Abstract
Ost4p is a minimembrane protein containing only 36 amino acids and is a subunit of oligosaccharyltransferase (OT) in Saccharomyces cerevisiae. It was found previously when amino acid residues 18–25 of Ost4p were mutated to ionizable amino acids and defects were observed in the interaction between Ost4p and either Stt3p or Ost3p, two other components of OT. The transmembrane segment of Ost4p is likely to extend from residues 10–25. This is consistent with the finding that α-helicity is estimated to be 36% by CD analysis of synthetic Ost4p in liposomes. This value is in reasonable agreement with the assumption that amino acids 10–25 (16 of 36 or 44%) are transmembrane. Therefore, the mutation-sensitive region (residues 18–25) is localized to only one half of the putative transmembrane domain of Ost4p. To learn where this region of Ost4p is situated in relation to the faces of endoplasmic reticulum (ER) membrane, we determined the membrane topology of Ost4p using an in vivo method and established that it is an Nlumen-Ccyto, type I membrane protein. These results indicate that the mutation-sensitive region of Ost4p is localized in the cytoplasmic leaflet of the ER membrane. In the current study, we also observed a loss of direct interaction between Ost3p and Stt3p in the presence of ost4 temperature-sensitive mutants, which indicates Ost4p, via interactions with amino acid residues in the cytosolic leaflet of the ER membrane, functions to bind these two proteins together in a subcomplex of OT.
Many membrane and secretory proteins having the consensus sequence Asn-Xaa-Thr/Ser (Xaa can be any amino acid except proline) become N-glycosylated cotranslationally in mammalian cells and both co- and posttranslationally in yeast (1–3). Despite the fact that the basic structural requirements of N-linked glycoproteins were elucidated more than three decades ago (4), very little has been learned about the functional role of the multiprotein components of oligosaccharyltransferase (OT). In recent years, it has become clear that nine membrane proteins (Ost1p, Wbp1p, Swp1p, Ost2p, Ost3p, Ost4p, Ost5p, Ost6p, and Stt3p) are components of the OT complex in Saccharomyces cerevisiae through the use of genetic tools such as overexpression and synthetic lethality as well as immunological techniques (5–13). Furthermore, it has been shown that eight of these nine proteins in OT exist in three subcomplexes (I, Ost1p–Ost5p; II, Wbp1p–Swp1p–Ost2p; III, Ost3p–Ost4p–Stt3p) in the endoplasmic reticulum (ER) membrane (2, 14, 15).
Compared with other glycosyltransferases that usually exist as monomers having one transmembrane-spanning segment, it is interesting that OT is a very large complex and that some of the yeast OT subunits such as Stt3p (10), Ost3p (8), and Ost6p (12) contain many transmembrane segments. In addition, two subunits, Ost4p (13) and Ost2p (9), are predicted to be almost entirely embedded in the ER membrane. The catalytic transfer of the oligosaccharide chain to the appropriate Asn is believed to occur in the lumen of the ER, 30–40 Å away from the plane of the membrane (16). However, many of the OT subunits (Ost2p, Ost3p, Ost4p, Ost5p, and Ost6p) have only very small portions of their polypeptide chains exposed at the luminal side of the ER membrane. These observations raise the question of why OT requires many membrane protein subunits that may not be directly involved in its catalytic activity. Therefore, elucidation of the function of each individual subunit is critical to the understanding of the mechanism and structure of OT.
Ost4p is a minimembrane protein subunit of yeast OT that contains only 36 amino acid residues. In an earlier study (17), we reported that the introduction of a single ionizable amino acid in residues 18–25 of Ost4p resulted a defect in cell growth, in vitro OT activity, and impaired interaction of Ost4p with each of two OT subunits, Stt3p and Ost3p. In contrast, no effect was observed when similar single mutations were made in residues 2–17. These findings suggested the functional importance of the mutation-sensitive region of Ost4p. Interestingly, this region was found to be limited to approximately one half of the putative transmembrane domain of Ost4p. Therefore, it seemed likely these seven residues are located either in the cytoplasmic leaflet or the luminal leaflet of the ER membrane. In the current studies, CD analysis revealed that Ost4p in phospholipid liposomes exhibited α-helicity of 36%, in reasonable agreement with the calculated value of 44%. To learn where the mutation-sensitive region of Ost4p is located in relation to the bilayer of the ER membrane, the topology of Ost4p was determined; the results showed that Ost4p has an Nlumen-Ccyto membrane orientation. Thus, the mutation-sensitive region of the hydrophobic domain of Ost4p that mediates formation of the subcomplex (Ost3p–Ost4p–Stt3p) is located in the leaflet of the ER membrane bilayer facing the cytosol.
Methods
Plasmid Construction. pJK90. From the plasmid pHP84OST4HA (17), OST4HA containing a triosephosphate isomerase (TPI) promoter was amplified by PCR with two primers (5′-GGGCCCGAGC TCGAAGTCGAC-3′ and 5′-GGGATCCTCGAGAGCGTAATC-3′) that contain SacI and XhoI restriction enzyme sites. PCR amplification of this fragment resulted in the elimination of the stop codon at the end of hemagglutinin (HA). The OST4HA PCR product was digested with SacI and XhoI and subcloned into SacI/XhoI-digested pR90 (18). The resulting vector contained OST4HASUC2HIS4C under the control of the TPI promoter.
pRS314OST4HASUC2HIS4C. pJK90 was digested with SacI and ApaI, and the fragment containing the TPI promoter with the OST4HASUC2HIS4C gene was subcloned into SacI/ApaI-digested pRS314. DNA sequencing confirmed that the sequence of the construct was correct.
pRS314TPIOST4HA. pHP84OST4HA was digested with SalI and NotI, and the fragment containing the TPI promoter and OST4HA was subcloned into a SalI/NotI-digested pRS314 vector.
Materials and CD Methods. 1,2-Dioleyl-sn-glycero-3-phosphocholine (diC18:1Δ9cPC) and 1,2-dioleyl-sn-glycero-3-phosphoglycerol (diC18:1Δ9cPG) were purchased from Avanti Polar Lipids. The concentrations of lipid solutions dissolved in chloroform were confirmed by dry weight. Stock solutions of lipids dissolved in chloroform were stored at –20°C.
Synthetic OST4 peptide was purchased from Genemed Biotechnologies (South San Francisco, CA). The peptide, 0.3 mg, was dissolved in a 7:3 ethanol/water mixture and stored at 4°C. The concentration of the peptide was determined by absorbance spectroscopy on a Beckman DU-650 spectrophotometer by using an extinction coefficient of 1,400 cm–1·M–1 at 275 nm.
Model membrane vesicles were prepared by using the ethanol-dilution method (19, 20). Peptide dissolved in ethanol/water and lipid dissolved in chloroform were mixed and then dried under a stream of N2. Samples then where dried under high vacuum for 1 h. After redissolving the dried samples in 10 μl of 70% ethanol, 490 μl of 0.1× PBS (1 mM sodium phosphate/15 mM NaCl, pH 7.1) was added while vortexing. Vesicles were composed of 80 mol% 1,2-dioleyl-sn-glycero-3-phosphocholine/20 mol% 1,2-dioleyl-sn-glycero-3-phosphoglycerol. Final concentrations were 4 μM peptide and 400 μM total lipid. CD spectra were recorded on a Jasco J-715 CD spectrophotometer at room temperature by using a 1-mm path length quartz cuvette. Spectra were the average of 150 scans, and backgrounds from samples lacking peptide were the subtracted. Overall α-helix content was analyzed by using three deconvolution programs: selcon3 (21), continll (22), and cdsstr (23). The three programs gave the same helical content within ±2%, and the average value is reported.
Cell Lysate Preparation and Endoglycosidase H (Endo H) Digestion. Cell lysates were prepared as described (17) from yeast transformants carrying pRS314OST4HASUC2HIS4C. Cell lysates were supplemented with a final concentration of 80 mM potassium acetate, pH 5.6, and 2 μl of Endo H (1 unit/200 μl, Roche, Mannheim, Germany) was added. Samples were incubated at 37°C for 1 h, and mock samples were prepared in the same way without the enzyme.
Assessment of Growth. pRS314, pRS314OST4H A, and pRS314OST4HASUC2HIS4C were transformed into an ost4Δ strain, JCY11 (13) (MATa ade2 ura3 his3 trp1 leu2 can1 ost4Δ::URA3), and transformants containing these plasmids were selected on –Trp plates. The transformants carrying each of these plasmids were serially diluted 10-fold, and 10 μl of each was spotted on two –Trp plates. One was incubated at 25°C and the other at 37°C to assess temperature sensitivity. These transformants were also streaked on –Trp and –His/+histidinol (6 mM histidinol) plates to assess their ability to grow on medium supplemented with histidinol. The plasmids pR90 (18), pJK90, and pJK92YDL212 (24) were transformed into a strain, STY50 (18, 25) (obtained from Dieter Wolf, Institute of Biochemistry and University of Stuttgart) (MATa, his4-401, leu2-3, -112, trp1-1, ura3-52, HOL1-1, SUC2::LEU2), and selected on –Ura plates. The transformants were streaked on –Ura and –His/+histidinol plates and grown at 30°C for 2–3 days.
Coimmunoprecipitation. Cell lysates were prepared from yeast cells carrying OST4HA, wild-type OST4, or various Lys mutants of ost4 as described (17) and subjected to coimmunoprecipitation by using anti-Stt3p antibody (a gift of Satoshi Yoshida, Kirin Brewery). The procedure for coimmunoprecipitation has been described (17). The samples were analyzed on SDS/7.5% PAGE followed by Western blotting with an antibody that was directed to an anti-c-myc epitope at the C terminus of Ost3p.
Results
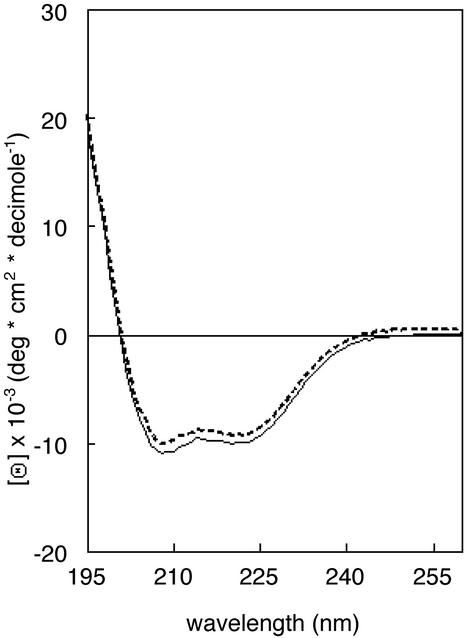
Membrane Topology of Ost4p. An important issue in interpreting the results of disruption of the Stt3p–Ost4p–Ost3p complex is the topological orientation of Ost4p in the membrane of the ER. Three available membrane-topology prediction programs have given contradicting results in that two of the three programs [hmmtop (26) and phd (27)] predict that the N terminus of Ost4p is in the lumen and the third [tmhmm (28)] predicts the opposite. Furthermore, by inspection we predict that a transmembrane span from residues 10 to 25 is much more likely than what the computer predicted, because the residues preceding residue 10 (and those succeeding residue 25) are not hydrophobic. CD measurements were used to estimate the amount of α-helical secondary structure (Fig. 1). Three different secondary-structure analysis programs predicted the percentage of helicity of synthetic Ost4p in phospholipid liposomes to be 36%, which is in reasonable agreement with the presence of a single transmembrane domain extending from residues 10–25, i.e., 44% of the 36-aa residue peptide.
Fig. 1.
CD of synthetic Ost4p in phospholipid residues liposomes (see Methods for details).
To determine the membrane topology of Ost4p experimentally, we took advantage of a construct containing both the SUC2 gene, which encodes for invertase, and HIS4C, which harbors the gene encoding histidinol dehydrogenase (18). OST4HA was fused 5′ to this construct, yielding Ost4HASuc2His4C fusion protein. If the C terminus of the Ost4p fusion protein localizes to the cytosol, invertase would not be glycosylated, whereas histidinol dehydrogenase would convert histidinol to histidine, thus allowing the cells to grow in the medium lacking histidine but supplemented with histidinol. In contrast, if the C terminus of Ost4HAp faces the lumen of the ER, invertase that contains eight N-glycosylation sites would be glycosylated.
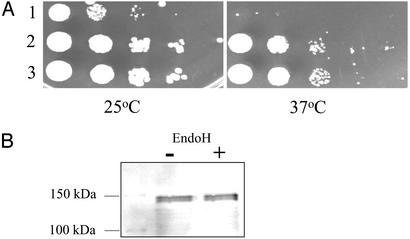
Because the fusion of Suc2His4C to the C terminus of Ost4p added ≈128 kDa in size to Ost4p, which is only 3.9 kDa, it was important to determine whether the Ost4HASuc2His4C fusion protein could serve the same function as Ost4p. pRS314, pRS314OST4HA, and pRS314OST4HASUC2HIS4C were transformed into an ost4Δ strain. Yeast transformants carrying each of these constructs were selected on –Trp plates at 25°C. A spotting assay was performed on two plates; one was incubated at 25°C and the other at 37°C (Fig. 2). The transformant carrying a vector alone did not grow at 37°C as shown in Fig. 2 A, row 1. The transformants carrying either pRS314OST4HA (Fig. 2 A, row 2) or pRS314OST4HASUC2HIS4C (Fig. 2A, row 3) rescued the growth defect of ost4Δ. This result showed that Ost4HASuc2His4C fusion protein could functionally replace wild-type Ost4p and therefore rescue the temperature-sensitive growth defect of ost4Δ. This finding strongly suggests that this fusion protein, at least with respect to topology and structure within the membrane, has the same structure as native Ost4p.
Fig. 2.
(A) Assessment of growth. The yeast strain JCY11 (ost4Δ) was transformed with a plasmid carrying pRS314 (row 1), pRS314OST4HA (row 2), or pRS314OST4HASUC2HIS4C (row 3). The yeast transformants carrying each of these plasmids were serially diluted 10-fold, and 10 μl of each was spotted on two –Trp plates. One was incubated at 25°C and the other at 37°C for 2 days. (B) Analysis of the N-glycosylation state of the Ost4HASuc2His4C fusion proteins. The whole-cell lysates prepared from cells carrying pRS314OST4HASUC2HIS4C were treated with Endo H. Samples were analyzed in SDS/7.5% PAGE.
To determine whether the C terminus of Ost4p is located in the luminal side of the ER membrane, the status of glycosylation of Ost4HASuc2His4C fusion protein was examined. Cell lysates were prepared from yeast transformants carrying pRS314OST4HASUC2HIS4C and digested with Endo H. A band at an apparent molecular mass of 135 kDa was observed, and there was no shift in molecular mass after Endo H digestion (Fig. 2B). Therefore, the fusion protein was not glycosylated, which indicates that the C terminus of Ost4p is not facing the luminal side of the ER membrane.
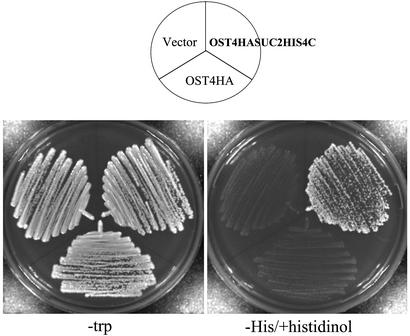
To confirm that the lack of significant glycosylation was due to the fact that the C terminus of Ost4 fusion protein faces the cytosol, the ability of the cells to grow on histidinol was assessed. pRS314, pRS314OST4HA, and pRS314OST4HASUC2HIS4C were transformed into STY50, a strain in which the chromosomal copy of the HIS4 gene is disrupted. As shown in Fig. 3 yeast transformants carrying the Ost4Suc2His4 fusion protein grew on medium supplemented with histidinol, confirming that the C terminus of the Ost4 fusion protein faces the cytosol.
Fig. 3.
Growth phenotypes of cells carrying Ost4 fusion construct in the medium containing histidinol. The yeast strain STY50 was transformed with a plasmid carrying pRS314, pRS314OST4HA, or pRS314OST4HASUC2HIS4C. Transformants were streaked on –Trp and the selective medium supplemented with histidinol and incubated at 30°C for 3 days.
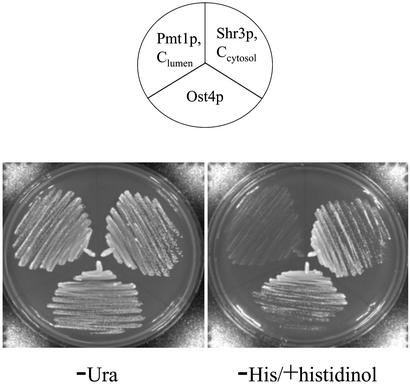
Moreover, we compared the growth on histidinol of cells carrying the Suc2His4 fusion construct with Ost4p and two other membrane proteins, Pmt1p and Shr3p. The membrane topology of Pmt1p (18) and Shr3p (29) were determined previously, thus we used these proteins as controls. Pmt1p has been determined to have the C terminus in the lumen of the ER, whereas the C terminus of Shr3p has been shown to be in the cytosol. Cells carrying OST4HA, PMT1, and SHR3 with 5′ fusion of SUC2HIS4C were streaked on histidinol medium lacking histidine. As shown in Fig. 4, cells having Ost4 fusion protein grew along with the control membrane protein with its C terminus in the cytosol. Therefore, these data further confirmed that the C terminus of Ost4p faces the cytosolic side of the ER membrane.
Fig. 4.
Comparison of growth phenotypes of cells carrying C-terminal fusion of Suc2His4C to proteins with known membrane topology or to Ost4p in the medium containing histidinol. The yeast strain STY50 was transformed with pR90 (Pmt1p, C terminus of the protein in the lumen of ER), pJKYDL212 (Shr3p, C terminus of the protein in the cytosolic side of the ER membrane), or pJK90 (Ost4p). Transformants were streaked on –Ura and the selective medium supplemented with histidinol and incubated at 30°C for 3 days.
Mutations in Residues 18–25 of Ost4p to Lys Caused a Disruption of Interaction Between Ost3p and Stt3p. Previously we showed that the mutation of each of the single amino acid residues from 18–25 of Ost4p to Lys or Asp caused disruption of the interaction of Ost4p with each of the two other OT subunits, Stt3p and Ost3p (17). To determine whether these mutations affected the formation of a subcomplex of Ost4p, Ost3p, and Stt3p, coimmunoprecipitation was carried out by using an anti-Stt3p antibody followed by Western blotting with an anti-c-myc antibody to detect Ost3mycp. In the coimmunoprecipitates from cells carrying either wild-type Ost4HAp or ost4M18LHAp, a mutant that did not exhibit a defect in growth and OT activity (17), Ost3mycp was found to be coimmunoprecipitated with Stt3p, indicating that Ost3p was binding to Stt3p in the subcomplex. This finding is in contrast to the findings that the immunoprecipitate of Stt3p prepared from cells carrying the ost4 Lys mutants ost4M19KHAp, ost4T20KHAp, ost4L21KHAp, ost4V23KHAp, and ost4I24KHAp did not contain Ost3mycp (Fig. 5). The expression of ost4I22KHAp was very low, and the immunoprecipitation experiment could not be done. These results, coupled with our earlier findings (17), indicate that single mutations to Lys of the residues from 19–24 of Ost4p caused the disruption of an interaction between Ost3p and Stt3p.
Fig. 5.
Mutations to Lys in residues 19–25 of Ost4p destabilizes interaction between Ost3p and Stt3p. Whole-cell lysates were prepared from cells carrying Ost4HAp or various ost4 Lys mutants and coimmunoprecipitated by using anti-Stt3p antibody. Cells carrying Ost4M18LHA, a mutant that did not cause a growth defect, was used as another control. The immunoprecipitates were resolved in SDS/7.5% PAGE, transferred to nitrocellulose membranes, and probed with antibody that was directed to the c-myc epitope on Ost3Mycp. The multiple lower molecular mass bands observed with Ost3mycp probably are the result of proteolytic degradation at the N terminus.
Discussion
Many multisubunit enzymes such as complexes of translocon, signal peptidase, and OT are present in the ER. Because many of these components are transmembrane proteins, it is important to understand the factors that mediate the interaction between them. Our objective is to learn about these factors in the ER membrane protein complex of OT. From our earlier study on Ost4p (17), we have found that mutations in residues 18–25 of Ost4p had a severe defect in stabilization of the subcomplex of Stt3p–Ost4p–Ost3p. We have proposed that these residues make up the mutation-sensitive region of Ost4p that is functionally important. Because these residues are predicted to span only the half of the transmembrane segment, it was important to determine (i) the length of the transmembrane segment and (ii) the membrane topology of Ost4p to find out where the mutation-sensitive region is oriented in relation to the ER membrane.
With respect to point i, the tmhmm program predicts the transmembrane domain to extend from residues 7–29, or 23 amino acid residues of Ost4p. However, as discussed earlier, our examination with the CD analysis showed that the transmembrane region is more likely to extend from residues 10–25, or 16 amino acids. Two observations support this idea. First, it is believed that ER and Golgi membranes are thinner than the plasma membrane and that helices in the ER and Golgi average closer to 15 residues in length (30). Second, our measurement of the α-helicity of Ost4p yields a value of 36%, in better agreement with the amino acid residue 10–25 transmembrane sequence than a residue 7–29 transmembrane sequence.
To deal with point ii, the N vs. C orientation issue, we used a dual Suc2His4C reporter construct and found that that the C terminus of Ost4p faces the cytosolic side of the ER membrane. Although none of the Ost4Suc2His4 fusion protein became glycosylated, which indicates that the C terminus is not oriented to the lumen of ER, the yeast transformant carrying this construct grew on the medium supplemented with histidinol lacking histidine, which indicates that the C terminus faces the cytosol. Although the C-terminal fusion of the Suc2His4C domain added ≈128 kDa in size to the 3.9-kDa Ost4p, this Ost4 fusion protein complemented the temperature-sensitive defect of ost4 deletion. Therefore, the C-terminal reporter fusion to Ost4p did not interfere with the function of Ost4p.
Although we have not directly determined the location of the N terminus of Ost4p, two lines of evidence support its luminal localization. First, the hydrophobic transmembrane segment in Ost4p is only ≈16 residues long, much shorter than the ≈30 hydrophobic residues required to form a hypothetical ``helical hairpin'' with two closely spaced transmembrane helices (31). Second, versions of Ost4p that have been N-terminally tagged with either the HA or a 6-His epitope cannot complement the defect of Ost4p deletion (data not shown), most likely because such modifications prevent translocation of the N-terminal modifications of other N-tail proteins (32, 33). Ost4p thus is a single-spanning type I membrane protein with Nout-Cin orientation.
Previously we showed that interaction between Ost4p and Stt3p or Ost4p and Ost3p was disrupted if residues 18–25 of Ost4p were mutated. We report here that we observed a loss of direct interaction between Ost3p and Stt3p in the presence of Ost4p mutations. This observation strongly supports the idea that Ost4p is a critical molecule that mediates interaction between Ost3p and Stt3p. In addition, we carried coimmunoprecipitation to determine the oligomeric status of Ost4p using two copies of Ost4p that were C-terminally tagged with two different epitopes, HA and c-myc (data not shown). The results that Ost4mycp was not coimmunoprecipitated with the HA-tagged Ost4p suggest that Ost4p exists as a monomer in the OT complex.
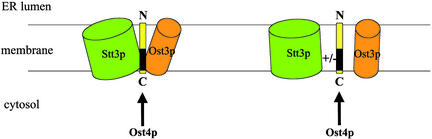
With knowledge of the membrane topology of Ost4p, we can pinpoint where the mutation-sensitive region in Ost4p is relative to the plane of the ER membrane. Interestingly, it has been noted that cells carrying mutations to an ionizable amino acid in the transmembrane region close to the cytosolic side of the ER membrane in another OT subunit, Ost2p (9), also exhibited temperature-sensitive growth phenotypes. Therefore, both of these subunits seem to have very little tolerance toward mutations in the region of the transmembrane helix close to the cytosolic face of the ER membrane. This leads us to speculate that, as shown in Fig. 6, the principal site of interaction of the three subunits is in the portion of their transmembrane helices that are close to the cytosolic leaflet of the ER membrane. Furthermore, our finding that Ost4p is present as a monomer strongly supports the idea that this minimembrane protein acts as a ``bridge molecule'' that holds the two other OT subunits together, thereby stabilizing the whole OT complex in the ER membrane.
Fig. 6.
Model of the role of Ost4p as a bridge molecule mediating the interaction between Ost3p and Stt3p. Interaction is postulated to occur in the membrane near the mutation-sensitive region of Ost4p located in the cytoplasmic leaflet of the ER membrane.
Acknowledgments
We thank Dr. Satoshi Yoshida for a generous gift of anti-Stt3p antibody and Dr. Dieter Wolf for providing the pR90 plasmid and STY50 strains. We also thank Dr. Erwin London for critical comments. This work was supported by National Institutes of Health Grant GM33185 (to W.J.L.). G.A.C. was the recipient of a Stony Brook Institute of Cell and Developmental Biology fellowship.
Abbreviations: OT, oligosaccharyltransferase; ER, endoplasmic reticulum; TPI, triosephosphate isomerase; HA, hemagglutinin; endo H, endoglycosidase H.
References
- 1.Schekman, R. (1996) Cell 87, 593–595. [DOI] [PubMed] [Google Scholar]
- 2.Knauer, R. & Lehle, L. (1999) Biochim. Biophys. Acta 1426, 259–273. [DOI] [PubMed] [Google Scholar]
- 3.Lyman, S. K. & Schekman, R. (1996) Experientia 52, 1042–1049. [DOI] [PubMed] [Google Scholar]
- 4.Marshall, R. D. (1972) Annu. Rev. Biochem. 41, 673–702. [DOI] [PubMed] [Google Scholar]
- 5.te Heesen, S., Janetzky, B., Lehle, L. & Aebi, M. (1992) EMBO J. 11, 2071–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.te Heesen, S., Knauer, R., Lehle, L. & Aebi, M. (1993) EMBO J. 12, 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silberstein, S., Collins, P. G., Kelleher, D. J., Rapiejko, P. J. & Gilmore, R. (1995) J. Cell Biol. 128, 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karaoglu, D., Kelleher, D. J. & Gilmore, R. (1995) J. Cell Biol. 130, 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silberstein, S., Collins, P. G., Kelleher, D. J. & Gilmore, R. (1995) J. Cell Biol. 131, 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zufferey, R., Knauer, R., Burda, P., Stagljar, I., te Heesen, S., Lehle, L. & Aebi, M. (1995) EMBO J. 14, 4949–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reiss, G., te Heesen, S., Gilmore, R., Zufferey, R. & Aebi, M. (1997) EMBO J. 16, 1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knauer, R. & Lehle, L. (1999) J. Biol. Chem. 274, 17249–17256. [DOI] [PubMed] [Google Scholar]
- 13.Chi, J. H., Roos, J. & Dean, N. (1996) J. Biol. Chem. 271, 3132–3140. [DOI] [PubMed] [Google Scholar]
- 14.Spirig, U., Glavas, M., Bodmer, D., Reiss, G., Burda, P., Lippuner, V., te Heesen, S. & Aebi, M. (1997) Mol. Gen. Genet. 256, 628–637. [DOI] [PubMed] [Google Scholar]
- 15.Karaoglu, D., Kelleher, D. J. & Gilmore, R. (1997) J. Biol. Chem. 272, 32513–32520. [DOI] [PubMed] [Google Scholar]
- 16.Nilsson, I. M. & von Heijne, G. (1993) J. Biol. Chem. 268, 5798–5801. [PubMed] [Google Scholar]
- 17.Kim, H., Park, H., Montalvo, L. & Lennarz, W. J. (2000) Proc. Natl. Acad. Sci. USA 97, 1516–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strahl-Bolsinger, S. & Scheinost, A. (1999) J. Biol. Chem. 274, 9068–9075. [DOI] [PubMed] [Google Scholar]
- 19.Ren, J., Lew, S., Wang, Z. & London, E. (1997) Biochemistry 36, 10213–10220. [DOI] [PubMed] [Google Scholar]
- 20.Caputo, G. & London, E. (2003) Biochemistry 42, 3275–3285. [DOI] [PubMed] [Google Scholar]
- 21.Sreerama, N., Venyaminov, S. Y. & Woody, R. W. (2000) Anal. Biochem. 287, 243–251. [DOI] [PubMed] [Google Scholar]
- 22.Provencher, S. W. & Glockner, J. (1981) Biochemistry 20, 33–37. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, W. C. (1999) Proteins 35, 307–312. [PubMed] [Google Scholar]
- 24.Kim, H., Melen, K. & von Heijne, G. (2003) J. Biol. Chem. 278, 10208–10213. [DOI] [PubMed] [Google Scholar]
- 25.Deak, P. M. & Wolf, D. H. (2001) J. Biol. Chem. 276, 10663–10669. [DOI] [PubMed] [Google Scholar]
- 26.Tusnady, G. E. & Simon, I. (1998) J. Mol. Biol. 283, 489–506. [DOI] [PubMed] [Google Scholar]
- 27.Rost, B., Fariselli, P. & Casadio, R. (1996) Protein Sci. 5, 1704–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. (2001) J. Mol. Biol. 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 29.Gilstring, C. F., Melin-Larsson, M. & Ljungdahl, P. O. (1999) Mol. Biol. Cell 10, 3549–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bretcher, M. & Munroe, S. (1993) Science 261, 1280–1281. [DOI] [PubMed] [Google Scholar]
- 31.Monné, M., Nilsson, I., Elofsson, A. & von Heijne, G. (1999) J. Mol. Biol. 293, 807–814. [DOI] [PubMed] [Google Scholar]
- 32.Monné, M., Gafvelin, G., Nilsson, R. & von Heijne, G. (1999) Eur. J. Biochem. 263, 264–269. [DOI] [PubMed] [Google Scholar]
- 33.Nilsson, I., Witt, S., Kiefer, H., Mingarro, I. & von Heijne, G. (2000) J. Biol. Chem. 275, 6207–6213. [DOI] [PubMed] [Google Scholar]