J., Marathe, R., Aravind, L., Mitchell, W., Olinger, L., Tatusov, R. L., Zhao, Q., et al. (1998) Science 282, 754-759. [DOI] [PubMed] [Google Scholar]
- 27.Slotte, J. P. (1999) Chem. Phys. Lipids 102, 13-27. [DOI] [PubMed] [Google Scholar]
- 28.Heino, S., Lusa, S., Somerharju, P., Ehnholm, C., Olkkonen, V. M. & Ikonen, E. (2000) Proc. Natl. Acad. Sci. USA 97, 8375-8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coxey, R. A., Pentchev, P. G., Campbell, G. & Blanchette-Mackie, E. J. (1993) J. Lipid Res. 34, 1165-1176. [PubMed] [Google Scholar]
- 30.Puglielli, L., Rigotti, A., Greco, A. V., Santos, M. J. & Nervi, F. (1995) J. Biol. Chem. 270, 18723-18726. [DOI] [PubMed] [Google Scholar]
- 31.Smart, E. J., Ying, Y., Donzell, W. C. & Anderson, R. G. (1996) J. Biol. Chem. 271, 29427-29435. [DOI] [PubMed] [Google Scholar]
- 32.Uittenbogaard, A., Ying, Y. & Smart, E. J. (1998) J. Biol. Chem. 273, 6525-6532. [DOI] [PubMed] [Google Scholar]
- 33.Futerman, A. H., Stieger, B., Hubbard, A. L. & Pagano, R. E. (1990) J. Biol. Chem. 265, 8650-8657. [PubMed] [Google Scholar]
- 34.Keller, P. & Simons, K. (1997) J. Cell Sci. 110, 3001-3009. [DOI] [PubMed] [Google Scholar]
- 35.Simons, K. & Ikonen, E. (2000) Science 290, 1721-1726. [DOI] [PubMed] [Google Scholar]
- 36.Urbani, L. & Simoni, R. D. (1990) J. Biol. Chem. 265, 1919-1923. [PubMed] [Google Scholar]
- 37.Koval, M. & Pagano, R. E. (1991) Biochim. Biophys. Acta 1082, 113-125. [DOI] [PubMed] [Google Scholar]
- 38.Jones, T. C. & Hirsch, J. G. (1972) J. Exp. Med. 136, 1173-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joiner, K. A., Fuhrman, S. A., Miettinen, H. M., Kasper, L. H. & Mellman, I. (1990) Science 249, 641-646. [DOI] [PubMed] [Google Scholar]
- 40.Mordue, D. G., Hakansson, S., Niesman, I. & Sibley, L. D. (1999) Exp. Parasitol. 92, 87-99. [DOI] [PubMed] [Google Scholar]
- 41.Sinai, A. P. & Joiner, K. A. (1997) Annu. Rev. Microbiol. 51, 415-462. [DOI] [PubMed] [Google Scholar]
- 42.Coppens, I., Sinai, A. P. & Joiner, K. A. (2000) J. Cell Biol. 149, 167-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lauer, S., VanWye, J., Harrison, T., McManus, H., Samuel, B. U., Hiller, N. L., Mohandas, N. & Haldar, K. (2000) EMBO J. 19, 3556-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Catron, D. M., Sylvester, M. D., Lange, Y., Kadekoppala, M., Jones, B. D., Monack, D. M., Falkow, S. & Haldar, K. (2002) Cell Microbiol. 4, 315-328. [DOI] [PubMed] [Google Scholar]
- 45.Byrne, G. I. & Kalayoglu, M. V. (1999) Am. Heart J. 138, S488-S490. [DOI] [PubMed] [Google Scholar]
- 46.Kalayoglu, M. V. & Byrne, G. I. (1998) Infect. Immun. 66, 5067-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalayoglu, M. V., Hoerneman, B., LaVerda, D., Morrison, S. G., Morrison, R. P. & Byrne, G. I. (1999) J. Infect. Dis. 180, 780-790. [DOI] [PubMed] [Google Scholar]
- 48.Kalayoglu, M. V. & Byrne, G. I. (1998) J. Infect. Dis. 177, 725-729. [DOI] [PubMed] [Google Scholar]
- 49.Netea, M. G., Dinarello, C. A., Kullberg, B. J., Jansen, T., Jacobs, L., Stalenhoef, A. F. & Van Der Meer, J. W. (2000) J. Infect. Dis. 181, 1868-1870. [DOI] [PubMed] [Google Scholar]
- 50.Hackstadt, T., Fischer, E. R., Scidmore, M. A., Rockey, D. D. & Heinzen, R. A. (1997) Trends Microbiol. 5, 288-293. [DOI] [PubMed] [Google Scholar]
- 51.Hackstadt, T. (2000) Traffic 1, 93-99. [DOI] [PubMed] [Google Scholar]
- 52.Scidmore, M. A., Fischer, E. R. & Hackstadt, T. (1996) J. Cell Biol. 134, 363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.