Abstract
Trypanosomosis, or sleeping sickness, is a major disease constraint on livestock productivity in sub-Saharan Africa. To identify quantitative trait loci (QTL) controlling resistance to trypanosomosis in cattle, an experimental cross was made between trypanotolerant African N'Dama (Bos taurus) and trypanosusceptible improved Kenya Boran (Bos indicus) cattle. Sixteen phenotypic traits were defined describing anemia, body weight, and parasitemia. One hundred seventy-seven F2 animals and their parents and grandparents were genotyped at 477 molecular marker loci covering all 29 cattle autosomes. Total genome coverage was 82%. Putative QTL were mapped to 18 autosomes at a genomewise false discovery rate of <0.20. The results are consistent with a single QTL on 17 chromosomes and two QTL on BTA16. Individual QTL effects ranged from ≈6% to 20% of the phenotypic variance of the trait. Excluding chromosomes with ambiguous or nontrypanotolerance effects, the allele for resistance to trypanosomosis originated from the N'Dama parent at nine QTL and from the Kenya Boran at five QTL, and at four QTL there is evidence of an overdominant mode of inheritance. These results suggest that selection for trypanotolerance within an F2 cross between N'Dama and Boran cattle could produce a synthetic breed with higher trypanotolerance levels than currently exist in the parental breeds.
African tsetse fly-transmitted trypanosomosis (ATT) affects a wide range of wild and domesticated animal species. The consequences of infection include anemia, weight loss, lymphadenopathy, infertility, and abortion. In susceptible animals, death may result within a few weeks to several months after infection (reviewed in refs. 1–3). In cattle alone, ATT is estimated to cost the sub-Saharan African economy billions of U.S. dollars annually (3). Trypanocidal drugs are expensive and may be rendered ineffective by drug resistance, vector control can be environmentally damaging and difficult to maintain, and there is no vaccine for ATT. In consequence, despite a century of research, ATT still ranks among the world's more serious livestock diseases.
A small number of indigenous African ruminant breeds that are ``trypanotolerant'' (i.e., relatively resistant to the consequences of infection) may provide a means to mitigate the ravages of the disease. Under ATT challenge, trypanotolerant animals are better able than trypanosusceptible animals to control parasite proliferation, maintain their packed red blood cell volume (PCV), mount an immune response, and maintain body weight. Among trypanotolerant cattle breeds, the West African N'Dama (Bos taurus) is the best characterized (4–11).
Trypanotolerance in N'Dama cattle was identified as a potentially rewarding area for quantitative trait loci (QTL) mapping (12). Construction of the required F2 population between trypanotolerant N'Dama and a susceptible breed was initiated in 1989 (13). Thus, the experiment described here was among the first to construct a livestock population specifically for QTL mapping.
Materials and Methods
Resource Families for Mapping. In 1983, with the assistance of the International Trypanotolerance Center (Banjul, The Gambia), embryos were collected from N'Dama cows in The Gambia. They were implanted into recipient cows of the trypanosusceptible improved Kenya Boran breed (Bos indicus; in all that follows, the term ``Boran'' will refer to ``improved Kenya Boran''), and five N'Dama males and five N'Dama females were born in 1984 (14). In 1989, construction of large full-sibling F2 families was initiated by crossing four of the five N'Dama males with four Boran cows by using multiple ovulation and embryo transfer (MOET) into surrogate Boran dams (13, 14).§§ Four large F1 families were produced and subsequently intercrossed to produce by MOET seven F2 families, comprising 177 offspring in total, that were genotyped and used for QTL analysis. Family size ranged from 21 to 39; families 1, 2, 5, and 6 had the same N'Dama and Boran grandparents; and families 3, 4, and 7 shared a different set of N'Dama and Boran grandparents. The F2 animals were born between November 1992 and September 1996. The structure of the pedigree can be found in Fig. 1, which is published as supporting information on the PNAS web site, www.pnas.org.
Challenge and Phenotypic Recording. Twenty-three groups of F2 animals, each containing 3–13 calves born over a period of 3 weeks, were reared on their Boran dams at Kapiti Plains Estate, a tsetse fly-free area near Nairobi. Weaning was at 8 mo of age, and after a further 2 mo the animals were moved to the International Livestock Research Institute (ILRI), also a tsetse fly-free area, for a 2-mo period of acclimatization. This was followed by ATT challenge at 12 mo of age with Trypanosoma congolense clone IL1180 (16, 17), delivered through the bites of eight infected tsetse flies Glossina morsitans centralis (18, 19). Phenotyping of the last group ended in January 1998.
Trait monitoring began 3 wk before challenge and continued until 150 days after challenge. PCV, level of parasitemia (estimated by the dark-ground/phase contrast buffy coat method) (20), and body weight were recorded at least once per week. Twenty-eight of the F2 offspring in which PCV declined to ≤12% were treated and removed from the experiment. The mean day of treatment was day 98 (minimum day 14, maximum day 146) after ATT challenge. For these animals, the value of the phenotypic traits on the last recording day before treatment was taken as the value of the trait for the remainder of the challenge period. Six N'Dama controls and six Boran controls were challenged with the F2 offspring to serve as positive controls for the challenge procedure.
Phenotypic Traits for QTL Analysis. We used a set of 16 phenotypic traits (Table 1). Some of these traits correspond to traits that were analyzed biometrically by van der Waaij (ref. 21; and see ref. 22). The two prechallenge traits, initial PCV (PCVI) and initial body weight (BWI), were considered unrelated to trypanotolerance. Mean body weight (BWM) in the postchallenge period is an ambiguous trait because it can be affected by both BWI, a nontrypanotolerance trait, and weight loss after challenge, a trypanotolerance trait. Thus, alleles with positive effects on BWM can derive from nontrypanotolerance loci through effects on BWI or from trypanotolerance loci through effects on weight loss after challenge. The remaining postchallenge traits were all directly related to trypanotolerance. These include traits related to changes in PCV, body weight after challenge, and parasitemia. It should be noted that for most of the trypanotolerance traits, a lower trait value is associated with higher trypanotolerance. The exceptions are minimum PCV (PCVM), final PCV (PCVF), PCVF minus PCVM (PCVFM), final body weight (BWF), BWI, and BWM. Spearman correlation coefficients between the 16 traits are presented as Table 4, which is published as supporting information on the PNAS web site. Means and SD were calculated by using the multiqtl software package (http://esti.haifa.ac.il/~poptheor).
Table 1. Phenotypic traits for QTL mapping analysis.
| Prechallenge (nontrypanotolerance) traits | |
| PCVI | PCVI (mean PCV days 21-0) before challenge |
| BWI | BWI (BWM days 21-0) before challenge |
| Ambiguous (trypanotolerance or nontrypanotolerance traits) | |
| BWM | BWM after challenge (days 0-150) |
| Postchallenge (trypanotolerance) traits | |
| PCV | |
| PCVF | PCVF (day 150 or day before treatment) |
| PCVM | PCVM recorded during the postchallenge period (days 0-150) |
| PCVI minus PCVF (PCVIF) | PCVI (day 0) minus PCVF |
| PCVI minus PCVM (PCVIM) | PCVI (day 0) minus PCVM |
| PCVFM | PCVF (day 150 or last day before treatment) minus PCVM |
| PCV variance (PCVV) | Variance of the PCV values postchallenge (days 0-150) |
| PCVD 150 | Percentage decrease in PCV up to day 150 after challenge [(Mean PCV days 0-11) - (mean PCV days 13-150)]/(mean PCV days 0-11) |
| PCVD 100 | Percentage decrease in PCV up to day 100 after challenge [(Mean PCV days 0-11) - (mean PCV days 13-100)]/(mean PCV days 0-11) |
| Body weight | |
| BWF/BWI | BWF scaled by BWI |
| BWD 150 | Percentage decrease in body weight up to day 150 after challenge [(BWM days 0-11) - (mean BWM days 13-150)]/(BWM days 0-11) |
| Parasitemia | |
| PARMLn | Mean of natural logarithm (ni + 1), ni = number of parasites at day i after challenge (days 11-150) |
| PARLnM | Natural logarithm of the mean number of parasites after challenge (days 11-150) |
| Detection rate (DR60-150) | Number of times an individual is detected to be infected (days 60-150) |
BWD, percentage decrease in body weight.
DNA Isolation and Molecular Typing. DNA was isolated from blood according to a standard protocol (23). PCR conditions for microsatellite amplification were as described (24, 25). Fluorescent PCR products were coloaded and separated on automated DNA sequencers (ABI PRISM 377, Applied Biosystems). Fragment length identifications were performed by using genescan 672 (Version 2.0.2, Applied Biosystems) and genotyper (Version 2.0, Applied Biosystems) software. Approximately half of the microsatellites were genotyped at ILRI and the remainder were genotyped at the Shirakawa Institute of Animal Genetics (Fukushima, Japan) and the University of Liverpool (Liverpool, U.K.). Nonmicrosatellite markers were genotyped at the Bernhard Nocht Institute of Tropical Medicine (Hamburg, Germany).
QTL Analyses. Four hundred seventy-seven markers, mainly microsatellites, distributed among the 29 cattle autosomes were genotyped in the 177 F2 animals, 14 F1 parents, and 8 F0 grandparents. By tracing alleles from the grandparent to the F2 generation, F2 marker genotypes were recorded as NN (both alleles of N'Dama origin), BB (both alleles of Boran origin), or H (NB or BN, with one allele of N'Dama origin and one of Boran origin). The genotyping data on the seven F2 families were pooled, assuming the grandparental generation F0 to be fixed for alternative susceptible or tolerant QTL alleles (26–28). The linkage map position of the markers is available from the U.S. Department of Agriculture (USDA) and Commonwealth Scientific and Industrial Research Organization (CSIRO) publicly available linkage maps (www.marc.USDA.gov/genome/genome.html and www.cgd.csiro.au, respectively). The markers in common with the USDA cattle linkage map were first placed along each autosome in accordance with the published order, then, by using the information of the CSIRO map, the remaining markers were placed into the linkage group. In the few cases where the position of the marker was ambiguous, the position of the marker showing the smaller number of single crossing-over with flanking markers was chosen. Genotyping data of markers <10 centimorgans (cM) apart were combined, filling in for the most informative marker the missing genotypes (NN, BB, or H), as inferred from the other markers when available. Data on markers or groups of markers showing <120 observations were not used in the analysis, with the exception of one marker on BTA22, n = 113, and two markers on BTA21, n = 117 and n = 119. The total number of markers or groups of markers was consequently reduced to 210 (genotyping data available on request from O.H.). The total length of the autosomal map, calculated by adding the size of each chromosomal linkage group, was 2,256 cM or 82% of the USDA linkage map.
All traits were corrected by least-squares analysis of variance for sex and time of challenge. With the exception of BWI, none of the traits were corrected for group effects, because the groups were of different sizes and with unequal representation of F2 individuals from different families. BWI was corrected for group effect, because this trait is subject to seasonal environmental factors, and the animals were reared for the first 10 mo of their lives at the ILRI Kapiti Plains Estate and only then moved to more controlled conditions at ILRI Kabete. Outliers (listed in Table 2) with trait values greater than ±3 SD of the mean of the trait were removed before QTL analysis. The distributions of some traits deviated from normality (namely, PCVFM, PCVM, PCVD150, PCVD100, and PARLnM). These traits were transformed and the QTL analysis was repeated. Very similar results were obtained with only a slight improvement in statistical significance (data not shown). Therefore, for consistency, only nontransformed traits were used for QTL mapping.
Table 2. Characteristics of the phenotypic traits of N'Dama (trypanotolerant) and Boran (trypanosusceptible) controls and F2 animals.
| Traits* | N'Dama control, mean ± SD (n = 6) | Boran control, mean ± SD (n = 6) | Interpretation | F2, mean ± SD | F2, maximum | F2, minimum | F2, outliers |
|---|---|---|---|---|---|---|---|
| PCVI | 36.60 ± 5.47 | 38.70 ± 3.98 | No breed difference in PCV | 36.63 ± 3.36 | 46.16 | 27.18 | — |
| BWI | 135.50 ± 24.67 | 253.90 ± 115.63 | Breed body weight greater in Boran | 156.31 ± 90.35 | 202.55 | 98.14 | 90.35 |
| BWM | 140.05 ± 26.38 | 241.40 ± 111.44 | Breed body weight greater in Boran | 154.51 ± 20.66 | 209.42 | 100.87 | 20.66 |
| PCVM | 20.57 ± 2.88 | 14.00 ± 1.37 | Lower decrease in PCV in N'Dama | 16.26 ± 7.91 | 24.45 | 10.85 | 25.27, 27.04 |
| PCVF | 28.80 ± 3.33 | 14.76 ± 2.43 | Faster PCV recovery in N'Dama | 21.78 ± 4.24 | 36.5 | 10.90 | — |
| PCVIF | 7.83 ± 6.06 | 23.98 ± 4.50 | Faster PCV recovery in N'Dama | 15.04 ± 3.62 | 33.45 | -1.55 | -6.94 |
| PCVIM | 15.98 ± 6.82 | 24.65 ± 3.73 | Lower decrease in PCV in N'Dama | 20.23 ± 6.24 | 29.34 | 9.09 | 5.44, 9.09, 32.39 |
| PCVFM | 8.20 ± 2.40 | 0.70 ± 1.40 | Little or no PCV recovery in Boran | 5.29 ± 2.06 | 16.16 | -0.04 | 17.45 |
| PCVV | 17.57 ± 7.09 | 35.60 ± 7.72 | Less variation of PCV in N'Dama | 26.95 ± 6.30 | 42.55 | 13.59 | 46.39 |
| PCVD150 | 26.68 ± 13.12 | 47.48 ± 5.45 | Lower decrease in PCV in N'Dama | 42.11 ± 9.36 | 62.41 | 14.16 | 13.01 |
| PCVD100 | 27.88 ± 14.07 | 44.49 ± 4.44 | Lower decrease in PCV in N'Dama | 40.99 ± 8.00 | 60.2 | 19.21 | 11.18 |
| BWF/BWI | 1.14 ± 0.09 | 0.88 ± 0.23 | Greater weight recovery in N'Dama | 0.97 ± 0.12 | 1.26 | 0.71 | — |
| BWD 150 | 16.14 ± 4.83 | 25.79 ± 4.74 | Less decrease in weight in N'Dama | 20.25 ± 0.45 | 21.52 | 18.78 | — |
| PARMLn | 2.66 ± 0.72 | 5.17 ± 0.43 | Fewer parasites in N'Dama | 3.37 ± 0.16 | 5.53 | 1.61 | 0.84 |
| PARLnM | 2.42 ± 0.86 | 4.89 ± 1.22 | Fewer parasites in N'Dama | 3.47 ± 0.75 | 5.51 | 1.60 | 5.95 |
| DR60-150 | 0.27 ± 0.09 | 0.46 ± 0.07 | Fewer parasites in N'Dama | 0.45 ± 0.16 | 0.85 | 0.02 | 0.97 |
See Table 1 for definitions of the traits.
Single-trait QTL mapping analysis was performed on the basis of the standard single-interval maximum likelihood model (29) by using the software package multiqtl. For each marker, the multiqtl software package uses ``virtual'' marker restoration by calculating the three probabilities (for NN, BB, and H) for each missing genotype based on available scores of the nearest-neighbor markers, and these probabilities enter the likelihood function. Logarithm-of-odds scores and P values were calculated for all trait-by-chromosome combinations with the significance of the QTL estimated after 10,000 chromosomewise permutation tests (30).
The multiqtl program was first run as a general model, with parameters d (main effect) and h (dominance) both fitted. The dominance ratio, D, was taken as 2h/d, where d is the difference between the means of the F2 homozygotes calculated as NN – BB, and h is the mean value of the F2 heterozygotes minus the mean of the NN and BB homozygotes. The trypanotolerant allele was assigned to the N'Dama grandparent when the sign of d or h was the same as the sign of the difference of the mean values of the trait in the control animals (N'Dama mean minus Boran mean). The trypanotolerant allele was assigned to the Boran grandparent when the reverse was true.
The genetic model for the QTL allele was then set as additive when –0.50 < D < 0.50, dominant when 0.50 < D < 3.00, recessive when –0.50 > D >–3.00, and overdominant when D > 3.00 or D < –3.00, and the analysis was repeated under the appropriate genetic model: additive (h = ±d/2), dominant or recessive (h = ±d), or overdominant (d = 0). The cut-off point for the additive model was set as half the distance from complete additivity (D = 0) and complete dominance or recessiveness (D = 1.0 or –1.0). An overdominant model was chosen when the main effect was very small relative to the dominance effect. There was a fairly continuous distribution of D values until D = ±3.0; then there was a series of very high values. Hence, D =±3.0 was taken as the effective cut-off value for overdominance. The multiqtl package does not estimate the power of discrimination between corresponding models. Therefore, only tests of significance between the models were performed. This was tested for situations where the logarithm of odds was rather high, so that one could expect some of the submodels to differ from the general model (see Table 5, which is published as supporting information on the PNAS web site, for the full results of the QTL analysis).
The false discovery rate (FDR) (31, 32) was used to set significance levels. Critical comparisonwise error rates (P values), estimated by permutation test for FDR 0.05, FDR 0.10, and FDR 0.20, were P < 0.0008, 0.0043, and 0.0185, respectively. The chromosomal location of the QTL was taken as the position with the highest logarithm-of-odds score; its SE was calculated after 5,000 boot-strappings (33). With the exception of BTA16, the results were consistent with a single QTL per chromosome affecting several correlated traits; hence, the estimated QTL location was calculated as the average location across traits weighted by their SE. The individual QTL effect for a given phenotypic trait was estimated as the proportion of the observed F2 phenotypic variance explained by the QTL.
Results
BWI of the Boran control animals were almost twice that of the N'Dama, whereas PCVI were similar. Under challenge, the superior trypanotolerance of the N'Dama was evident in all traits (Table 2). None of the six N'Dama controls needed treatment at any point during or after the challenge period, whereas five of the six Boran controls had to be removed from the experiment when their PCV values fell to 12% or less and treatment was required to prevent probable death. The Boran suffered a greater proportional loss of body weight than the N'Dama (BWD 150). By the end of the experiment, the N'Dama had recovered their body weight, whereas the Boran had not (BWF/BWI). The decrease in PCV (PCVI minus PCVM) was less for N'Dama than for Boran, and the recovery in PCV (PCVFM) was greater for N'Dama than for Boran, which showed little recovery. Mean number of parasites after challenge (PARMLn and PARLnM) and parasite detection rate (DR60–150) in the N'Dama were about half those in the Boran.
All F2 animals showed a decrease in PCV and loss of body weight after challenge. By the end of the experiment, none of the PCV values in the F2 offspring had reached prechallenge values, although some of the animals had recovered their body weight (Table 2, BWF/BWI). Typically, the level of parasites decreased over the course of the experiment. A proportion of F2 animals had become aparasitemic based on the visual scoring method used. It is possible that these animals were truly free of parasites. However, this would be somewhat surprising, because persistence of infection, albeit at very low levels, is common even in the N'Dama. A more plausible alternative is that apparently self-curing animals were parasitemic, but at levels below the detection limit of the method used.
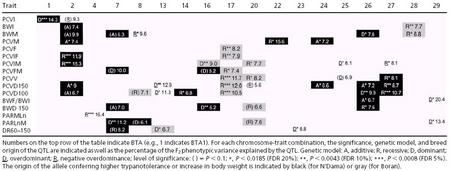
Table 3 shows the traits with significant effects at FDR ≤0.20, according to chromosomes. In addition, because of the strong correlations among trypanotolerance traits (see Table 4), if one of the trypanotolerance traits on a chromosome was significant at FDR ≤0.20, then all trypanotolerance traits having comparisonwise error rates of P < 0.10 are also shown, on the assumption that they all reflect effects of the same underlying QTL. For each listed trait, Table 3 also shows the breed origin and dominance status of the chromosomal haplotype with positive effects on trait value for the nontrypanotolerance traits, and degree of trypanotolerance for the trypanotolerance traits. The full details of the QTL analysis are in Table 5.
Table 3. Characteristics of the mapped QTL.
All 16 defined phenotypic traits were associated with putative QTL, with 40 significant trait–chromosome combinations on 18 different chromosomes (FDR ≤0.20). All but four traits (PCVI, PCVF, PCVFM, and PARMLn) were associated with significant effects on more than one chromosome.
For the nontrypanotolerance traits, highly significant effects were found for PCVI on BTA1 and for BWI on BTA28. The effect on PCVI was not accompanied by an effect on the trypanotolerance traits. The positive effect on BWI was associated with the haplotype derived from the Boran grandparent, as expected on the basis of the breed difference. This positive effect was recessive.
With respect to BWM, there was also a significant effect on BTA28 with the positive effect derived from the Boran. This is probably a secondary consequence of the effect on BWI of the putative QTL on this chromosome. Hence, we do not include this effect among the positive effects on trypanotolerance. In addition, there was a significant negative overdominance effect on BTA8, which was not associated with significant effects on either BWI or the trypanotolerance traits. Its interpretation is unclear. Three remaining effects on BWM on BTA2, -7, and -26 (one significant, the other two at P < 0.10) were all associated with strong effects on one or more other trypanotolerance traits, without a significant effect on BWI. In all three cases, the haplotype with positive effect derives from the N'Dama. Thus, the positive effect on BWM in these instances is probably a secondary effect of the trypanotolerance locus.
Fifteen chromosomes had effects on the trypanotolerance traits. Of these, seven chromosomal haplotypes of N'Dama origin (BTA2, -7, -14, -22, -24, -26, and -27) and two chromosomal haplotypes of Boran origin (BTA17 and -20) were associated with positive effects on trypanotolerance. In addition, the positive haplotype of BTA16 had a mixed origin with two positive effects of N'Dama origin and one of Boran origin, and BTA13 had a positive effect of Boran origin and two due to overdominance. Four chromosomes (BTA4, -23, -25, and -29) showed significant effects due to overdominance.
QTL were identified on BTA4, -14, -22, and -23 through only one of the defined trypanotolerance traits. QTL on the remaining 11 chromosomes were identified through more than one trait (i.e., at least one significant effect at FDR ≤0.20 and one or more effects that were either significant or at P < 0.10). For two of these chromosomes (BTA4 and -23), the effect was due to overdominance, and it involved a parasitemia trait. On the other two chromosomes (BTA14 and -22), the effects were recessive with the positive haplotype derived from the N'Dama.
A conservative interpretation of these results is that, with the exception of BTA16, a single QTL has been identified per chromosome and that differences in the spectrum of effects presented are due to sampling variation.
Of the 40 significant effects (FDR ≤0.20), 22 showed a recessive or negative overdominance mode of action, 12 showed a dominant or overdominant mode of action, and 6 showed an additive mode of action. The majority of the recessive effects was concentrated among the correlated PCV trypanotolerance traits, whereas for the parasitemia trypanotolerance traits, there was a slight excess of dominant compared with recessive effects. The genetic model for the inheritance of the trypanotolerance allele was the same for all significant trait–chromosome combinations on BTA13, -16, -17, -20, -24, -27, and -29. However, on BTA2, there were additive and recessive effects for trypanotolerance, and on BTA26 there were recessive, additive, and dominance effects for trypanotolerance. The overall consistency of dominance effects on a given chromosome supports the hypothesis that all effects detected are associated with a single QTL.
When a higher trait value is associated with higher trypanotolerance, dominance for trait value is also dominance for trypanotolerance. As noted above, however, for most of the trypanotolerance traits, a lower trait value is associated with higher trypanotolerance. For these traits, a recessive effect on trait value is a dominant effect on trypanotolerance. Therefore, when coded to express effects on trypanotolerance, 17 of the 35 significant effects on trypanotolerance traits showed a dominant (or overdominant) mode of action, 6 showed additive action, and 12 showed a recessive (or negative overdominant) mode of action. Thus, there is a slight preponderance of dominant effects at the trypanotolerance level. In this case, the dominant effects were concentrated among the PCV traits, body weight traits were equally divided among dominant and recessive effects, and parasitemia traits showed mostly recessive effects.
Table 3 indicates also the proportion of phenotypic variance of the traits in the F2 population explained by individual QTL identified at P < 0.1, for chromosomes with at least one effect significant at FDR ≤0.20. The proportion explained for individual chromosome–trait combinations ranges from 6% to 20%. Effects averaged ≈10% and were similar among categories (PCV, body weight, and parasitemia). The total proportion of the phenotypic variance explained across all chromosomes carrying putative QTL ranges from 8.2% (PCVF, one chromosome) to 63% (PCVD150, seven chromosomes).
Discussion
The results strongly support a multilocus model for the inheritance of trypanotolerance. Several QTL on several chromosomes contribute to the three major tolerance indicators: anemia, body weight, and parasitemia controls. Summing the individual gene effects at a single trait, the proportion of the phenotypic variance of the traits explained by these QTL remains relatively low (Table 3). It should also be noted that estimated effects of QTL that are detected in experiments of only moderate power tend to be overestimated. This suggests the presence of (i) other QTL affecting trypanotolerance that are segregating in the N'Dama; (ii) other QTL with effects too small to be detected in this experiment; and/or (iii) environmental or complex epistatic effects affecting the traits under consideration.
At four chromosomes (BTA13, -16, -17, and -20) showing significant effects on trypanotolerance, the trypanotolerance haplotype appears to derive from the susceptible Boran. The origin of these Boran trypanotolerant haplotypes is unclear. The improved Boran at Kapiti Plains Estate that were the source of ``Boran'' genes in this study have not been exposed to ATT, because the herd was established many decades ago. Moreover, the four Boran founders of the F2 population required treatment when challenged with the same clone of parasite as that used in the F2 study, whereas the N'Dama founders did not. However, all existing evidences point to the Boran breed having evolved in a broad region from southern Sudan across southern Ethiopia to southern Somalia, and tsetse flies were present in parts of this region until relatively recent times. Thus, it is possible that the forebears of the Kapiti Plains Estate Boran had been exposed to ATT and, as a result, may have evolved a degree of tolerance. There are also anecdotal reports that the Boran, although susceptible when compared with the N'Dama, withstand ATT better than exotic European breeds. In this context, the Orma Boran, a breed with the same origin as the Kenya Boran but which, unlike the latter, has been maintained in a high-challenge tsetse-infested area (34), has been reported to be relatively trypanotolerant (35, 36). Moreover, positive heritability estimates for trypanotolerance were found in East African B. indicus cattle in Ethiopia (37). Alternatively, the improved Kenya (Kapiti) Borans have probably always faced, and are relatively resistant to, tick-borne diseases. These are caused by hemoprotozoa that induce severe anemia in susceptible individuals. Although not sufficient on their own to provide protection against ATT, the loci involved might contribute to anemia control after ATT infection in crossbred N'Dama × Boran animals. Indeed, three of the four Boran trypanotolerance QTL (BTA16, -17, and -20) exerted their primary positive effects on control of PCV.
Although the two parental breeds differ strongly in body weight and trypanotolerance, the experiment uncovered (excluding those with ambiguous effects) only a single chromosome (BTA28) affecting BWI with a QTL explaining ≈10% of the F2 phenotypic variance, whereas 15 chromosomes affecting trypanotolerance were revealed. This seemingly anomalous result is readily accommodated by the different evolutionary histories of the two traits. Trypanotolerance is a trait for which an extreme phenotype is optimal. Hence, alleles of strong positive effect will tend to reach fixation rapidly. The experiment was of a magnitude that can detect alleles of large effect (26, 38). In contrast, body weight is a trait for which an intermediate phenotype is optimal. In this case, alleles of large effect are eliminated by stabilizing selection, and genetic variation is due to alleles generally of too small effect to be detected in an experiment of the present size.
Of the five chromosomes affecting parasitemia, only two (BTA13 and -29) show a significant effect on PCV or body weight. Furthermore, in the case of BTA13, the parasitemia effect maps as an outlier with respect to the PCV effects. It should also be noted that a recessive model applies to most of the PCV QTL, whereas a dominant or overdominant (dominant) model is associated with four of five of the QTL controlling parasitemia. Thus, the genetic control of anemia and parasitemia after infection may be distinct.
The use of only measures of change in body weight under challenge to select for trypanotolerance in a synthetic population, although convenient, might miss many of the relevant QTL. Indeed, although in a number of chromosomes (BTA2, -7, and -26) body-weight effects under challenge were associated with trypanotolerance as reflected in PCV and parasitemia, there was an isolated high-body-weight effect on BTA28. Moreover, on most chromosomes, positive PCV and/or parasitemia effects were not associated with effects on body weight. Selection for trypanotolerance in N'Dama populations or in synthetic populations formed by crossing N'Dama and susceptible breeds might, however, be based jointly on parasitemia and PCV traits, because control of anemia is often unaccompanied by control of parasitemia and vice versa (refs. 2 and 21; see Table 5). Also, the finding that BTA28 carries a QTL with a strong positive effect on body weight without any effects on trypanotolerance suggests that marker-assisted introgression of this chromosome from the Boran to the N'Dama may result in an increase in body weight while preserving trypanotolerance.
The finding of trypanotolerance QTL not present in the N'Dama but present in the Boran (BTA17, -20, and -16) raises the exciting possibility that selection for trypanotolerance within an F2 cross between N'Dama and Boran cattle would produce a synthetic breed that would exceed either parental breed in trypanotolerance terms. This is supported by the presence of some F2 animals showing higher levels of trypanotolerance than any of the N'Dama controls (Table 2). Also, chromosomal regions carrying positive QTL alleles identified in this study and heavily marked by microsatellites and single-nucleotide polymorphisms could be introduced into more productive breeds to provide a degree of trypanotolerance. However, the large number of chromosomes carrying trypanotolerance QTL identified suggests that in most instances marker-assisted selection from an F2 population, rather than marker-assisted introgression, would be the most effective way to produce synthetic breeds combining high productivity and trypanotolerance.
The map locations of the trypanotolerance QTL also provide a platform for a search for population-wide linkage disequilibrium between marker loci and QTL affecting trypantolerance in purebred N'Dama cattle and, particularly, in admixture populations produced in the recent or more distant past by crosses between N'Dama and susceptible breeds. Population-wide linkage disequilibrium of this nature can be a powerful tool for within-breed selection.
Recently, trypanosomosis-resistance QTL have been mapped in a mouse model (15, 39) after infection with the same clone of T. congolense as that used in the cattle study reported here. Ongoing comparative mapping has so far revealed a region of homology between one of the cattle chromosomal regions containing a trypanotolerance QTL identified in this study (conferring control of parasitemia on BTA7) and a chromosomal region in the C57BL/6 mouse containing a major QTL linked to survival time after infection (40). Thus, the mouse model may enable the genes corresponding to the mapped cattle QTL affecting trypanotolerance to be identified. This will enable much more effective use of marker-assisted selection and introgression for improvement of trypanotolerance in domestic livestock.
Supplementary Material
Acknowledgments
We thank T. Jordt, D. Kennedy, B. King, S. Leak, J. Mwakaya, D. Mwangi, M. Ogugo, H. ga'Thuo, and the farm personnel at ILRI headquarters and Kapiti Plains Estate for help in producing ILRI F2 resources, blood collection, phenotypic recording of the raw data, and/or technical help in producing the genotyping data. We also acknowledge Max Murray, Ivan Morrison, Jack Doyle, and Ross Gray [all formerly of the International Laboratory for Research on Animal Diseases (ILRAD)], and John Trail (formerly of the International Livestock Center for Africa), for the vision and support that led to the establishment of the N'Dama cattle resource in the former ILRAD, without which the work described here would not have been possible. Leyden Baker and John Rowlands provided useful comments on the manuscript. ILRI research is principally funded by program grants from the United Kingdom, Japan, the European Union, Ireland, Germany (Bundesministerium für wirtschaftliche Zusammenarbeit und Entwicklung), and France, and unrestricted funding from other donors to the Consultative Group on International Agricultural Research. This is ILRI publication 2002-41.
Abbreviations: ATT, African tsetse fly-transmitted trypanosomosis; PCV, packed red blood cell volume; QTL, quantitative trait loci; ILRI, International Livestock Research Institute; PCVI, initial PCV; BWI, initial body weight; BWM, mean body weight; BWF, final body weight; FDR, false discovery rate; PCVM, minimum PCV; PCVF, final PCV; PCVFM, PCVF minus PCVM; PCVD, percentage decrease in PCV; PARMLn, mean of natural logarithm of parasites; PARLnM, natural logarithm of mean number of parasites.
Footnotes
Kennedy, D. J., Leak, S. G. A., Kemp, S. J. & Teale, A. J. (1994) Theriogenology 41, 226 (abstr.).
References
- 1.Urquhart, G. M. (1980) Trans R. Soc. Trop. Med. Hyg. 74, 726–729. [DOI] [PubMed] [Google Scholar]
- 2.d'Ieteren, G., Authié, E., Wissocq, N. & Murray, M. (1999) in Breeding for Disease Resistance in Farm Animals, eds. Axford, R. F. E., Bishop, S. C., Nicholas, F. W. & Owen, J. B. (CABI, Wallingford, U.K.), 2nd Ed., pp. 195–216.
- 3.Kristjanson, P. M., Swallow, B. M., Rowlands, G. J., Kruska, R. L. & de Leeuw, P. N. (1999) Agric. Syst. 59, 79–98. [Google Scholar]
- 4.Stewart, J. L. (1937) Vet. Rec. 49, 1289–1297. [Google Scholar]
- 5.Roberts, C. J. & Gray, A. R. (1973) Trop. Anim. Health Prod. 5, 220–233. [DOI] [PubMed] [Google Scholar]
- 6.Roelants, G. E. (1986) Parasite Immunol. 8, 1–10. [DOI] [PubMed] [Google Scholar]
- 7.Doko, A., Guedegbe, B., Baelmans, R., Demey, F., N'Diaye, A., Pandey, V. S. & Verhulst, A. (1991) Vet. Parasitol. 40, 1–7. [DOI] [PubMed] [Google Scholar]
- 8.Trail, J. C., d'Ieteren, G. D. & Teale, A. J. (1989) Genome 31, 805–812. [DOI] [PubMed] [Google Scholar]
- 9.Paling, R. W., Moloo, S. K., Scott, J. R., Gettinby, G., McOdimba, F. A. & Murray, M. (1991) Parasite Immunol. 13, 427–445. [DOI] [PubMed] [Google Scholar]
- 10.Paling, R. W., Moloo, S. K., Scott, J. R., McOdimba, F. A., Logan-Henfrey, L. L., Murray, M. & Williams, D. J. (1991) Parasite Immunol. 13, 413–425. [DOI] [PubMed] [Google Scholar]
- 11.Murray, M., Stear, M. J., Trail, J. C. M., d'Ieteren, G. D. M., Ageymang, K. & Dwinger, R. H. (1991) in Breeding for Disease Resistance in Farm Animals, eds. Owen, J. B. & Axford, R. F. E. (CABI, Wallingford, U.K.), 1st Ed., pp. 203–223.
- 12.Soller, M. & Beckmann, J. S. (1987) Toward an Understanding of the Genetic Basis of Trypanotolerance in the N'Dama Cattle of West Africa (Consultation Report submitted to Food and Agriculture Organization, Rome, March 1987).
- 13.Teale, A. (1993) Bioscience 43, 475–483. [Google Scholar]
- 14.Jordt, T., Mahon, G. D., Touray, B. N., Ngulo, W. K., Morrison, W. I., Rawle, J. & Murray M. (1986) Trop. Anim. Health Prod. 18, 65–75. [DOI] [PubMed] [Google Scholar]
- 15.Iraqi, F., Clapcott, S. J., Kumari, P., Haley, C. S., Kemp, S. J. & Teale, A. J. (2000) Mamm. Genome 11, 645–648. [DOI] [PubMed] [Google Scholar]
- 16.Geigy, R. & Kauffmann, M. (1973) Acta Trop. 30, 12–23. [PubMed] [Google Scholar]
- 17.Nantulya, V. M., Musoke, A. J., Rurangirwa, F. R. & Moloo, S. K. (1984) Infect. Immun. 43, 735–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emery, D. L. & Moloo, S. K. (1981) Acta Trop. 38, 15–28. [PubMed] [Google Scholar]
- 19.Dwinger, R. H., Murray, M. & Moloo, S. K. (1987) Parasite Immunol. 9, 353–362. [DOI] [PubMed] [Google Scholar]
- 20.Paris, J., Murray, M. & McOdimba, F. (1982) Acta Trop. 39, 307–316. [PubMed] [Google Scholar]
- 21.van der Waaij, E. H. (2001) Ph.D. thesis (Wageningen University, Wageningen, The Netherlands).
- 22.Falconer, D. S. & MacKay, T. F. C. (1996) Introduction to Quantitative Genetics (Longman, New York), 4th Ed. [DOI] [PMC free article] [PubMed]
- 23.Sambrook, J. E. F., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 24.Hirano, T., Nakane, S., Mizoshita, K., Yamakuchi, H., Inoue-Murayama, M., Watanabe, T., Barendse, W. & Sugimoto, Y. (1996) Anim. Genet. 27, 365–368. [DOI] [PubMed] [Google Scholar]
- 25.Kemp, S. J., Hishida, O., Wambugu, J., Rink, A., Longeri, M. L., Ma, R. Z., Da, Y., Lewin, H. A., Barendse, W. & Teale, A. J. (1995) Anim. Genet. 26, 299–306. [DOI] [PubMed] [Google Scholar]
- 26.Beckmann, J. S. & Soller, M. (1988) Theor. Appl. Genet. 76, 228–236. [DOI] [PubMed] [Google Scholar]
- 27.Andersson, L., Haley, C. S., Ellegren, H., Knott, S. A., Johansson, M., Andersson, K., Andersson-Eklund, L., Edfors-Lilja, I., Fredholm, M., Hansson I., et al. (1994) Science 263, 1771–1774. [DOI] [PubMed] [Google Scholar]
- 28.Haley, C. S., Knott, S. A. & Elsen, J. M. (1994) Genetics 136, 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knott, S. A. & Haley, C. S. (1992) Genetics 132, 1211–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Churchill, G. A. & Doerge, R. W. (1994) Genetics 138, 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini, Y. & Hochberg, Y. (1995) J. R. Stat. Soc. B 57, 289–300. [Google Scholar]
- 32.Mosig, M. O., Lipkin, E., Khutoreskaya, G., Tchourzyna E., Soller, M. & Friedmann, A. (2001) Genetics 157, 1683–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visscher, P. M., Thompson, R. & Haley, C. S. (1996) Genetics 143, 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rege, J. E. O., Kahi, A. K., Okomo-Adhiambo, M., Mwacharo, J. & Hanotte, O. (2001) Zebu Cattle of Kenya: Uses, Performance, Farmer Preferences, Measures of Genetic Diversity and Options for Improved Use (Animal Genetic Resources Research 1, International Livestock Research Institute, Nairobi).
- 35.Njogu, A. R., Dolan, R. B., Wilson, A. J. & Sayer, P. D. (1985) Vet. Rec. 117, 632–636. [DOI] [PubMed] [Google Scholar]
- 36.Dolan, R. B. (1998) The Orma Boran: A Trypanotolerant East African Breed (Kenya Trypanosomiasis Research Institute, Kikuyu, Kenya).
- 37.Rowlands, G. J., Mulatu, W., Nagda, S. M., Dolan, R. B. & d'Ieteren, G. D. M. (1995) Livestock Prod. Sci. 43, 75–84. [Google Scholar]
- 38.Soller, M., Brody, T. & Genezi, A. (1976) Theor. Appl. Genet. 47, 35–39. [DOI] [PubMed] [Google Scholar]
- 39.Kemp, S. J., Iraqi, F., Darvasi, A., Soller, M. & Teale, A. J. (1997) Nat. Genet. 16, 194–196. [DOI] [PubMed] [Google Scholar]
- 40.Kang'a, S. (2003) Ph.D. thesis (Kenyatta Univ., Nairobi).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.