Abstract
In plants, the formation of isopentenyl diphosphate and dimethylallyl diphosphate, the central intermediates in the biosynthesis of isoprenoids, is compartmentalized: the mevalonate (MVA) pathway, which is localized to the cytosol, is responsible for the synthesis of sterols, certain sesquiterpenes, and the side chain of ubiquinone; in contrast, the recently discovered MVA-independent pathway, which operates in plastids, is involved in providing the precursors for monoterpenes, certain sesquiterpenes, diterpenes, carotenoids, and the side chains of chlorophylls and plastoquinone. Specific inhibitors of the MVA pathway (lovastatin) and the MVA-independent pathway (fosmidomycin) were used to perturb biosynthetic flux in Arabidopsis thaliana seedlings. The interaction between both pathways was studied at the transcriptional level by using GeneChip (Affymetrix) microarrays and at the metabolite level by assaying chlorophylls, carotenoids, and sterols. Treatment of seedlings with lovastatin resulted in a transient decrease in sterol levels and a transient increase in carotenoid as well as chlorophyll levels. After the initial drop, sterol amounts in lovastatin-treated seedlings recovered to levels above controls. As a response to fosmidomycin treatment, a transient increase in sterol levels was observed, whereas chlorophyll and carotenoid amounts decreased dramatically when compared with controls. At 96 h after fosmidomycin addition, the levels of all metabolites assayed (sterols, chlorophylls, and carotenoids) were substantially lower than in controls. Interestingly, these inhibitor-mediated changes were not reflected in altered gene expression levels of the genes involved in sterol, chlorophyll, and carotenoid metabolism. The lack of correlation between gene expression patterns and the accumulation of isoprenoid metabolites indicates that posttranscriptional processes may play an important role in regulating flux through isoprenoid metabolic pathways.
Keywords: 1-deoxy-d-xylulose 5-phosphate, fosmidomycin, lovastatin, mevalonate
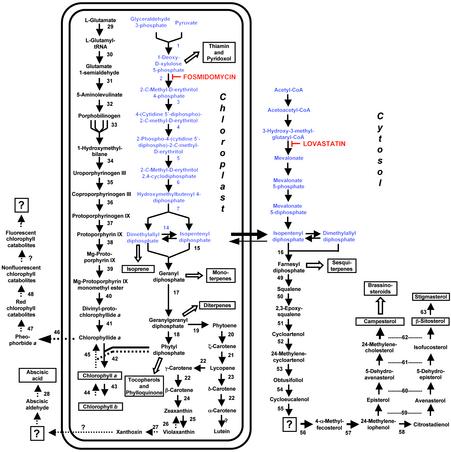
The isoprenoids, which constitute the most diverse group of natural products, serve numerous biochemical functions in plants. They play important roles as quinones in electron transport chains, as components of membranes (sterols), in subcellular targeting and regulation (prenylation of proteins), as photosynthetic pigments (carotenoids, side chain of chlorophyll), as hormones (gibberellins, brassinosteroids, abscisic acid, cytokinins), and as plant defense compounds as well as attractants for pollinators (monoterpenes, sesquiterpenes, and diterpenes) (1). Isoprenoids are synthesized ubiquitously among prokaryotes and eukaryotes through condensation of the five-carbon intermediates isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) (2). In higher plants, two distinct biosynthetic routes to IPP and DMAPP exist (Fig. 1). The cytosolic pathway, which starts from acetyl-CoA and proceeds through the intermediate mevalonate (MVA), provides the precursors for sterols and ubiquinone (3). The plastidial MVA-independent pathway, which involves a condensation of pyruvate and glyceraldehyde-3-phosphate via 1-deoxy-D-xylulose 5-phosphate as a first intermediate, is used for the synthesis of isoprene, carotenoids, abscisic acid, and the side chains of chlorophylls and plastoquinone (4–8). Although this subcellular compartmentation allows both pathways to operate independently in plants, there is evidence that they cooperate in the biosynthesis of certain metabolites. For example, the chamomile sesquiterpenes are composed of two C5 isoprenoid units formed via the MVA-independent pathway, with a third unit being derived from the MVA-independent pathway (9). An interaction of both pathways has also been reported for monoterpene and sesquiterpene volatiles emitted by lima beans (10).
Fig. 1.
Overview of isoprenoid metabolic pathways localized to the cytosol and to plastids in plants, with an emphasis on the metabolism of chlorophylls, carotenoids, and sterols. The symbol for anabolic reactions is a solid arrow, and catabolic reactions are indicated by arrows with dotted lines. Open arrows depict multiple enzymatic steps, and key metabolites are boxed. Known enzymes involved in isoprenoid metabolism are numbered, and question marks indicate steps for which an enzymatic activity has not yet been demonstrated. Evidence for the branching of the plastidial MVA-independent pathway to yield IPP and dimethylallyl diphosphate independently has recently been reported (27). The following enzymes are represented: 1, 1-deoxy-d-xylulose 5-phosphate synthase; 2, DXR; 3, 2-C-methyl-d-erythritol 4-phosphate cytidyltransferase; 4, 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol kinase; 5, 2,4-C-methyl-d-erythritol cyclodiphosphate synthase; 6, 1-hydroxy-2-methyl-2-(E)-butenyl-4-phosphate synthase; 7, 1-hydroxy-2-methyl-2-(E)-butenyl-4-phosphate reductase; 8, acetoacetyl-CoA thiolase; 9, 3-hydroxy-3-methylglutaryl-CoA synthase; 10, HMGR; 11, MVA kinase; 12, phospho-MVA kinase; 13, MVA diphosphate decarboxylase; 14, IPP: dimethylallyl diphosphate isomerase; 15, geranyl diphosphate synthase; 16, farnesyl diphosphate synthase; 17, geranylgeranyl diphosphate synthase; 18, geranylgeranyl reductase; 19, phytoene synthase; 20, phytoene desaturase; 21, ζ-carotene desaturase; 22, lycopene β-cyclase; 23, lycopene ε-cyclase; 24, β-carotene hydroxylase; 25, zeaxanthin epoxidase; 26, violaxanthin de-epoxidase; 27, epoxycarotenoid (neoxanthin) cleavage enzyme; 28, abscisic aldehyde oxidase; 29, glutamyl tRNA synthetase; 30, glutamyl tRNA reductase; 31, glutamate 1-semialdehyde aminotransferase; 32, aminolevulinate dehydratase; 33, porphobilinogen deaminase; 34, uroporphyrinogen synthase; 35, uroporphyrinogen decarboxylase; 36, coproporphyrinogen III oxidase; 37, protoporphyrinogen IX oxidase; 38, Mg-protoporphyrinogen IX chelatase; 39, Mg-protoporphyrinogen IX methyltransferase; 40, protoporphyrinogen cyclase (enzyme activity not yet characterized in plants); 41, protochlorophyllide reductase; 42, chlorophyll synthetase; 43, chlorophyll a oxygenase; 44, chlorophyll b reductase (gene not cloned yet); 45, chlorophyllase; 46, de-chelatase (gene not cloned yet); 47, pheophorbide a oxygenase (gene not yet cloned); 48, red chlorophyll catabolite reductase; 49, squalene synthase; 50, squalene monooxygenase; 51, cycloartenol synthase; 52, cycloartenol C24 methyltransferase; 53, 24-methylenecycloartenol C-4 methyl oxidase; 54, cycloeucalenol cycloisomerase; 55, sterol C14 reductase; 56, obtusifoliol 14-demethylase; 57, C-8,7 sterol isomerase; 58, sterol C-methyltransferase 2; 59, sterol C-4 methyl oxidase; 60, sterol C5-desaturase; 61, sterol Δ7 reductase; 62, sterol C24-reductase; and 63, sterol C22-desaturase.
Here we report the interactions between the cytosolic and the plastidial pathways of isoprenoid biosynthesis in Arabidopsis thaliana as probed by treatment with either an inhibitor of the MVA pathway (lovastatin; ref. 11) or the MVA-independent pathway (fosmidomycin; ref. 12). After inhibitor addition, key metabolites of the plastidial pathway (carotenoids and chlorophylls) and the cytosolic pathway (sterols) were assayed, and the expression patterns of >8,200 genes were monitored by using oligonucleotide microarray technology. The dramatic inhibitor-mediated changes in end-product levels were not reflected in changes in gene expression levels of the biosynthetic enzymes, thus indicating that posttranscriptional events play a major role in the regulation of these pathways.
Materials and Methods
Plant Material, Growth Conditions, and Application of Inhibitors. WT A. thaliana seeds (ecotype Columbia) were surface-sterilized and sown on filter paper (Whatman) saturated with Murashige and Skoog (MS) medium (13) in Petri dishes. The Petri dishes were preincubated at 4°C in the dark for 2 d, and seedlings were grown in a growth chamber at 23°C with a 12-h photoperiod provided by white fluorescent light (150 μEm-2·s-1). The inhibitors were stored as 5 mM stock solutions; lovastatin (A.G. Scientific, San Diego) in ethanol at 4°C and fosmidomycin (Molecular Probes) in water at -20°C. Lovastatin is an inactive lactone; at physiological pH values in the cytosol, the lactone form of lovastatin gradually enters the cells and is slowly converted into the biologically active β-hydroxy-acid form. Media containing the inhibitors were prepared by adding aliquots from the stock solution to a final concentration of 50–100 μM (lovastatin) or 50 μM to 10 mM (fosmidomycin). After germination, seedlings were grown for 5 d on filter disks saturated with MS medium and then transferred either to fresh MS medium (controls) or to MS medium containing the respective inhibitor. Pooled seedling samples were harvested at 3, 12, 48, and 96 h after inhibitor addition, frozen in liquid nitrogen, and stored at -80°C until further analysis. Metabolite assays (carotenoids, chlorophylls, and sterols) were performed with tissue from at least three independent experiments, whereas pooled seedling samples from at least two independent experiments were used to extract RNA for microarrays and RT-PCR.
Extraction and Analysis of Chlorophylls and Carotenoids. Frozen tissue samples were homogenized in 1.5-ml Eppendorf reaction vials by using polypropylene pestles, and chlorophylls and carotenoids were extracted with methanol (500 μl per 20 mg of frozen tissue) at 4°C in dim light. After centrifugation (10,000 × g, Eppendorf bench-top centrifuge) and filtration (4-mm syringe filters, Whatman), chlorophylls and carotenoids were separated by capillary diode-array HPLC (Agilent, Wilmington, DE, 1100 series) with a reverse-phase C18 column [ZORBAX (Agilent) SB-C18; 5 μm; 0.5 × 150 mm]. The solvent system consisted of methanol/ water (3:1, vol/vol) containing 10 mM ammonium acetate (solvent A) and methanol/dichloromethane (4:1, vol/vol) (solvent B). A linear gradient from solvent A (100%) to solvent B (80%) was applied over a period of 5 min with a flow-rate of 20 μl·min-1. This solvent composition was held for 7 min, solvent B was increased to 100% within 2 min, and metabolites were separated isocratically for 11 min. Eluting compounds were monitored using a diode-array detector at 430, 520, and 650 nm. Compounds were identified and quantified by comparison with corresponding reference samples. The purity of eluting HPLC peaks was examined by recording UV/VIS and mass spectra (data not shown). Retention times were as follows: xanthophylls (violaxanthin, zeaxanthin, lutein), 8.4–9.9 min; chlorophyll a, 14.3 min; chlorophyll b, 17.7 min; and β-carotene, 21.6 min.
Extraction and Analysis of Sterols. Seedling samples were extracted according to Bligh and Dyer (14), with some modifications. Briefly, the seedlings were incubated in CHCl3/MeOH (1:2, vol/vol) for 30 min at 70°C. The extract was then transferred to a new vial, and the seedlings were extracted again with CHCl3 by vortexing for 30 s. Both extracts were pooled, the solvents were evaporated, and the dried residue was saponified with 6% (wt/vol) KOH in methanol at 90°C for 1 h to release the sterol moiety of steryl esters. Sterols were then extracted with 1.5 volumes of n-hexane and evaporated to dryness under N2. The dried residue was redissolved in n-hexane, and the sterols were analyzed by gas chromatography (Agilent 6890 Series) equipped with a flame ionization detector and a bonded-phase GC column developed for the analysis of underivatized sterols (SAC-5 capillary column, 30 m × 0.25 mm, ID 0.25 μm, Supelco) by using helium as carrier gas (2 ml·min-1) and split injection. Separation was carried out at 300°C with a run time of 12 min. Compounds were identified and quantified by comparison with corresponding reference samples (GC-MS; data not shown). Retention times: epi-cholesterol (internal standard), 4.4 min; campesterol, 5.4 min; stigmasterol, 5.6 min; and β-sitosterol, 6.2 min.
RNA Isolation, cRNA Synthesis, Microarray Hybridization, and RT-PCR. For A. thaliana GeneChip (Affymetrix) experiments and RT-PCR assays, mRNA was extracted by using Qiagen (Chatsworth, CA) RNeasy columns according to the manufacturer's instructions. Subsequent cRNA synthesis for microarray hybridization, cDNA synthesis for quantitative PCR, and data analysis were performed as described (15). RT-PCR reactions were monitored by using an ABI Prism 7700 Sequence Detection System with the SYBR green PCR Mastermix (Applied Biosystems, P/N 4309155). For RT-PCR genes encoding 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR), 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were PCR-amplified in separate tubes using the same amount of cDNA retrieved from the same sample as described by the kit manufacturer (Applied Biotechnology). The following primers were used: HMGR (HMG1 gene; forward primer: TTCCATCGTACTCGCTTGAATCT; reverse primer: CGCCTCACGACGAATCG), DXR (forward primer: TCATCTGTGCTTGCTCAATTGG; reverse primer: ATCGGGCCATGACATGGT) and GAPDH (forward primer: CTCCCTTGGAAGGAGCTAGG; reverse primer: TTCTTGGCACCAGCTTCAAT). Thermal cycling consisted of an initial step at 50°C for 2 min and denaturation for 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. The adjustment of baseline and threshold was performed according to the manufacturer's instructions. The relative abundance of HMG1 and DXR transcripts was normalized to the constitutive expression level of glyceraldehyde 3-phosphate dehydrogenase mRNA. The data were analyzed by using the comparative threshold cycle method as described in the Applied Biotechnology User Bulletin.
Results and Discussion
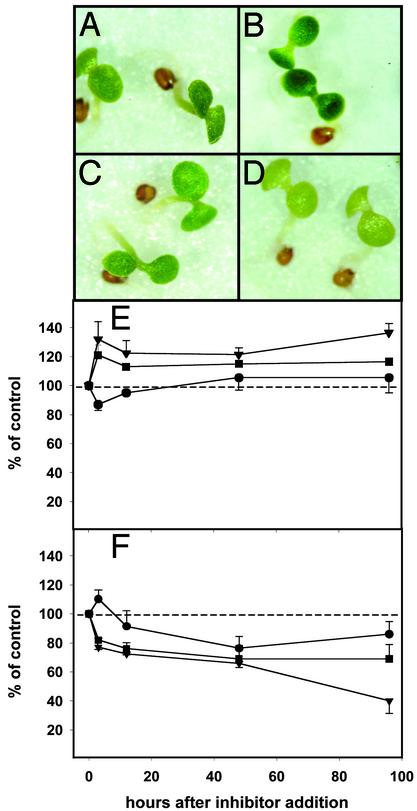
Experimental Design and Phenotyping of Inhibitor-Mediated Responses. An A. thaliana-based model system was developed to evaluate the effects of flux perturbations, caused by subjecting seedlings to inhibitors of isoprenoid biosynthesis, on global gene expression patterns and the accumulation of isoprenoid metabolites. A. thaliana seedlings were precultivated on filter disks saturated with MS medium for 5 d under a 12-h photoperiod. Then, parallel seedling populations were either grown on MS medium in the presence of inhibitor (lovastatin or fosmidomycin) or kept on MS medium (controls) before being harvested 48 h after inhibitor addition. Inhibition of the cytosolic MVA pathway enzyme HMGR, by cultivation of seedlings in media containing lovastatin (concentration range 50–100 μM), led to a visible change toward a dark green color at 100 μM within 48 h (Fig. 2 A and B). This color change was confirmed by an increase in absorption of methanolic extracts at wavelength ranges characteristic of carotenoids (370–520 nm) and chlorophylls (430–460 and 640–670 nm) (ref. 16 and data not shown). These results provided evidence for an increased flux through the plastidial MVA-independent pathway, resulting in an increased production, or alternatively reduced catabolism, of carotenoids and chlorophylls as a response to inhibition of the cytosolic MVA pathway. An increased accumulation of the carotenoid lycopene has been reported for lovastatin-treated tomato fruits (17), indicating that crosstalk between the cytosolic and the plastidial pathway of isoprenoid biosynthesis may be a common phenomenon in plants. Treatment of seedlings with fosmidomycin (concentration range 50 μM to 10 mM), an inhibitor of the plastidial MVA-independent pathway DXR, resulted in severe leaf bleaching within 48 h when fosmidomycin concentrations >100 μM were used (Fig. 2 C and D). An analysis of methanolic extracts from fosmidomycin-treated seedlings indicated a reduction of absorbance compared with controls at the carotenoid- and chlorophyll-specific wavelengths mentioned above (data not shown). These intriguing initial results encouraged us to launch an in-depth study of crosstalk between the cytosolic and the plastidial pathway at the metabolite and gene expression levels.
Fig. 2.
Effects of inhibitor treatments on A. thaliana seedlings; phenotypic characteristics of control seedlings for lovastatin experiments (A); lovastatin-treated seedlings (B); control seedlings for fosmidomycin experiments (C); fosmidomycin-treated seedlings (D); levels of sterols (•), chlorophylls (▾), and carotenoids (▪) (± standard deviation) in the presence of lovastatin, and fosmidomycin (E and F). Values were calculated on 1 μg/g fresh-weight basis and were normalized (100% for mock control). Fresh weight per seedling did not change significantly with the various treatments.
Fosmidomycin and Lovastatin Exert Opposite Effects on Chlorophyll, Carotenoid, and Sterol Levels. To refine the effects of pathway flux perturbations on specific isoprenoid end-product classes, the inhibitor mediated changes of major metabolites derived from the MVA pathway (campesterol, β-sitosterol, and stigmasterol) and the MVA-independent pathway (chlorophyll a, chlorophyll b, and the carotenoids β-carotene, lutein, zeaxanthin, and violaxanthin) and were monitored by GC and HPLC, respectively. Because the individual representatives for each compound class exhibited a very similar time course under a variety of experimental conditions (data not shown), only the total content of carotenoids, chlorophylls, and sterols was used to calculate the changes caused by inhibitor treatments.
In the presence of lovastatin (100 μM), an inhibitor of the cytosolic MVA pathway, the levels of MVA-derived sterols decreased rapidly within the first 3 h after inhibitor addition to 87 ± 4% of controls (Fig. 2E). During this time period (phase I), a pronounced inhibitor-mediated increase in the levels of carotenoids (121 ± 7% of controls) and chlorophylls (132 ± 12% of controls), which are both synthesized via the plastidial MVA-independent pathway, was observed. Between 3 and 12 h after inhibitor addition, carotenoid and chlorophyll levels dropped to 113 ± 3% and 123 ± 7% of controls, respectively. Subsequently (phase II), the carotenoid levels increased slowly to 116 ± 1% of controls by 96 h, whereas chlorophyll levels remained constant until 48 h after inhibitor addition (122 ± 5% of controls), before rising again to 137 ± 7% of controls at 96 h. After the initial drop at 3 h (phase I), sterol levels recovered to 106 ± 9% of controls at 48 h and remained constant through 96 h (phase II).
Within the first 3 h after A. thaliana seedlings were subjected to fosmidomycin (400 μM), an inhibitor of the plastidial MVA-independent pathway, a dramatic drop in the levels of carotenoids (82 ± 1% of controls) and chlorophylls (77 ± 1% of controls) was detected (Fig. 2F; phase I). In contrast, the total amount of cytosolic sterols increased to 110 ± 6% above controls. The carotenoid and chlorophyll levels in fosmidomycin-treated seedlings continued to drop, although at a slower rate, until 48 h after inhibitor addition (carotenoids: 69 ± 9% of controls; chlorophylls: 40 ± 6% of controls). During the same time period (phase II), the sterol levels decreased more rapidly to 76 ± 7% of controls. At 96 h after inhibitor addition, the carotenoid levels had stayed constant at 69 ± 10% of controls, the chlorophyll levels had dropped to 40 ± 6% of controls, and the sterol levels had increased to 86 ± 8% of controls.
Both inhibitor treatments caused a very rapid response of A. thaliana seedlings, as evidenced by the changes in the steady-state levels of isoprenoid metabolites during phase I. The biphasic inhibitor-mediated changes in metabolite levels suggest that separate regulatory mechanisms may exist for short- and midterm responses to inhibitor treatment. Furthermore, the divergent time courses of cytosolic and plastidial metabolites indicate that A. thaliana seedlings are able to synchronize changes in metabolite pools in different subcellular compartments. At 96 h after inhibitor addition, the levels of cytosolic as well as plastidial metabolites were above control levels in lovastatin-treated seedlings, whereas treatment with fosmidomycin resulted in a substantial decrease of the levels of all assayed metabolites. Thus, the ability to compensate for metabolic imbalances appears to be more effective for the cytosolic pathway of isoprenoid biosynthesis than for the plastidial pathway, indicating that metabolite sharing between both pathways may be a unidirectional process (export from the plastid, not import from the cytosol) under the conditions tested.
Inhibitor-Mediated Changes in the Expression of Genes Involved in Isoprenoid Metabolism. After the profiles for isoprenoid metabolites as a response to inhibitor treatments had been established, we examined the expression of known structural and regulatory genes involved in A. thaliana isoprenoid biosynthesis. To characterize global gene expression patterns specifically regulated in response to lovastatin and fosmidomycin treatments, we used oligonucelotide microarrays covering >8,200 genes, thus representing ≈one-third of the genome (15). To ensure that inhibitor-mediated changes in gene expression patterns could be correlated objectively with changes in metabolite profiles, RNA and metabolites were extracted from the same tissue samples. cRNA probes representing RNA from populations of either control or inhibitor-treated seedlings were hybridized individually to separate oligonucleotide chips, and the hybridization signal intensity was compared for each set of corresponding control and inhibitor-treated samples. After data normalization (15), genes were identified as being regulated by inhibitor treatment if their expression level exceeded the baseline of 35 and if the expression changed at least 2-fold when compared with the expression levels in the appropriate control samples.
As a response to lovastatin (100 μM) treatment, the expression of 329 genes changed within the first 3 h, the expression of 187 genes was changed at 12 h, and the expression levels of 169 genes differed significantly from controls at 48 h after inhibitor addition (see Fig. 4, which is published as supporting information on the PNAS web site, www.pnas.org). After fosmidomycin (400 μM) treatment, 156 genes showed a change in expression levels compared with controls at 3 h, the expression levels of 163 genes were found to be different from controls at 12 h, and at 48 h, the expression levels of 659 genes were either up- or down-regulated when compared with controls (see Fig. 4).
To assess the effect of the inhibitor treatments on the expression levels of isoprenoid metabolic genes, key word and sequence-based searches (18) were used to generate a database representing all known genes coding for plant enzymes involved in the metabolism of carotenoids, chlorophylls, and sterols. Sequence similarity searches at the Institute for Genomic Research A. thaliana genome database (www.tigr.org./tdb/e2k1/ath1/) revealed the likely A. thaliana homologues. In general, relatively high levels of constitutive gene expression (mean of controls for the lovastatin and fosmidomycin experiments >300 at all time points with average expression level set to 100) were detected for enzymes at pathway branchpoints (e.g., 1-deoxy-D-xyulose 5-phosphate synthase, IPP isomerase, geranylgeranyl diphosphate synthase, geranylgeranyl reductase, squalene synthase, and squalene monooxygenase) (Table 1). Interestingly, within several gene families (e.g., those encoding HMGR, geranylgeranyl diphosphate synthase, and squalene synthase), only one gene was expressed consistently at levels above the baseline, whereas within other gene families (e.g., those encoding IPP isomerase, glutamyl tRNA reductase, and squalene monooxygenase), two or more isogenes showed expression levels above background. After lovastatin treatment, the expression of genes for epoxycarotenoid cleavage enzyme and chlorophyllase, enzymes involved in the catabolism of carotenoids and chlorophylls, respectively, were found to increase slightly above control levels, whereas the gene coding for protochlorophyllide reductase, an enzyme involved in the late steps of chlorophyll biosynthesis, showed a moderate decrease in expression when compared with control levels. The expression of all other genes known to encode enzymes involved in isoprenoid metabolism did not change significantly compared with control levels. In fosmidomycin-treated seedlings, two isogenes encoding squalene monooxygenase increased above control levels, whereas the expression levels of all other genes coding for enzymes involved in isoprenoid metabolism remained unchanged.
Table 1. Expression patterns of genes encoding enzymes involved in the metabolism of isoprenoids (carotenoids, chlorophylls, and sterols) in A. thaliana seedlings.
| Expression levels in controls, h
|
Fold change lovastatin, h
|
Fold change fosmidomycin, h
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | 3 | 12 | 48 | 3 | 12 | 48 | 3 | 12 | 48 | Fold change CLA1:WT |
| 1 | 496 | 400 | 467 | 1.08 | 1.16 | 1.06 | 1.00 | 0.85 | 1.76 | n.d. |
| 2 | 263 | 214 | 229 | 0.71 | 1.11 | 0.97 | 1.34 | 1.54 | 0.71 | 0.15 |
| 3 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| 4 | 58 | 75 | 72 | 0.53 | 1.08 | 0.85 | 1.35 | 1.42 | n.d. | 0.2 |
| 5 | 336 | 193 | 576 | 0.97 | 1.16 | 1.03 | 1.09 | 0.94 | 1.30 | 1.42 |
| 10 | 44 | 47 | 50 | 0.96 | n.d. | 0.71 | n.d. | 0.86 | 0.89 | 2.52 |
| 10 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 6.81 |
| 11 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 1.19 | 1.17 | n.d. | 0.81 |
| 14 | 140 | 138 | 107 | 0.91 | 1.09 | 0.99 | 1.11 | 1.06 | 0.89 | 0.67 |
| 14 | 387 | 434 | 351 | 1.06 | 1.01 | 1.15 | 1.44 | 1.09 | 1.11 | 0.56 |
| 15 | 78 | n.d. | 93 | 1.23 | 1.08 | 0.86 | 0.76 | n.d. | 0.95 | 1.48 |
| 16 | 78 | 69 | 50 | 0.82 | 0.77 | 1.04 | 1.15 | 1.22 | 0.77 | 0.32 |
| 17 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 1.49 |
| 17 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 0.66 |
| 17 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| 17 | 347 | 322 | 367 | 0.92 | 0.99 | 0.97 | 0.91 | 0.92 | 0.97 | 1 |
| 17 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| 17 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| 18 | 1158 | 950 | 1595 | 1.11 | 1.22 | 1.01 | 1.02 | 0.88 | 1.20 | 0.33 |
| 19 | 168 | 147 | 242 | 1.41 | 1.16 | 1.07 | 1.15 | 0.99 | 0.82 | 0.21 |
| 20 | 122 | 121 | 159 | 0.88 | 1.20 | 1.02 | 1.27 | 1.09 | 1.04 | 0.73 |
| 21 | 187 | 214 | 304 | 1.12 | 1.32 | 0.88 | 0.97 | 0.82 | 1.16 | 0.49 |
| 22 | 267 | 271 | 402 | 1.07 | 1.04 | 1.03 | 1.14 | 0.89 | 1.57 | 0.69 |
| 23 | 123 | 89 | 148 | 0.67 | 1.56 | 0.89 | 1.47 | 0.96 | 0.78 | 0.23 |
| 24 | 236 | 154 | 481 | 0.96 | 1.33 | 1.29 | 1.06 | 1.55 | 1.06 | 0.4 |
| 26 | 82 | 121 | 130 | 0.66 | 0.81 | 1.30 | 1.37 | 0.96 | 0.96 | 1.19 |
| 27 | 289 | 262 | 528 | 0.71 | 1.44 | 1.04 | 1.11 | 1.25 | 1.02 | 2.95 |
| 27 | 63 | n.d. | n.d. | 2.17 | n.d. | 1.62 | n.d. | n.d. | n.d. | 0.57 |
| 29 | 173 | 169 | 139 | 0.86 | 0.84 | 0.91 | 0.86 | 0.94 | 0.78 | 0.84 |
| 30 | 44 | 81 | 54 | n.d. | 0.84 | 1.24 | 0.95 | 1.00 | 0.83 | 4.12 |
| 30 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| 30 | 256 | 160 | 377 | 1.00 | 0.88 | 1.04 | 0.81 | 1.12 | 1.87 | 0.89 |
| 31 | 218 | 251 | 215 | 1.02 | 1.18 | 0.70 | 1.20 | 0.82 | 0.62 | 0.17 |
| 35 | 110 | 185 | 109 | 0.82 | 1.05 | 0.89 | 1.00 | 1.06 | 0.85 | 0.52 |
| 36 | 310 | 299 | 420 | 0.79 | 1.01 | 0.91 | 1.51 | 1.10 | 0.77 | 0.33 |
| 41 | 385 | 157 | 365 | 0.48 | 0.94 | n.d. | 0.79 | 1.09 | 0.55 | 0.28 |
| 42 | 167 | 161 | 267 | 0.91 | 1.09 | 1.03 | 1.19 | 1.02 | 0.73 | 0.17 |
| 44 | n.d. | 57 | 50 | 2.07 | 1.34 | 1.26 | n.d. | n.d. | 1.53 | 0.03 |
| 46 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 0.95 | n.d. | n.d. | 1.16 |
| 46 | 315 | 277 | 358 | 1.38 | 0.91 | 0.97 | 0.83 | 0.71 | 0.93 | 1.33 |
| 47 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 1.05 |
| 47 | 232 | n.d. | 370 | 1.70 | 1.46 | 1.45 | 1.45 | n.d. | 2.14 | 0.06 |
| 47 | 269 | 134 | 444 | 1.06 | 1.38 | 1.26 | 0.84 | 0.75 | 2.06 | 1.22 |
| 48 | 105 | 67 | 93 | 0.74 | 0.96 | 0.90 | 0.64 | 0.90 | 0.58 | 1.04 |
| ? | 292 | 274 | 174 | 0.67 | 0.67 | 0.53 | 0.96 | 0.96 | 1.21 | 0.47 |
| ? | n.d. | 110 | n.d. | 0.55 | n.d. | 0.61 | n.d. | 0.86 | n.d. | 0.78 |
| 57 | 115 | 109 | 89 | 1.08 | 1.08 | 0.92 | 1.21 | 1.00 | 1.21 | 0.46 |
| ? | 109 | 105 | 87 | 0.96 | 1.01 | 1.05 | 1.12 | 0.97 | 0.84 | 0.09 |
| 58 | 150 | 172 | 115 | 0.78 | 0.76 | 0.88 | 1.10 | 1.27 | 0.80 | 0.76 |
n.d., not detectable (expression level below detection limit). The gene numbers refer to the encoded enzymes as depicted in Fig. 1. The “fold change” was calculated by dividing the expression level of a gene in an inhibitor-treated seedling population (or in the case of the cla1 mutant, of a mutant seedling population) by the expression level of the gene in a control population.
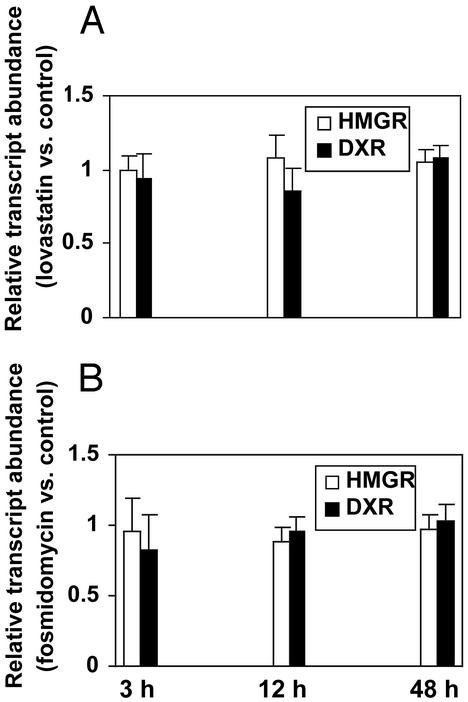
In addition to the utilization of GeneChips, RT-PCR was used as an independent method for the analysis of gene expression patterns for selected genes. The lovastatin (100 μM) and fosmidomycin (400 μM) treatments of A. thaliana seedlings were repeated, and tissue of treated as well as control seedling populations was harvested at 3, 12, and 48 h. The expression level of the DXR and HMGR (HMG1) genes in inhibitor-treated seedlings did not differ significantly from the abundance of these transcripts in the corresponding control seedlings (Fig. 3), thus confirming the gene expression patterns obtained using GeneChip technology. This striking lack of correlation between the isoprenoid metabolite levels, which changed rapidly as a response to the inhibitor treatments, and the expression levels of the genes encoding metabolic enzymes, which did not show changes when subjected to the inhibitors, indicates that inhibitor-mediated alterations in flux through the cytosolic and plastidial pathways of isoprenoid metabolism are not transcriptionally regulated. Thus, rapid changes of the enzymatic activities involved in isoprenoid metabolism, e.g., by increased translation, posttranslational enzyme modification, or other posttranscriptional regulatory processes, appear to play an important role in modulating flux, especially during the first few hours after inhibitor addition.
Fig. 3.
Effects of lovastatin (A) and fosmidomycin (B) treatments on the expression of the genes encoding HMGR (HMG1 gene) and DXR (± standard deviation) in A. thaliana seedlings as indicated by RT-PCR analysis.
One protein modification commonly affecting enzyme activity includes phosphorylation (19, 20, 21). In the presence of lovastatin, the expression levels of 60 protein kinase genes differed from controls, 49 of which (82%) were up- or down-regulated as early as 3 h after inhibitor addition. In fosmidomycin-treated seedlings, the expression levels of 58 protein kinase genes changed compared with controls, with 26 of the genes (45%) showing altered expression patterns at3hafterinhibitor addition (see Fig. 5, which is published as supporting information on the PNAS web site).
In chloroplasts, flux through metabolic pathways is dramatically redirected during day/night transitions. The ferredoxin/ thioredoxin system plays a key role in regulating the activity of several plastidial enzymes in response to changes in thylakoid redox state (22). Using affinity column chromatography with a subsequent proteomic detection of thioredoxin-interacting proteins, three enzymes of the MVA-independent pathway and four enzymes involved in chlorophyll biosynthesis have been identified as previously unrecognized thioredoxin f targets in stromal extracts of spinach (23). Redox regulation may therefore mediate the activity of enzymes involved in plastidial pathways, including isoprenoid metabolism.
Besides posttranslational modifications of biosynthetic enzymes, flux through pathways of isoprenoid metabolism may also be regulated by precursor availability. Inhibition of the cytosolic MVA pathway enzyme HMGR by lovastatin treatment caused only a transient reduction of sterol levels, indicating that the plastidial MVA-independent pathway may compensate for the lack of cytosolic IPP needed for the biosynthesis of cytosolic sterols. Such a crosstalk between cytosolic and plastidial pathways of isoprenoid biosynthesis has also been discussed to occur in Ginkgo biloba (24). Only a limited number of putative transporters of the plastidial envelope membrane are represented on the A. thaliana oligonucleotide microarray and, based on the expression characteristics, the dataset presented here does not reveal a promising candidate gene for a transporter responsible for the exchange of isoprenoid intermediates across the plastidial envelope membrane (see Fig. 6, which is published as supporting information on the PNAS web site). Nonetheless, our data are highly suggestive of the existence of a mechanism for trafficking of isoprenoid intermediates from the plastid to the cytosol, which could be mediated by a specific, as-yet-undiscovered metabolite transporter. Our future efforts will be focused on the characterization of this transport system and on efforts to obtain global metabolite profiles that may help to identify the processes involved in modulating flux through pathways of isoprenoid biosynthesis.
Gene Expression Patterns in a Mutant Impaired in the MVA-Independent Pathway. The albino mutant cla1 was isolated from an A. thaliana T-DNA insertion mutant collection (25). In cla1 plants, chloroplast development is arrested at an early stage, and genetic and molecular analyses have established that the disruption of a single gene, encoding 1-deoxy-D-xylulose 5-phosphate synthase, is responsible for the phenotype (26). We grew WT and cla1 mutant seeds on sucrose-containing media for 7 d and compared the global gene expression patterns of both seedling populations by using oligonucleotide microarrays. In cla1 mutant seedlings, the expression levels for most genes involved in isoprenoid metabolism were drastically reduced compared with WT levels (Table 1; see also supplemental material at www.pb.ethz.ch). The only genes whose expression was elevated in cla1 were the two isogenes encoding HMGR, the gene coding for epoxycarotenoid cleavage enzyme, and the gene encoding glutamyl-tRNA reductase. The drastic changes in gene expression might be due to the fact that in the cla1 mutant the gene encoding 1-deoxy-D-xylulose 5-phosphate synthase is never functionally expressed. Because the cla1 mutant shows a near-albino phenotype, it is evident that genetic disruption of plastidic isoprenoid biosynthesis cannot be efficiently compensated by the cytosolic pathway. This supports, in addition to the results shown above, the hypothesis that communication between the two pathways occurs preferentially from the chloroplast to the cytoplasm.
Supplementary Material
Acknowledgments
We thank Dr. N. Provart (Torrey Mesa Research Institute; presently at University of Toronto) and M. Hirsch-Hoffmann (Swiss Federal Institute of Technology Zürich) for providing valuable bioinformatic support for the analysis of the gene expression datasets. We also thank the Functional Genomics Center Zurich for technical and financial support.
Abbreviations: DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; IPP, isopentenyl diphosphate; MVA, mevalonate; MS, Murashige and Skoog.
References
- 1.Harborne, J. B. (1991) in Ecological Chemistry and Biochemistry of Plant Terpenoids, eds. Harborne, J. B. & Tomas-Barberan, R. A. (Clarendon, Oxford), pp. 399-426.
- 2.Lange, B. M., Rujan, T., Martin, W. & Croteau, R. (2000) Proc. Natl. Acad. Sci. USA 97, 13172-13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Disch, A., Hemmerlin, A., Bach, T. J. & Rohmer, M. (1998) Biochem. J. 331, 615-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arigoni, D., Sagner, S., Latzel, C., Eisenreich, W., Bacher, A. & Zenk, M. H. (1997) Proc. Natl. Acad. Sci. USA 94, 10600-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lichtenthaler, H. K., Schwender, J., Disch, A. & Rohmer, M. (1997) FEBS Lett. 400, 271-274. [DOI] [PubMed] [Google Scholar]
- 6.Schwender, J., Zeidler, J., Groner, R., Muller, C., Focke, M., Braun, S., Lichtenthaler, F. W. & Lichtenthaler, H. K. (1997) FEBS Lett. 414, 129-134. [DOI] [PubMed] [Google Scholar]
- 7.Milborrow, B. & Lee, H. (1998) Aust. J. Plant Physiol. 25, 507-512. [Google Scholar]
- 8.Hirai, N., Yoshida, R., Todoroki, Y. & Ohigashi, H. (2000) Biosci. Biotechnol. Biochem. 64, 1448-1458. [DOI] [PubMed] [Google Scholar]
- 9.Adam, K. P., Thiel, R. & Zapp, J. (1999) Arch. Biochem. Biophys. 369, 127-132. [DOI] [PubMed] [Google Scholar]
- 10.Piel, J., Donath, J., Bandemer, K. & Boland, W. (1998) Angew. Chem. 37, 2478-2481. [DOI] [PubMed] [Google Scholar]
- 11.Bach, T. J. & Lichtenthaler, H. K. (1982) Z. Naturforsch. 37, 46-50. [DOI] [PubMed] [Google Scholar]
- 12.Schwender, J., Müller, C., Zeidler, J. & Lichtenthaler, H. K. (1999) FEBS Lett. 455, 140-144. [DOI] [PubMed] [Google Scholar]
- 13.Murashige, T. & Skoog, F. (1962) Physiol. Plant 15, 473-497. [Google Scholar]
- 14.Bligh, E. & Dyer, W. (1959) Can. J. Biochem. Physiol. 37, 911-919. [DOI] [PubMed] [Google Scholar]
- 15.Zhu, T., Budworth, P., Han, B., Brown, D., Chang, H. S., Zou, G. & Wang, X. (2001) Plant Physiol. Biochem. 39, 221-242. [Google Scholar]
- 16.Fraser, P. D., Pinto, M. E., Holloway, D. E. & Bramley, P. M. (2000) Plant J. 24, 551-558. [DOI] [PubMed] [Google Scholar]
- 17.Rodríguez-Concepción, M. & Gruissem, W. (1999) Plant Physiol. 119, 41-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romeis, T. (2001) Curr. Opin. Plant Biol. 4, 407-414. [DOI] [PubMed] [Google Scholar]
- 20.Joubes, J., Chevalier, C., Dudits, D., Heberle-Bors, E., Inzé, D., Umeda, M. & Renaudi, J. P. (2000) Plant Mol. Biol. 43, 607-620. [DOI] [PubMed] [Google Scholar]
- 21.McCarty, D. R. & Chory, J. (2000) Cell 103, 201-209. [DOI] [PubMed] [Google Scholar]
- 22.Schürmann, P. & Jacquot, J. P. (2000) Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 371-400. [DOI] [PubMed] [Google Scholar]
- 23.Balmer, Y., Koller, A., del Val, G., Manieri, W., Schürmann, P. & Buchanan, B. B. (2002) Proc. Natl. Acad. Sci. USA 100, 370-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarz, M. & Arigoni, D. (1999) in Comprehensive Natural Product Biochemistry, ed. Cane, D. E. (Pergamon, Oxford), Vol. 2, pp. 367-399. [Google Scholar]
- 25.Mandel, M. A., Feldmann, K. A., Herrera-Estrella, L., Rocha-Sosa, M. & Léon, P. (1996) Plant J. 9, 649-658. [DOI] [PubMed] [Google Scholar]
- 26.Estévez, J. M., Cantero, A., Romero, C., Kawaide, H., Jimenez, L. F., Kuzuyama, T., Seto, H., Kamiya, Y. & Leon, P. (2000) Plant Physiol. 124, 95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoeffler, J. F., Hemmerlin, A., Grosdemange-Billiard, C., Bach, T. J. & Rohmer, M. (2002) Biochem. J. 366, 573-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.