Directing proteins to specific subcellular addresses is a central problem in cell biology. Historically, perhaps because of their general lack of compartmentalized organelles, bacteria were viewed as relatively uniform at the subcellular level. However, with advances in fluorescence microscopy came the realization that bacteria, like their eukaryotic counterparts, segregate their proteins to different cellular regions (1). Recent studies have extended this principle by demonstrating that bacteria not only localize intracellular proteins to distinct regions of the cell, but that within these regions they can also organize proteins into discrete ordered structures. For example, the actin homologs MreB and Mbl form spirals along the long axis of Bacillus subtilis (2), and the cytokinetic tubulin homolog FtsZ localizes to a ring that goes through a spiral intermediate to reposition itself during B. subtilis sporulation (3). The work of Shih et al. (4) in this issue of PNAS demonstrates that the ability of intracellular proteins to organize into ordered structures is not exclusive to bacterial cytoskeletal elements. The authors make the exciting observation that proteins that were once thought to diffusely oscillate between the cell poles are in fact present in ordered spirals.
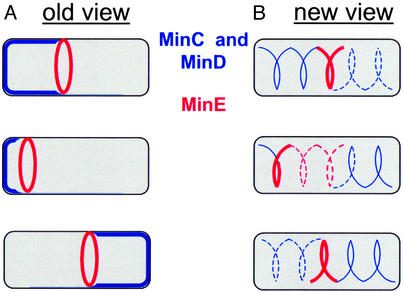
Almost all cells divide at a highly reproducible location within the cell. This regularity requires the cell to first find its appropriate division site and then localize its division machinery to that site. This process is perhaps best characterized in the rod-shaped bacterium, Escherichia coli, where three proteins, MinC, MinD, and MinE, collaborate to position the cell division protein FtsZ at midcell (5, 6). FtsZ is a cytosolic tubulin homolog that can polymerize in vitro and assembles into a ring-like structure (the Z ring) in vivo (7, 8). The Z ring assembles at the middle of the cell, which is also the future division site, and together with its accessory factors executes the process of cytokinesis (reviewed in ref. 9). The formation of Z rings at the cell poles is prevented by the polar presence of MinC, an inhibitor of FtsZ polymerization (10). MinC localization is dynamic and completely dependent on MinD (11, 12). Remarkably, the original description of their localization revealed that both MinC and MinD rapidly oscillate from one pole to the other in the span of ≈20 s, with the consequence that the time-averaged concentration of MinC is lowest in the middle of the cell (11–13). The oscillations of MinC and MinD depend on MinE, which forms what has been described as a ring-like structure. MinE avoids the cell poles, but is also dynamic, oscillating from side to side about the midcell position, sweeping MinD out of its path (Fig. 1A) (14, 15).
Fig. 1.
The old view of the organization of MinC, MinD, and MinE proteins within E. coli and a new view based on the observations of Shih et al. (4) in this issue of PNAS. (A) In the old view, MinC and MinD (shown in blue) were thought to be diffusely present at one pole at a time and to oscillate between poles. MinE (shown in red) was thought to be a ring that oscillated around the midcell position. (B) In the new view, MinC and MinD (blue) are organized into spiral structures, and, although the bulk of MinC and MinD oscillates from pole to pole, a small amount is always present at both poles. MinE (red) is also organized into a spiral that can be seen to extend to the poles.
This view of Min protein dynamics has now been refined by the work of Shih et al. (4). Fluorescence microscopy images are blurred due to the fact that in any given optical section of a three-dimensional structure, one observes both the in-focus light from that plane, as well as the out-of-focus light from other planes. Deconvolution microscopy is a method by which the out-of-focus light is computationally subtracted from the image by using a calculated image model (16). Shih et al. used deconvolution microscopy to examine fusions of MinC, MinD, and MinE to yellow fluorescent protein (YFP) or GFP. Surprisingly, the authors found that MinC and MinD, which were previously thought to be diffusely associated with the membrane at the poles, are in fact organized into extended spirals that wind around the cell. Although the bulk of MinC and MinD is assembled into spirals at one pole, faint spirals of MinC and MinD can also be detected at the opposite pole. A closer examination of MinE-YFP also revealed surprises: the MinE ring appears to consist of one to two loops of a coiled structure rather than a closed ring, and faint MinE spirals can be detected extending from the midcell to the cell pole (Fig. 1B). Based on the similarity of the MinC, MinD, and MinE spirals and their known biochemical associations, the authors suggest that all three Min proteins are part of the same higher-order structure and that their oscillations reflect a redistribution of subunits across a permanent scaffold of Min proteins, rather than de novo assembly and disassembly of Min structures with each cycle. In the future it will be intriguing to see how the MinC, MinD, and MinE spirals colocalize or interact with one another within the same cell.
It has been previously hypothesized that MinCD could assemble into linear filaments or rings (12, 17). In vitro, MinD has been shown to polymerize into linear filaments in the presence of ATP and phospholipids (18, 19). It has also been noted that there are not enough MinD molecules in a cell to form a coherent mono-layer covering the entire pole region, such that one must invoke an organized MinD structure to explain the area observed to be encompassed by MinD-GFP fluorescence (17). Shih et al. (4) report that MinD spirals do not form in minE mutants, and that MinE spirals do not form in minD mutants. The relationship between MinE and MinD has also been studied in vitro (18, 19). The addition of MinE can depolymerize MinD filaments, seemingly in contradiction with the observation that MinD spirals depend on the presence of MinE. However, when MinE is added to MinD filament preparations, some filaments remain that are longer and thicker (19). It is possible that a dynamically regulated interaction between MinD and MinE results in two MinE activities: organizing MinD into spirals and depolymerizing MinD, thereby driving its oscillation from pole to pole.
Based on the biochemical properties of MinD and MinE, three independent computer simulations have demonstrated that under certain parameters and given a defined container, the self-assembly of MinD and its inhibition by MinE are sufficient to establish and maintain the type of oscillations seen for both proteins in vivo (20–22). Although other factors might be involved as well, this finding is exciting in that it represents a description of a biological system that can self-organize to achieve subcellular localization without the need to invoke any previously localized components. Indeed, studies on spherical and irregularly shaped E. coli mutants demonstrate that the Min system can be used to find the long axis of the cell (23). However, these models may need to be revisited now that it appears that MinE does not completely remove MinD from the pole, but rather leaves a scaffold behind on which the oscillating MinD could later reassemble.
Clearly it will be of great interest to determine the mechanism by which Min spirals form. It is possible that some dynamic interaction between MinD and MinE results in their polymerization into coiled filaments. Alternatively, the interaction between MinD and MinE may allow them to associate with an existing spiral-shaped scaffold. One excellent candidate for such a scaffold is the prokaryotic actin homolog, MreB. MreB has a structure strikingly similar to that of actin, polymerizes in an ATP-dependent fashion, is required for maintaining the rod-like shape of E. coli, and forms spirals along the length of B. subtilis (2, 24). Shih et al. (4) take the first steps toward investigating the possibility that MinCDE spirals assemble on an MreB scaffold by imaging a YFP fusion to MreB. They find that, as is the case for B. subtilis, E. coli MreB forms spirals that coil along the long axis of the cell. However, the pitch of the YFP–MreB spirals is significantly greater than that of the MinCDE spirals. The discrepancy between the shapes of their spirals suggests that there is not a simple relationship between MinCDE and MreB. However, some caveats remain as MinCDE and MreB were not imaged in the same cell, the yfp–mreB fusion does not fully rescue an mreB mutant, and the endogenous MreB has not been visualized, such that it is still possible that the YFP fusion alters the pitch of the MreB spiral. This issue could perhaps best be addressed by examining the fate of the MinCDE spirals in mreB mutants.
Another interesting possibility raised by the presence of Min spirals is that the Min proteins could act as additional cytoskeletal elements and play cellular roles in addition to Z ring placement. MinCDE are not essential, but they have been implicated in chromosome segregation (25–27). Moreover, the localization of SpoIIIE to the forespore septum during sporulation is impaired in B. subtilis minC and minD mutants (28). The minE gene is not found in B. subtilis, and B. subtilis constitutively localizes MinC and MinD to both cell poles (they do not oscillate) (9). By determining whether B. subtilis MinC and MinD are also found in spirals, one could exploit their differences to learn more about both systems.
Regardless of how they form and what they do, it is clear that E. coli Min proteins are organized into spirals along the long axis axis of the cell (4). This expands the list of bacterial proteins found in spiral-shaped structures. B. subtilis has three actin homologs, and the two that have been examined, MreB and Mbl, assemble into differently shaped spirals (2). Although FtsZ normally appears to be a medially positioned ring, during B. subtilis sporulation it has been reported to go through a spiral intermediate on its way to becoming a polarly positioned ring (3). In addition, FtsZ has been shown to take on a spiral shape under certain conditions in E. coli (29). These results lend support to the idea that the Z ring, like MinE, is not normally a closed ring, but rather a tightly coiled spiral (3). Spirals can also be found outside of the cytoplasm, as flagella and pili have been shown to be helical (30, 31). Finally, it is worth noting that several bacteria, such as Helicobacter and Borrelia, are themselves spiraled structures. Why do bacterial proteins form spirals? Is the spiral a general organizing principle for prokaryotes? Could eukaryotic structures such as the contractile ring actually be spirals? Some of these questions will be harder to answer than others, but at least now we know that they should be asked.
Perhaps the most important lesson to be learned from the work by Shih et al. (4) is that the more closely we look, the more order we see within bacterial cells. The fact that the phrase ``bacteria are not just small bags of enzymes'' has become cliché is a sign that bacterial cell biology is coming of age. This field will continue to mature and grow as imaging technology improves even further. Soon the simplicity and experimental power of working with bacteria will far outweigh the obstacles associated with their diminutive size. Such advances, coupled with the ever-increasing recognition of the profound similarities between prokaryotes and eukaryotes, promises to make bacterial cell biology an exciting field to follow in the next decade.
See companion article on page 7865.
References
- 1.Shapiro, L. & Losick, R. (2000) Cell 100, 89–98. [DOI] [PubMed] [Google Scholar]
- 2.Jones, L. J., Carballido-Lopez, R. & Errington, J. (2001) Cell 104, 913–922. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Yehuda, S. & Losick, R. (2002) Cell 109, 257–266. [DOI] [PubMed] [Google Scholar]
- 4.Shih, Y.-L., Le, T. & Rothfield, L. (2003) Proc. Natl. Acad. Sci. USA 100, 7865–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Boer, P. A., Crossley, R. E. & Rothfield, L. I. (1992) J. Bacteriol. 174, 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi, E. & Lutkenhaus, J. (1993) J. Bacteriol. 175, 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukherjee, A. & Lutkenhaus, J. (1998) EMBO J. 17, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bi, E. F. & Lutkenhaus, J. (1991) Nature 354, 161–164. [DOI] [PubMed] [Google Scholar]
- 9.Errington, J., Daniel, R. A. & Scheffers, D. J. (2003) Microbiol. Mol. Biol. Rev. 67, 52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu, Z., Mukherjee, A., Pichoff, S. & Lutkenhaus, J. (1999) Proc. Natl. Acad. Sci. USA 96, 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raskin, D. M. & de Boer, P. A. (1999) J. Bacteriol. 181, 6419–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu, Z. & Lutkenhaus, J. (1999) Mol. Microbiol. 34, 82–90. [DOI] [PubMed] [Google Scholar]
- 13.Raskin, D. M. & de Boer, P. A. (1999) Proc. Natl. Acad. Sci. USA 96, 4971–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hale, C. A., Meinhardt, H. & de Boer, P. A. (2001) EMBO J. 20, 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu, X., Shih, Y. L., Zhang, Y. & Rothfield, L. I. (2001) Proc. Natl. Acad. Sci. USA 98, 980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wallace, W., Schaefer, L. H. & Swedlow, J. R. (2001) BioTechniques 31, 1076–1078. [DOI] [PubMed] [Google Scholar]
- 17.Rothfield, L. I., Shih, Y. L. & King, G. (2001) Cell 106, 13–16. [DOI] [PubMed] [Google Scholar]
- 18.Hu, Z., Gogol, E. P. & Lutkenhaus, J. (2002) Proc. Natl. Acad. Sci. USA 99, 6761–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suefuji, K., Valluzzi, R. & RayChaudhuri, D. (2002) Proc. Natl. Acad. Sci. USA 99, 16776–16781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruse, K. (2002) Biophys. J. 82, 618–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howard, M., Rutenberg, A. D. & de Vet, S. (2001) Phys. Rev. Lett. 87, 278102–278104. [DOI] [PubMed] [Google Scholar]
- 22.Meinhardt, H. & de Boer, P. A. (2001) Proc. Natl. Acad. Sci. USA 98, 14202–14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corbin, B. D., Yu, X. C. & Margolin, W. (2002) EMBO J. 21, 1998–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Ent, F., Amos, L. A. & Lowe, J. (2001) Nature 413, 39–44. [DOI] [PubMed] [Google Scholar]
- 25.Jaffe, A., D'Ari, R. & Hiraga, S. (1988) J. Bacteriol. 170, 3094–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulder, E., El'Bouhali, M., Pas, E. & Woldringh, C. L. (1990) Mol. Gen. Genet. 221, 87–93. [DOI] [PubMed] [Google Scholar]
- 27.Akerlund, T., Gullbrand, B. & Nordstrom, K. (2002) Microbiology 148, 3213–3222. [DOI] [PubMed] [Google Scholar]
- 28.Sharp, M. D. & Pogliano, K. (2002) EMBO J. 21, 6267–6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun, Q. & Margolin, W. (1998) J. Bacteriol. 180, 2050–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahn, E., Wild, P., Hermanns, U., Sebbel, P., Glockshuber, R., Haner, M., Taschner, N., Burkhard, P., Aebi, U. & Muller, S. A. (2002) J. Mol. Biol. 323, 845–857. [DOI] [PubMed] [Google Scholar]
- 31.Li, C., Corum, L., Morgan, D., Rosey, E. L., Stanton, T. B. & Charon, N. W. (2000) J. Bacteriol. 182, 6698–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]