Herpes simplex virus type 1 (HSV-1) is the prototype and best-studied virus of the α-herpesvirus group. HSV-1 undergoes a rapid productive replication cycle in host epithelial cells in vivo and in susceptible cultured cells in vitro. However, HSV-1 is also neurotropic and establishes life-long latent infections in the sensory neurons of the host, where the genome is in a nonreplicating chromatin-associated state (1), and viral gene expression is largely repressed (2). Although the mechanisms for the establishment of latency are poorly understood, reactivation requires an immediate early protein, termed ICP0, which is also required for efficient initiation of lytic infection (3–7).
The 152-kb linear double-stranded HSV-1 genome is divided into long and short regions of unique sequences, termed UL and US, which are bounded by regions of inverted repeats, termed internal repeats. The genome termini are bounded by regions of direct repeats, termed terminal repeats (Fig. 1). Furthermore, the HSV-1 genome undergoes inversions that result from recombination events mediated by the viral DNA replication machinery, which yield four genomic isomers in equimolar amounts (8, 9). In vivo, the HSV-1 genome has been found to exist in at least three different states: linear, circular, and concatemeric. In the virion, genomes are linear, but within hours after infection, end joining has been found to occur, resulting in ``endless genomes'' that have been interpreted to represent circles (10–12). Although the mechanism of genome circularization has not been established, one model predicts that circularization may involve recombination of the terminal repeats, resulting in a θ replication mode for the initial round of replication (13), providing the template for a rolling circle mode of replication (14). Thus, genome circularization has been thought to be a prerequisite for viral DNA replication, although there is no direct proof for θ replication intermediates in vivo (15). In this issue of PNAS, Jackson and DeLuca (16) provide strong evidence that genome circularization does not occur in productive infection but instead may occur during the establishment of latency. These authors determined that HSV-1 DNA circles are formed only in cells infected with viruses that do not express ICP0. Using Gardella gels to separate circular DNA molecules from linear genomes, Jackson and DeLuca did not observe circular genomes during productive infection or under conditions where genome replication was inhibited but ICP0 was expressed. Furthermore, using a mutant virus that is deleted for all immediate early genes (17), which results in the complete absence of viral gene expression, only circular genomes persisted. This is a condition that mimics the latent state of the virus. Thus, circles were detected only under conditions where DNA replication was inhibited or impaired, which would eliminate circles as an obligatory template for HSV-1 DNA replication during lytic infection.
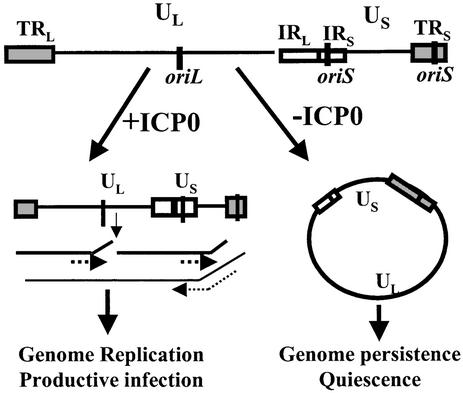
Fig. 1.
Schematic representation of the HSV-1 genome showing the unique long region (UL) bounded by a terminal repeat region (TRL) and an inverted internal repeat region (IRL). The unique short region (US) is similarly bounded by a terminal repeat region (TRS) and an inverted internal repeat region (IRS). The three origins of DNA replication, oriL and two copies of oriS, are shown. Jackson and DeLuca (16) have shown that, in the absence of ICP0, the genome circularizes and persists. When ICP0 is expressed very early during infection, the genome replicates and both linear and concatemeric replication forms are detected. Thus, these authors suggest that ICP0 regulates circle formation, and that circle formation may lead to latency, whereas DNA replication during productive infection prefers a linear template.
Previous studies that have reported ``endless'' DNA molecules concluded that these molecules represented circles or concatenated forms, because terminal restriction enzyme fragments, characteristic of linear molecules, were underrepresented relative to fragments arising from the joints (12). Furthermore, in a study using pulsed-field gel electrophoresis, the authors concluded that ``well hybridization'' represented circles. However, these authors also stated it was possible that molecules other than circles might serve as the initial templates for HSV-1 replication (11). Both the finding of endless genomes and the well hybridization observed in these studies are consistent with the formation of concatemeric molecules, which need not arise from circles. In fact, Jackson and DeLuca (16) also observed these replicative forms, which accumulated in the wells of the Gardella gels. These authors have postulated that genome replication may proceed from the three origins of HSV-1 DNA replication, oriL and two copies of oriS, to produce highly branched structures that might then be resolved by recombination or during packaging of viral DNA. Clearly, the results presented in this study warrant that other models of HSV-1 DNA replication be considered and tested.
One of the most intriguing aspects of the study by Jackson and DeLuca (16) is the finding that the formation of circles appears to be regulated by ICP0. ICP0 was first described as a promiscuous transactivator of viral gene expression (18), which is required for efficient initiation of viral lytic infection (6, 19) and reactivation from latency (3–5). ICP0 is an E3 ubiquitin ligase (20–22), which induces degradation of cellular proteins associated with ND10 bodies or promyeolytic leukemia structures through the ubiquitin proteasome pathway (23, 24). HSV-1 genomes appear to localize at these sites early in infection (25–27). Although all of the functional activities of the ND10 bodies have yet to be resolved, it was shown recently that these foci appear to be sites of DNA double-strand break repair (28). Jackson and DeLuca (16) suggest that the ends of the HSV-1 linear genome could be treated as double-strand breaks, which could be repaired as circular genomes. Conversely, the expression of ICP0 very early during lytic infection would preclude this repair process, because the cellular repair proteins present in the ND10 bodies would be degraded through the ubiquitin proteasome pathway. Thus, circles would not be formed. It is also possible that circle formation results from homologous or nonhomologous recombination of the direct repeats in the genome termini. Nonhomologous end joining requires a DNA-dependent protein kinase that has also been shown to be degraded through the action of ICP0 (29, 30). The finding that ICP0 appears to prevent the formation of circular molecules (16) may explain why replication of HSV-1 is greatly reduced in mutants defective in ICP0 at low multiplicities of infection (6, 19). In the present study (16), circular genomes accumulated early in infection then persisted during low multiplicity of infection with mutants defective in ICP0, whereas linear and concatemeric forms increased greatly in abundance, suggesting that linear molecules serve as better templates than circular genomes. In contrast, linear genomes did not persist in cells infected with a virus that was transcriptionally quiescent, whereas circular genomes were found to persist for >3 wk in these cells (16). Importantly, viral genomes derived from trigeminal ganglia of latently infected animals or in quiescently infected neuronal cells in culture appear to be maintained in circular or concatenated forms (31–33). Under the latter conditions, linear genomes were found to be unstable and subject to exonucleolytic degradation.
The expression of ICP0 very early during lytic infection would preclude repair.
The study by Jackson and DeLuca (16) has tackled two aspects of HSV-1 biology that are far from being fully understood: the mechanism of HSV-1 genome replication and the role of ICP0 in latency. These authors have provided evidence that during productive infection, it is unlikely that there is a requirement for genome circularization. In fact, in this study, circular genomes were not detected during infection with wild-type replication-competent virus. Only under conditions in which ICP0 was not expressed were circular genomes found. These data, coupled with the fact that θ intermediates have not been detected in vivo or in vitro (15), indicate that other models for HSV-1 DNA replication will have to be developed. Possibilities include recombination events that result in endless genomes. Second, Jackson and DeLuca (16) have shed more light on the possible role of ICP0 in latency by demonstrating that ICP0 regulates the formation of HSV-1 genome circles. Because circular forms persist in quiescently and latently infected cells, but the current findings suggest that circles are not the preferred template during lytic infection, the formation of circles may steer the infection toward latency. In neuronal cells, levels of ICP0 may be decreased, leading to the formation of circles, which in turn would lead to the establishment of the latent state. Again, further testing of this very intriguing model will be required.
In summary, the data presented by Jackson and DeLuca (16) have challenged the current models of HSV-1 DNA replication, while at the same time unfolding an elegant mechanism whereby the viral protein ICP0, which plays an essential role in latency, may act to shift infection from the productive lytic infection to the quiescent latent state.
See companion article on page 7871.
References
- 1.Deshmane, S. L. & Fraser, N. W. (1989) J. Virol. 63, 943–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Preston, C. M. (2000) J. Gen. Virol. 81, 1–19. [DOI] [PubMed] [Google Scholar]
- 3.Cai, W., Astor, T. L., Liptak, L. M., Cho, C., Coen, D. M. & Schaffer, P. A. (1993) J. Virol. 67, 7501–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clements, G. B. & Stow, N. D. (1989) J. Gen. Virol. 70, 2501–2506. [DOI] [PubMed] [Google Scholar]
- 5.Leib, D. A., Coen, D. M., Bogard, C. L., Hicks, J. B., Yager, D. R., Knipe, D. M., Tyler, K. L. & Schaffer, P. A. (1989) J. Virol. 63, 759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stow, N. D. & Stow, E. C. (1986) J. Gen. Virol. 67, 2571–2585. [DOI] [PubMed] [Google Scholar]
- 7.Everett, R. D. (2000) BioEssays 22, 761–770. [DOI] [PubMed] [Google Scholar]
- 8.Mocarski, E. S., Post, L. E. & Roizman, B. (1980) Cell 22, 243–255. [DOI] [PubMed] [Google Scholar]
- 9.Weber, P. C., Chalberg, M. D., Nelson, N. J., Glorioso, J. C. & Levine, M. (1988) Cell 54, 369–381. [DOI] [PubMed] [Google Scholar]
- 10.Deshmane, S. L., Raengsakulrach, B., Berson, J. F. & Fraser, N. W. (1995) J. Neurovirol. 1, 165–176. [DOI] [PubMed] [Google Scholar]
- 11.Garber, D. A., Beverley, S. M. & Coen, D. M. (1993) Virology 197, 459–462. [DOI] [PubMed] [Google Scholar]
- 12.Poffenberger, K. L. & Roizman, B. (1985) J. Virol. 53, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roizman, B. (1979) Cell 16, 481–494. [DOI] [PubMed] [Google Scholar]
- 14.Skaliter, R., Makhov, A. M., Griffith, J. & Lehman, I. R. (1996) J. Virol. 70, 1132–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehman, I. R. & Boehmer, P. E. (1999) J. Biol. Chem. 274, 28059–28062. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, S. A. & DeLuca, N. A. (2003) Proc. Natl. Acad. Sci. USA 100, 7871–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samaniego, L. A., Neiderhiser, L. & DeLuca, N. A. (1998) J. Virol. 72, 3307–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D., Preston, C. M. & Stow, N. D. (1991) in Herpesvirus Transcription and Its Regulation, ed. Wagner, E. K. (CRC, Boca Raton, FL), pp. 49–73.
- 19.Sacks, W. R. & Schaffer, P. A. (1987) J. Virol. 61, 829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boutell, C., Sadis, S. & Everett, R. D. (2002) J. Virol. 76, 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Sant, C., Hagglund, R., Lopez, P. & Roizman, B. (2001) Proc. Natl. Acad. Sci. USA 98, 8815–8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagglund, R., Van Sant, C., Lopez, P. & Roizman, B. (2002) Proc. Natl. Acad. Sci. USA 99, 631–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maul, G. G., Guldner, H. H. & Spivack, J. G. (1993) J. Gen. Virol. 74, 2679–2690. [DOI] [PubMed] [Google Scholar]
- 24.Everett, R. D. & Maul, G. G. (1994) EMBO J. 13, 5062–5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burkham, J., Coen, D. M. & Weller, S. K. (1998) J. Virol. 72, 10100–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishov, A. M. & Maul, G. G. (1996) J. Cell Biol. 134, 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukonis, C. J. & Weller, S. K. (1997) J. Virol. 71, 2390–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carbone, R., Pearson, M., Minucci, S. & Pelicci, P. G. (2002) Oncogene 21, 1633–1640. [DOI] [PubMed] [Google Scholar]
- 29.Lees-Miller, S. P., Long, M. C., Kilvert, M. A., Lam, V., Rice, S. A. & Spencer, C. A. (1996) J. Virol. 70, 7471–7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parkinson, J., Lees-Miller, S. P. & Everett, R. D. (1999) J. Virol. 73, 650–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Efstathiou, S., Minson, A. C., Field, H. J., Anderson, J. R. & Wildy, P. (1986) J. Virol. 57, 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rock, D. & Fraser, N. W. (1983) Nature 302, 523–525. [DOI] [PubMed] [Google Scholar]
- 33.Su, Y.-H., Moxley, M. J., Ng, A. K., Lin, J., Jordan, R., Fraser, N. W. & Block, T. M. (2002) J. Gen. Virol. 83, 2943–2950. [DOI] [PubMed] [Google Scholar]