Abstract
Glycosphingolipids form glycosphingolipid signaling microdomains. Here, we report an unrecognized type of phosphatidylglucoside (PhGlc)-based lipid microdomain in HL60 cells. Treatment of cells with rGL-7, which preferentially reacts with PhGlc, induced differentiation of HL60 cells. This was manifested by the appearance of nitroblue tetrazolium-positive cells together with CD38 expression and c-Myc down-regulation. We determined the molecular mechanisms underlying early stages of signal transduction. rGL-7 treatment induced rapid tyrosine phosphorylation of Src family protein kinases Lyn and Hck. Reduction of endogenous cholesterol after application of methyl-β-cyclodextrin suppressed rGL-7-stimulated tyrosine phosphorylation. Phosphorylated proteins and PhGlc colocalized in the Triton X-100 insoluble, light buoyant density fraction after sucrose gradient ultracentrifugation of HL60 cell lysates. This suggests PhGlc-based microdomain is involved in GL-7 signaling. Ligation of known components of microdomains, such as sphingomyelin and ganglioside GM1, with corresponding antibodies failed to induce differentiation and tyrosine phosphorylation. These results show that PhGlc constitutes a previously undescribed lipid signaling domain, and the glucose residue of PhGlc is critical for organization of the carbohydrate-dependent signaling domain involved in cellular differentiation of HL60 cells.
In the majority of vertebrate cells, most glycosphingolipids (GSLs) are localized in the outer leaflet of the plasma membrane, forming microdomains (1–3). The microdomain concept evolved from numerous studies over the course of two decades. Four lines of study were particularly significant: (i) clustering of GSL (4), (ii) detergent-insoluble properties of clustered GSLs (5), (iii) association of signal transducers with complex of glycosylphosphatidylinositol (GPI)-anchored proteins, GSLs, and caveolin (6, 7), and (iv) polarized presence of microdomains at the apical cell surface (8). Mixtures of microdomains are variously termed detergent-insoluble membrane (DIM) (5), glycolipid-enriched membrane (9), caveolae (10), or raft (1).
GSLs in microdomains have multiple functions (e.g., toxin receptors, cell adhesion, cell growth modulators, and signal transduction initiators) (11–14). Interestingly, Src-family kinases in the inner leaflet of the microdomain can be activated via surface GSL domains (for review, see refs. 13 and 14).
Because there is enormous molecular diversity among membrane lipids, the existence of non-GSL lipid domains is quite possible (15). Despite the existence of many studies dealing with the characterization of membrane lipid domains, the precise composition of these domains is still not fully elucidated. In the present study, we describe a glycolipid, phosphatidylglucoside (PhGlc), that is present in DIM fractions from the promyelocytic cell line HL60. This lipid was first isolated from human umbilical cord red blood cells (RBCs) as an antigen detected by a human monoclonal antibody (GL-2) against blood group antigen, i. Studies using GL-2 unexpectedly revealed that one of the target antigens was not GSL, but rather an alkaline-labile lipid. We isolated this lipid from human cord RBCs and identified it as PhGlc (16). To analyze the roles of PhGlc, we prepared a recombinant Fab fragment from GL-2 that specifically reacts with PhGlc. This monovalent protein (rGL-7) provides a powerful tool to examine the distribution and biological functions of PhGlc. Indeed, using rGL-7, we identified PhGlc as a member of lipid signaling domains in HL60 cells. Stimulation of PhGlc with rGL-7 in HL60 cells resulted in rapid phosphorylation of Src family kinases, leading to up- and down-regulation of CD38 and c-Myc expression. However, stimulation of lipid domains with antibodies against GM1 and sphingomyelin (SM) failed to induce cellular differentiation. The importance of the glucose residue of PhGlc in formation of functional lipid domains is discussed.
Materials and Methods
Cells. Cells from the human promyelocytic cell line HL60 were cultured in complete serum-free E-RDF medium (Kyokuto Pharmaceutical, Tokyo) supplemented with insulin–transferrin–sodium selenite media (Sigma), 100 μg/ml streptomycin, and 100 units/ml penicillin. Cells were cultured at 37°C in a 5% CO2 humidified atmosphere.
Immunofluorescence. Cells were fixed with 3% paraformaldehyde in PBS for 20 min at room temperature, quenched with 50 mM NH4Cl, and then blocked with 0.2% gelatin in PBS. Cells were incubated overnight with mAb GL-2 at 4°C, then with FITC-conjugated anti-human IgM (EY Laboratories) and Alexa 594-conjugated cholera toxin (Molecular Probes) or monoclonal anti-SM, Vj41 (17). SM was visualized by incubating the cells with Alexa 546-conjugated anti-mouse IgM (Molecular Probes). Cells were examined with a Zeiss LSM 510 confocal microscope equipped with a Plan-Apochromat ×100 oil DIC objective.
Production of Recombinant GL-2 Fab Fragment rGL-7. Monoclonal human anti-GL-2 antibody was prepared by using in vitro transformation of normal human peripheral lymphocytes with Epstein–Barr virus (18). Total RNA was isolated from these transformed cells, and the cDNA of the Fab fragment was amplified by RT-PCR using primers for the μ and κ Ig chains (19, 20). Amplified DNAs were ligated into a pFab1-His-2 vector (21). The resulting crude plasmid DNA was transformed into competent Escherichia coli JM109 cells, and all resulting clones were assessed with ELISA for anti-PhGlc activity. The clone that exhibited the strongest ELISA signal was selected and termed rGL-7.
Bacterial cells were grown in 200 ml of SOB medium, harvested, and then lysed by sonication in 2 ml of bacterial protein extract reagent (B-PER, Pierce) solution supplemented with 1 mM phenylmethylsulfonyl fluoride and 100 μg/ml leupeptin. Fab protein in the B-PER lysate was purified by using an Immunoassist MGPP gel column (Kanto Kagaku, Tokyo). Anti-PhGlc activity was checked with ELISA using partially purified cord RBC extract as the antigen. Positive fractions were pooled and used as recombinant Fab antibody rGL-7. The yield of purified rGL-7 was ≈38–63 μg/ml. SDS/PAGE analysis showed that the purified Fab fragment was essentially homogeneous (not shown). Amino acid sequences of variable regions within the heavy and light chains are presented in Fig. 1. For TLC immunostaining, biotinylated rGL-7 was prepared by using the BIOTINTAG microbiotinylation kit (Sigma) according to the manufacturer's instructions.
Fig. 1.
Deduced amino acid sequences of the variable regions within the heavy (H) and light (L) chains of the reconstructed Fab antibody rGL-7. FR, framework region; CDR, complementarity-determining region.
Purification and Characterization of PhGlc. Glycolipids were isolated from DIM fractions of HL60 cells (22). Specificity of PhGlc was confirmed by using TLC or two-dimensional TLC immunostaining with biotinylated rGL-7 and 3,3′-diaminobenzidine tetrahydrochloride (ICN) as the chromagen (16, 23). Immunoreactive spots were transferred onto PVDF membranes and analyzed with a TSQ 70 triple quadruple mass spectrometer (Finnigan-MAT, San Jose, CA; ref. 24). The amount of PhGlc was calculated according to phosphorus content determined by Bartlett, and glucose content was determined by gas chromatography (16).
Stimulation of HL60 Cells with Antibodies and Retinoic Acid. HL60 cells (106 cells in E-RDF medium) were stimulated with various concentrations of rGL-7 (≈0.4–4 μg/ml corresponding to ≈8–80 pmol/ml), equivalent molecules of anti-GM1 (DH59B; IgG), anti-SM (Vj41; IgM), or 0.1 μM all-trans retinoic acid (ATRA). Fc receptor (Fc-R) was blocked by pretreating HL60 cells with 1 μg per 106 cells of human myeloma protein (for IgM-Fc-R) or mAb TAPC301 C1–4 [Epstein–Barr virustransformed B cell line-derived anti-hepatitis B virus surface antigen (HB) mAb for IgG-Fc-R]. Fab fragments of Vj41 and DH59B were prepared via digestion with immobilized papain (Pierce), followed by gel filtration of Sephacryl S-300 (for Vj41) or affinity separation using protein A agarose (for DH59B). These fragments were used for cell stimulation. CD38 expression was determined by using mAb IOB6. NAD+ glycohydrolase activity in RA- or rGL-7-treated HL60 cells was measured as before (25). HL60 cells were cultured for 4 days, then 100-μl aliquots of cell suspension (107 cells per ml) were stimulated with rGL-7 (1 μg per 106 cells) for ≈10 s–10 min in the presence or absence of 10 μM 4-amino-5-(4-methylphenyl)-7-(t-butyl)pyrazolo[3,4-d]pyrimidine or 1 mM methyl-β-cyclodextrin. As a control, HL60 cells were simultaneously treated with rGL-7 and an unrelated recombinant Fab antibody (human monoclonal rFab-305 or -308) against HB. Treatment was terminated by adding an equal volume of SDS sample buffer (2×).
Immunoblotting and Immunoprecipitation. Immunoblotting analysis was performed (26), using horseradish peroxidaseconjugated PY20 (Amersham Biosciences) for protein tyrosine phosphorylation, anti-transferrin receptor antibody (Zymed), or polyclonal antibodies against Lyn, Hck, Cbp, and LAT (Santa Cruz Biotechnology). For immunoprecipitation, antibodies were conjugated to protein A agarose (Oncogene Research Products). rGL-7 was conjugated to avidin agarose (Oncogene Research Products). Agarose gels were incubated with rGL-7-treated HL60 cell lysates (107 cells per gel) overnight at 4°C. After washing, each gel was treated with SDS sample buffer and subjected to SDS/PAGE for immunoblotting analysis.
DIM Preparation. DIM fractions were prepared as described (26, 27) with slight modifications. Untreated and rGL-7-treated HL60 cells (108 cells) were lysed with 1 ml of lysis buffer consisting of 1% Triton X-100 and TN buffer (25 mM Tris-HCl, pH 7.5/150 mM NaCl/0.1 mM phenylmethylsulfonyl fluoride/1 mM NaHSO3/1 mM benzamidene/2 mM CaCl2/5 mM MgCl2/0.15 mM spelmin/0.3 mM spelmidine). Lysates were transferred into centrifugal tubes (Ultra Clear, 14 × 95 mm, Beckman), kept on ice for 30 min, and then mixed with 1 ml of 80% sucrose in TN buffer. Sucrose solutions of varying concentrations (5.5 ml of 30% sucrose, 3.5 ml of 5% sucrose) were subsequently layered onto the lysate in 40% sucrose and then centrifuged in a SW41Ti swinging rotor (Beckman) at 39,000 rpm at 4°C for 22 h. Light-scattering bands (i.e., DIM fraction), located at the 5% and 30% sucrose interface, were recovered mainly into two fractions, 4 and 5, using a Piston Gradient Fractionator (BioComp, Fredericton, Canada).
Expression Profiling. Total RNA from HL60 cells (108) stimulated with freshly prepared rGL-7 (1 μg per 106 cells) for 0, 3, and 24 h was extracted by using an Atlas glass total RNA isolation kit (CLONTECH). Total RNA (20 μg) was reverse transcribed, amplified, and labeled per the manufacturer's instructions (CLONTECH). cDNA was labeled with fluorescent Cy3 dye (Amersham Biosciences) and hybridized to oligonucleotide arrays (Atlas Array containing probes for 4,881 human genes). Arrays were washed, scanned, and then analyzed with Atlas Iris software (CLONTECH) to determine the expression of each gene.
Results
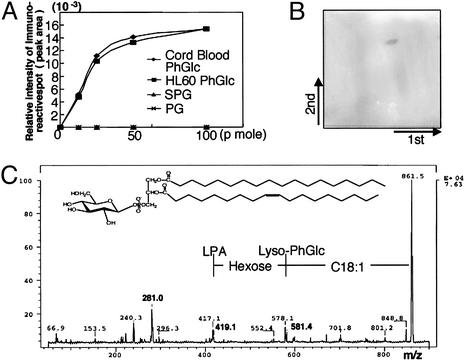
PhGlc in HL60 Cells. To examine the distribution of PhGlc in mammalian cells, we isolated lipids from several cell lines (107 cells) derived from a variety of tissues. One-tenth of total lipid extracts were analyzed by TLC immunostaining. For immunostaining, direct labeling with avidin-labeled recombinant Fab antibody (rGL-7) was used because immunostaining with a secondary antibody against human IgM resulted in nonspecific staining. rGL-7 reacted preferentially with PhGlc isolated from cord RBCs, but not with GSLs such as paragloboside and sialylparagloboside (Fig. 2A). We found that PhGlc was present in various cell lines, including HL60, B95-8 (marmoset B cell line), Cos 7, PC12, and Madin–Darby canine kidney. We used HL60 cells in the present study.
Fig. 2.
Occurrence of PhGlc in HL60 cells. (A) rGL-7 reacts with antigens from both cord RBCs and HL60 cells. PhGlc obtained from umbilical cord RBCs and HL60 cells (≈12.5–100 pmol) was separated via TLC, and spots were visualized through sequential incubation with biotinylated rGL-7, horseradish peroxidase-conjugated avidin, and the chromagen Ni-intensified DAB. Immunoreactive spots were measured by a dual wavelength TLC scanner (Shimadzu CS-930). The peak areas were plotted against amounts of PhGlc applied. (B)A single immunoreactive spot arose from the PBA-retained fraction from HL60 cells. PhGlc-containing spots were visualized by using the same method described in A.(C) Secondary ion mass spectrometry–collision-induced dissociation spectrum of PhGlc from HL60 cells. The immunoreactive spot shown in B was transferred onto a PVDF membrane and subjected to secondary ion mass spectrometry–collision-induced dissociation mass spectrometric analysis.
To confirm the chemical structure of the antigen in HL60 cells recognized by rGL-7, total lipids were isolated and partially purified by using phenylboronate agarose column chromatography. TLC revealed a single rGL-7-positive spot with mobility identical to that of PhGlc isolated from cord RBCs (Fig. 2B). This compound was further purified by using two-dimensional TLC blotting and analyzed with secondary ion mass spectrometry–collision-induced dissociation spectrometry. Its mass spectrum was identical to that of PhGlc and phosphatidylinositol (Fig. 2C). Because PhGlc and phosphatidylinositol have the same mass number, sugar composition of the rGL-7-positive lipid was analyzed by using gas chromatography and compared with those of PhGlc and phosphatidylinositol. The lipid from HL60 cells contained only glucose, not inositol, indicating that the rGL-7-positive lipid isolated from HL60 cells was PhGlc, not phosphatidylinositol. The fatty acid species component of PhGlc from HL60 cells differed significantly from the fatty acid component of PhGlc from cord RBCs. The major lipid from cord RBCs contained polyunsaturated fatty acid C20:4 at C-2 of glycerol (16). In contrast, PhGlc from HL60 cells possessed C18:1 at C-2 of glycerol.
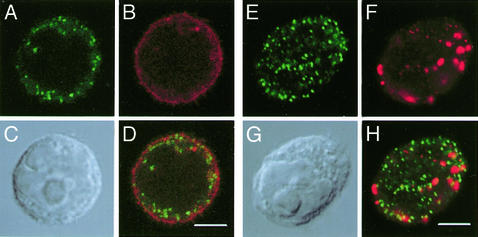
Next, we examined the cellular localization of PhGlc. Immunofluorescent confocal images of GL-2-stained cells showed that the antigen was localized in both plasma and intracellular membranes. To differentiate PhGlc from ganglioside GM1 and SM, cells were double stained with GL-2 and either cholera toxin to visualize GM1 or Vj41 to visualize SM. PhGlc localization was distinct from that of GM1 and SM (Fig. 3).
Fig. 3.
Immunofluorescence staining of HL60 cells with GL-2. Antibody binding was detected as described in the text. PhGlc (A and E), GM1 (B), sphingomyelin (F), and Nomarski images (C and G) are shown. D and H are merged images of A and B and E and F, respectively. (Scale bar, 5 μm.)
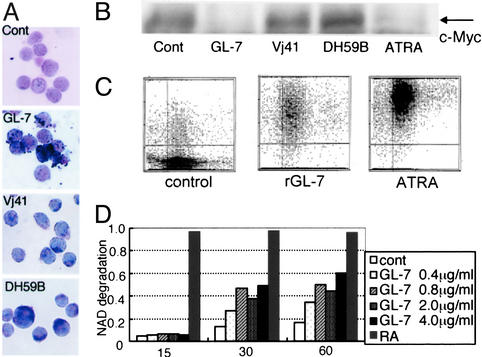
Induction of Granulocytic Differentiation of HL60 Cells by Fab Antibody Against PhGlc. HL60 cells changed morphologically when cultured in the presence of rGL-7. HL60 cells have the potential to differentiate into granulocytes or monocytes when exposed to various stimuli, such as ATRA, DMSO, etc. First, we performed a nitroblue tetrazolium (NBT) reduction test. rGL-7 treatment induced differentiation into NBT-positive cells (Fig. 4A). Expression of the protooncogene c-Myc, which is suppressed by differentiation (28, 29), was down-regulated in rGL-7-treated cells (Fig. 4B). Recent studies showed that HL60 cells strongly express CD38 when granulocytic differentiation is induced by retinoic acid (30). Munshi et al. (31) reported that CD38 has a causal role in granulocytic differentiation of HL60 cells. These findings prompted us to test whether rGL-7 also induces CD38 expression in HL60 cells. rGL-7 enhanced CD38 expression, although the degree of surface expression was less than that in cells treated with ATRA (Fig. 4C). rGL-7 also activated the enzymatic activity of CD38 (i.e., NAD+ glycohydrolase activity) in a dose-dependent manner (Fig. 4D). Granulocytic differentiation was further evidenced by gene expression profiling using microarray analysis. Of the 4,881 genes in the array, expression of human eosinophil granule major basic protein mRNA increased the most (5.8-fold after a 24-h treatment with rGL-7). Sialyltransferase ST3Gal-V (CMP-NeuAc:lactosylceramide α2,3-sialyltransferase; GM3 synthase) mRNA expression also increased (3.8-fold after 24 h). Sialyltransferase is activated during granulocytic differentiation (32).
Fig. 4.
Phenotypic changes in HL60 cells resulting from long-term rGL-7 treatment. (A) NBT reduction by cells treated with rGL-7 or Fab fragments of Vj41 and DH59B. Cells in each treatment were reacted with 0.5% NBT solution at 37°C for 30 min and then stained with the May–Grünwald–Giemsa method. (B) Down-regulation of C-Myc expression. HL60 cells were treated with rGL-7, Vj41, DH59B, or ATRA for 36 h as described in the text. Nuclear fraction from HL60 cells was examined by Western blotting with the anti-c-Myc, 9E-10. (C) Surface expression of CD38 determined by mAb IOB-6 immunofluorescence. HL60 cells were stimulated with rGL-7 (1 μg per 106 cells) or 0.1 μM ATRA for 48 h, stained with mAb IOB-6, followed by FITC-conjugated anti-mouse IgG, then analyzed by using a flowcytometer CytoAce 150. (D) Induction of NAD+ glycohydrolase activity by rGL-7. HL60 cells were stimulated with ≈0.4–4μg/ml rGL-7. Aliquots (105 cells, 100 μl) of cell suspensions were used for analysis of NAD+ glycohydrolase activity.
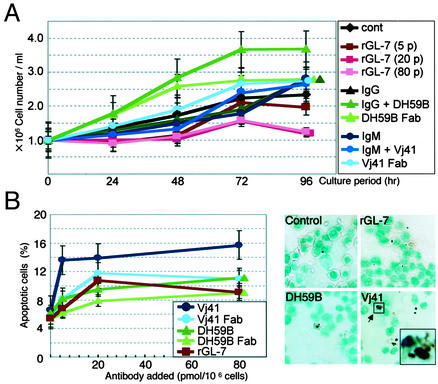
To examine whether these effects were specific to rGL-7, we stimulated HL60 cells with monoclonal antibodies against ganglioside GM1 (DH59B) and SM (Vj41). Interestingly, neither DH59 nor Vj41 induced cellular differentiation, as indicated by c-Myc expression (Fig. 4B) or the NBT reduction test (Fig. 4A). In contrast to rGL-7, treatment with both DH59B and its Fab fragment caused a slight, but distinct, increase in growth rate (Fig. 5A). This is consistent with findings that the B subunit of cholera toxin, which binds GM1, stimulates increased 3T3 cell growth (33).
Fig. 5.
Effects of antibody treatment on molecules that localize to the DIM fraction of HL60 cells. (A) Antibody stimulation affects HL60 cell growth. Cells were cultured in the presence of rGL-7 (≈5–80 pmol per 106 cells), GL-2 (20 pmol per 106 cells), or the Fab fragments of Vj41 or DH59B. To block Fc-R, cells were pretreated with an unrelated human IgM (for Vj41) or IgG (for DH59B), then treated with GL-2 (i.e., entire antibody). Results are means ± SEM, with statistical significance (P < 0.05) determined by Student's t test. (B) SM stimulation increased apoptosis. (Left) HL60 cells were treated with rGL-7 (≈5–80 pmol), GL-2, or Fab fragments of Vj41 or DH59B for 48 h, and annexin-positive cells were counted by using a flow cytometer. (Right) Cells treated with 20 pmol per 106 cells of rGL-7, Vj41 (IgM) after pretreatment with human IgM and DH59B (IgG) after pretreatment with human IgG for 48 h were stained with the terminal deoxynucleotidyltransferase-mediated dUTP endlabeling method. Arrow points to a typical cell with apoptotic bodies.
Interestingly, apoptosis significantly increased when the cells were treated with Vj41. Apoptotic cells were stained with both FITC-conjugated annexin V and the terminal deoxynucleotidyltransferase-mediated dUTP end-labeling method (Fig. 5B). However, the Fab fragment prepared from Vj41 did not induce phenotypic changes in these cells. These results suggest that antibody stimulation of granulocytic differentiation is highly specific to PhGlc and distinct from GM1 (DH59B)- or SM (Vj41)-mediated stimulation.
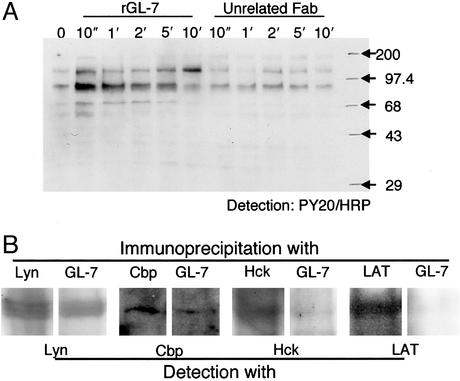
Induction of Rapid Protein Tyrosine Phosphorylation by Fab Antibody Against PhGlc. We analyzed biochemically the early responses of HL60 cells to stimulation with rGL-7. When HL60 cells were treated with rGL-7 (1 μg per 106 cells) for 0 or 10 s or 1, 2, 5, or 10 min, protein tyrosine phosphorylation occurred almost immediately (see the 58- to 60-, 65-, and 89-kDa bands in the immunoblot of Fig. 6A). These bands were identified as Src family protein kinases Lyn and Hck, and transmembrane protein Cbp, respectively, based on immunoblotting with corresponding specific antibodies (data not shown). The Src kinase inhibitor PP1 completely inhibited protein tyrosine phosphorylation (data not shown). Phosphorylation was absent when HL60 cells were treated with DH59B, Vj41 (data not shown), or unrelated Fab antibodies (e.g., r-Fab 305) (Fig. 6A).
Fig. 6.
(A) Short-term stimulation of HL60 cells with rGL-7 induces protein tyrosine phosphorylation. After incubation of HL60 cells (2 × 107 cells per ml) for 4 days, 50-μl aliquots were transferred to microtubes, and an equal volume of rGL-7 (20 μg/ml) was added. After 10 s–10 min, stimulation was terminated by adding 50 μl of SDS sample buffer. For the control, cells were treated with an unrelated Fab antibody (anti-HBs). Protein tyrosine phosphorylation was assessed by immunoblotting using horseradish peroxidase-conjugated PY20. (B) Lyn, Hck, and Cbp coexist with PhGlc in the DIM fraction. Lysates from rGL-7-treated HL60 cells were incubated with Lyn-, Hck-, and Cbp-conjugated protein A agarose gel or biotinylated rGL-7-conjugated avidin agarose gel. The resulting protein complexes were subjected to SDS/PAGE and analyzed by immunoblotting with antibodies against corresponding proteins.
PhGlc in DIM Domain Fraction. PhGlc was expressed on the membrane surfaces of HL60 cells. As described above, rGL-7 activated several Src family protein kinases. This activation was cholesterol dependent, as pretreatment of HL60 cells with 1 mM methyl-β-cyclodextrin, which reduces endogenous cholesterol (34), impaired the effects of rGL-7 treatment (data not shown). These results indicate that the lipid signaling domain is crucially involved in rGL-7-induced tyrosine phosphorylation of Src protein family kinases. This finding prompted us to examine whether PhGlc and these kinases colocalize in the DIM fraction, even though PhGlc is not a sphingolipid.
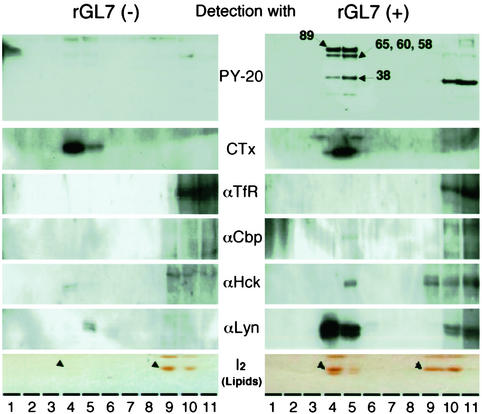
DIMs from untreated (control) and rGL-7-treated (24 h) HL60 cells were isolated by stepwise sucrose gradient centrifugation. DIMs were concentrated into two fractions (4 and 5), those in which GM1 distributes as a DIM or raft marker. In contrast, one surface protein, TfR, a known non-raft marker, did not distribute to DIM fractions of rGL-7-treated and untreated cells. Fractions 4 and 5 were designated as ``light'' and ``heavy'' DIMs, respectively. Cholesterol, the marker used to identify lipid rafts, was more concentrated in the ``light'' DIM (not shown). Surprisingly, five bands (89, 64, 58–60, 38, and 25 kDa) corresponding to phosphotyrosine proteins were present in fractions (i.e., 4 and 5) derived from cells after 48-h treatment (Fig. 7). In contrast, these phosphotyrosine protein bands were barely detectable in fractions from control cells. Western blotting using specific anti-phosphotyrosine antibodies identified the 89-, 64-, and 58- to 60-kDa bands as Cbp, Hck, and Lyn, respectively. In particular, Lyn protein significantly and specifically increased in the DIM fraction (fractions 4 and 5) after treatment with rGL-7.
Fig. 7.
Preparation and analysis of the DIM fraction. HL60 cells (108 cells, 106 cells per ml) were treated with rGL-7 (1 μg per 106 cells) for 18 h and separated by sucrose density gradient centrifugation. The gradient solutions were separated into 12 fractions (fraction 12 was omitted in this figure). GM1 (CTx), DIM marker; TfR, non-DIM marker. For TLC analysis of PhGlc, lipids were visualized by exposure to iodine vapor. Arrowheads indicate PhGlc bands.
Interestingly, rGL-7 treatment significantly promoted PhGlc production. The PhGlc content in untreated HL60 cells was ≈2 nmol per 107 cells (based on glucose content). Incubation of HL60 cells for 48 h in the presence of rGL-7 (4–40 μg/ml) resulted in an ≈2–10-fold increase in PhGlc; the extent of this increase depended on rGL-7 concentration (data not shown). The dramatic increase in PhGlc was evident in the DIM fraction (Fig. 7). NAD+ glycohydrolase activity in rGL7-treated cells was also detected in fraction 5 (i.e., ``heavy'' DIM) and 6, whereas no significant activity was observed in DIM fractions from untreated cells (not shown).
In the next series of experiments, we examined whether the tyrosine-phosphorylated proteins colocalized within the PhGlc-rich DIM fractions. Lyn protein immunoprecipitated in an rGL-7-immobilized gel (Fig. 6B). Cbp and Hck proteins also immunoprecipitated in this gel; however, LAT protein did not. This finding suggests that tyrosine phosphorylation of LAT found in the DIM fraction may be induced by an indirect interaction between PhGlc and LAT protein. This hypothesis is consistent with the observation that rGL-7 induced tyrosine phosphorylation of the 58- to 60-, 65-, and 89-kDa proteins that corresponded to Lyn, Hck, and Cbp, respectively, but did not induce the phosphorylation of the 38-kDa protein that corresponded to LAT (Fig. 6A).
Discussion
PhGlc, an Unrecognized Type of Glycolipid. We found that a new type of glycolipid, PhGlc, was widely distributed not only in human cells but also in cells of non-human origin. We isolated PhGlc from HL60 cells and determined the structure as phosphatidyl-β-d-glucoside. Our observation that the TLC migration pattern of PhGlc was identical to a major phospholipid, phosphatidylcholine, explains why the presence of this lipid may have been previously overlooked. Moreover, because PhGlc and phosphatidylinositol have an identical mass number, it is difficult to identify PhGlc solely by mass spectrometric analysis (16). At present, the use of recombinant protein rGL-7 is the only way to specifically identify PhGlc.
It is interesting to note that most lipids present in biological membranes can be modified by carbohydrates. As a typical example, ceramide is modified with glucose and galactose to form GlcCer and GalCer, respectively (35). Diacylglycerol is glycosylated to form galactosyldiacylglycerol. Most recent studies show that cholesterol can also be modified with glucose (36). These glycosylated lipids play significant roles in a variety of biological processes, including cellular proliferation, development, differentiation, and stress responses (35, 37). Thus, we expected that PhGlc might also have potential roles in mammalian cells. Indeed, here we demonstrated that PhGlc forms glycolipid domains in surface membranes of HL60 cells, and stimulation of PhGlc with the monovalent Fab antibody rGL-7 generated membrane signaling through the lipid microdomain.
Monovalent Fab Antibody as a Specific Probe for PhGlc. In the present study, we generated a Fab antibody from the human monoclonal antibody GL-2. Originally, GL-2 was established as an antibody against i antigen (16). However, recombinant rGL-7 failed to bind i-active GSLs, but specifically bound PhGlc. At present, we cannot explain why rGL-7 lost reactivity only to i antigen. According to the amino acid sequence of reconstructed rGL-7 (Fig. 1), its heavy chain variable region belongs to the VH-4 family and the light chain variable region to the Vk-4 family.
Until now, multivalent anti-carbohydrate antibodies have been used to stimulate cells (for review, see ref. 13). The activation mechanism is thought to depend on the divalent or multivalent antibody-mediated cross-linking of membrane lipids. In fact, in the case of Vj41 or DH59B, Fab fragments prepared from these antibodies barely induced phenotypic changes in HL60 cells, whereas treatment with intact antibodies (after blocking Fc-R) induced cell growth or apoptosis. However, to our surprise, rGL-7 stimulated HL60 cells. This demonstrates that antibody stimulation without cross-linking can successfully generate signals. Although the exact mechanism of the rGL-7-mediated activation of Src family protein kinases is largely unknown, the binding of rGL-7 to glucose residues may affect the formation of PhGlc within the microdomain. Such perturbation may be enough to initiate a cascade of reactions starting with the tyrosine phosphorylation of Src family protein kinases. Alternatively, because various lipid-binding proteins that play key roles in lipid domains localize to microdomains (for review, see ref. 38), it is also possible that an unidentified PhGlc-binding protein is responsible for mediating PhGlc-dependent signaling. Although mechanisms underlying this signaling are not yet clear, phenomena observed in the present study may play roles in certain biological processes. Immunoglobulins belonging to the VH-4 and Vk-4 families are abundant, and a single gene, VH4–21, which encodes anti-blood group antigen i antibody, is found in ≈6% of normal human peripheral blood cells (39). Therefore, anti-i antibody itself may possibly be a signal generator that regulates hematopoietic cell differentiation.
PhGlc, a Member of Glycolipid Signaling Domains. Recent studies propose that GSL clustering forms GSL signaling domains (14). GSL clusters are detergent insoluble and are isolated in a light buoyant density fraction in sucrose density gradients. Even though PhGlc does not belong to the GSL class of lipids, it was also found in the DIM fraction, thus indicating that PhGlcenriched domains or clusters were formed. A major molecular species of PhGlc contains stearyl oleoyl chains. Review of the literature indicates that phosphatidylcholine, which contains the same fatty acyl chains as PhGlc, is mostly excluded from microdomains or lipid rafts. Thus, this suggests that the head group is likely to be responsible for the partition of PhGlc to the lipid domain. Although no empirical evidence is available at present, we propose that the glucose residue (for hydrogen bonding), as well as both acyl chains (for van der Waals interactions), is important in glycolipid domain formation and maintenance.
Membrane microdomains are heterogeneous, and each domain mediates different functions (14). In the case of B16 melanoma cells, for example, GSL-enriched domains are involved in cell adhesion and signaling, whereas raft-like domains are involved in other pathways. Non-adhesive cells, such as HL60 cells, may contain functionally different GSL- or glycolipid-rich domains. This is supported by our finding that PhGlc domains, not GSL-rich domains, mediate granulocytic differentiation of HL60 cells (Fig. 4 C and D). PhGlc is readily degraded by enzymes, such as phospholipases, suggesting that PhGlc-based domains are more labile and flexible than GSL-rich domains. This characteristic may confer a unique and distinct function for PhGlc in microdomains or clusters.
Synthesis of PhGlc. Stimulation of HL60 cells with rGL-7 induced the enhancement of PhGlc production (Fig. 7). DIM fractions isolated from rGL-7-treated cells contained a significant amount of tyrosine-phosphorylated proteins, such as Lyn, Hck, and LAT, compared with those from nontreated cells (Fig. 7). This is especially the case for Lyn protein (Fig. 7). ATRA-induced cell differentiation did not induce changes in PhGlc content within the lipid domain fraction (data not shown). These results suggest that the up-regulation of PhGlc production served as a typical signal generated in the lipid domain, and that PhGlc enhanced recruitment of signaling proteins, such as Lyn, into the membrane domains. This type of positive-feedback circuit may promote and strengthen PhGlc-mediated protein tyrosine phosphorylation in the lipid domains. To test this hypothesis, it is essential to understand where and how PhGlc is biosynthesized and how its synthesis is regulated in the cell. However, details of the biosynthetic pathway of PhGlc are entirely unknown. Molecular cloning of the PhGlc synthase gene is a key step for understanding the physiological roles of PhGlc.
Acknowledgments
We thank Dr. K. Nakamura of the Institute of Physical and Chemical Research for the kind gift of paragloboside. We also thank the Brain Science Institute (Y.H.) and Frontier Research System (T.K.) of the Institute of Physical and Chemical Research for financial support. This work was supported in part by Grants-in-Aid 14370753 (to T.K.) and 12140201 (Scientific Research on Priority Areas B to Y.H.) and a special grant for the High-Tech Research Center to Nihon University (to Y.N.) from the Ministry of Education, Culture, Sports, Science and Technology.
Abbreviations: GSL, glycosphingolipid; PhGlc, phosphatidylglucoside; DIM, detergent-insoluble membrane; SM, sphingomyelin; ATRA, all-trans retinoic acid; NBT, nitroblue tetrazolium; HBs, hepatitis B virus surface antigen.
Data deposition. The sequences reported in this article have been deposited in the DNA Data Bank of Japan [accession nos. AB110646 (for H chain) and AB110647 (for L chain)].
References
- 1.Simons, K. & Ikonen, E. (1997) Nature 387, 569–572. [DOI] [PubMed] [Google Scholar]
- 2.Brown, D. A. & London, E. (1998) Annu. Rev. Cell Dev. Biol. 14, 111–135. [DOI] [PubMed] [Google Scholar]
- 3.Brown, D. A. & London, E. (2000) J. Biol. Chem. 275, 17221–17224. [DOI] [PubMed] [Google Scholar]
- 4.Tillack, T. W., Allietta, M., Moran, R. E. & Young, W. W. J. (1983) Biochim. Biophys. Acta 733, 15–24. [DOI] [PubMed] [Google Scholar]
- 5.Okada, Y., Mugnai, G., Bremer, E. G. & Hakomori, S. (1984) Exp. Cell Res. 155, 448–456. [DOI] [PubMed] [Google Scholar]
- 6.Stefanova, I., Horejsi, V., Ansotegui, I. J., Knapp, W. & Stockinger, H. (1991) Science 254, 1016–1019. [DOI] [PubMed] [Google Scholar]
- 7.Brown, D. A. & Rose, J. K. (1992) Cell 68, 533–544. [DOI] [PubMed] [Google Scholar]
- 8.Simons, K. & van Meer, G. (1988) Biochemistry 27, 6197–6202. [DOI] [PubMed] [Google Scholar]
- 9.Sorice, M., Parolini, I., Sansolini, T., Garofalo, T., Dolo, V., Sargiacomo, M., Tai, T., Peschle, C., Torrisi, M. R. & Pavan, A. (1997) J. Lipid Res. 38, 969–980. [PubMed] [Google Scholar]
- 10.Anderson, R. G. W. (1998) Annu. Rev. Biochem. 67, 199–225. [DOI] [PubMed] [Google Scholar]
- 11.Hakomori, S. & Igarashi, Y. (1995) J. Biochem. (Tokyo) 118, 1091–1103. [DOI] [PubMed] [Google Scholar]
- 12.Hakomori, S. (2002) Proc. Natl. Acad. Sci. USA 99, 225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasahara, K. & Sanai, Y. (2000) Glycoconj. J. 17, 153–162. [DOI] [PubMed] [Google Scholar]
- 14.Hakomori, S. (2001) Trends Glycosci. Glycotech. 13, 219–230. [Google Scholar]
- 15.Kobayashi, T. & Hirabayashi, Y. (2000) Glycoconj. J. 17, 163–171. [DOI] [PubMed] [Google Scholar]
- 16.Nagatsuka, Y., Kasama, T., Uzawa, J., Ohashi, Y., Ono, Y. & Hirabayashi, Y. (2001) FEBS Lett. 497, 141–147. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto, K., Ono, K., Nakahara, K., Yagita, H. & Okumura, K. (1988) Cancer Res. 48, 3148–3152. [PubMed] [Google Scholar]
- 18.Nagatsuka, Y., Watarai, S., Yasuda, T., Higashi, H., Yamagata, T. & Ono, Y. (1995) Immunol. Lett. 46, 93–100. [DOI] [PubMed] [Google Scholar]
- 19.Takekoshi, M., Maeda, F., Tachibana, H., Inoko, H., Kato, S., Takakura, I., Kenjo, T., Hiraga, S., Ogawa, Y., Horiki, T. & Ihara, S. (1998) J. Virol. Methods 74, 89–98. [DOI] [PubMed] [Google Scholar]
- 20.Takekoshi, M., Maeda, F., Nagatsuka, Y., Aotsuka, S., Ono, Y. & Ihara, S. (2001) J. Biochem. (Tokyo) 130, 299–303. [DOI] [PubMed] [Google Scholar]
- 21.Maeda, F., Nagatsuka, Y., Ihara, S., Aotsuka, S., Ono, Y., Inoko, H. & Takekoshi, M. (1999) J. Med. Virol. 58, 338–345. [PubMed] [Google Scholar]
- 22.Higashi, H., Hirabayashi, Y., Fukui Y., Naiki, M., Matsumoto, M., Ueda, S. & Kato, S. (1985) Cancer Res. 45, 3796–3802. [PubMed] [Google Scholar]
- 23.Higashi, H., Fukui, Y., Ueda, S., Kato, S., Hirabayashi, Y., Matsumoto, M. & Naiki, M. (1984) J. Biochem. (Tokyo) 95, 1517–1520. [DOI] [PubMed] [Google Scholar]
- 24.Taki, T., Kasama, T., Handa, S. & Ishikawa, D. (1994) Anal. Biochem. 223, 232–238. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama, M. H., Nagatsuka, Y., Katsumata, O., Irie, F., Kontani, K., Hoshino, S., Katada, T., Ono, Y., Yoshigaki, J. F., Sugiya, H., et al. (2001) Biochemistry 40, 888–895. [DOI] [PubMed] [Google Scholar]
- 26.Katsumata, O., Yokoyama, M. H., Fridman, C. S., Nagatsuka, Y., Katada, T., Hirabayashi, Y., Shimizu, K., Yoshigaki, J. F., Sugiya, H. & Furuyama, S. (2001) J. Immunol. 167, 5814–5823. [DOI] [PubMed] [Google Scholar]
- 27.Rodgers, W. & Rose, J. K. (1996) J. Cell Biol. 135, 1515–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westin, E. H., Wong-Staal, F., Gelmann, E. P., Favera, R. D., Papas, T. S., Lautenberger, J. A., Eva, A., Reddy, E. P., Tronick, S. R., Aaronson, S. A. & Gallo, R. C. (1982) Proc. Natl. Acad. Sci. USA 79, 2490–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leglise, M. C., Dent, G. A., Ayscue, L. H. & Ross, D. W. (1988) Blood Cells 13, 319–337. [PubMed] [Google Scholar]
- 30.Kontani, K., Nishina, H., Ohoka, Y., Takahashi, K. & Katada, T. (1993) J. Biol. Chem. 268, 16895–16898. [PubMed] [Google Scholar]
- 31.Munshi, C. B., Graeff, R. & Lee, H. C. (2002) J. Biol. Chem. 277, 49453–49458. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura, M., Tsunoda, A., Sakoe, K., Gu, J., Nishikawa, A., Taniguchi, N. & Saito, M. (1992) J. Biol. Chem. 267, 23507–23514. [PubMed] [Google Scholar]
- 33.Spiegel, S. & Panagiotopoulos, C. (1988) Exp. Cell Res. 177, 414–427. [DOI] [PubMed] [Google Scholar]
- 34.Kilsdonk, E. P., Yancey, P. G., Stoudt, G. W., Bangerter, F. W., Johnson, W. J., Phillips, M. C. & Rothblat, G. H. (1995) J. Biol. Chem. 270, 17250–17256. [DOI] [PubMed] [Google Scholar]
- 35.Ichikawa, S. & Hirabayashi, Y. (1998) Trends Cell Biol. 8, 198–202. [DOI] [PubMed] [Google Scholar]
- 36.Kunimoto, S., Murofushi, W., Kai, H., Ishida, Y., Uchiyama, A., Kobayashi, T., Kobayashi, S., Murofushi, H. & Murakami-Murofushi, K. (2002) Cell Struct. Funct. 27, 157–162. [DOI] [PubMed] [Google Scholar]
- 37.Yamashita, T., Wada, R., Sasaki, T., Deng, C., Bierfreund, U., Sandhoff, K. & Proia, R. L. (1999) Proc. Natl. Acad. Sci. USA 96, 9142–9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson, R. G. W. & Jocobson, K. (2002) Science 296, 1821–1825. [DOI] [PubMed] [Google Scholar]
- 39.Schutte, M. E. M., van Es, J. H., Silberstein, L. E. & Logentberg, T. (1995) J. Immunol. 151, 6569–6576. [PubMed] [Google Scholar]